Abstract
The use of genetically engineered microorganisms such as bacteria or yeasts as live vehicles to carry out bioconversion directly in the digestive environment is an important challenge for the development of innovative biodrugs. A system that mimics the human gastrointestinal tract was combined with a computer simulation to evaluate the survival rate and cinnamate 4-hydroxylase activity of a recombinant model of Saccharomyces cerevisiae expressing the plant P450 73A1. The yeasts showed a high level of resistance to gastric and small intestinal secretions (survival rate after 4 h of digestion, 95.6% ± 10.1% [n = 4]) but were more sensitive to the colonic conditions (survival rate after 4 h of incubation, 35.9% ± 2.7% [n = 3]). For the first time, the ability of recombinant S. cerevisiae to carry out a bioconversion reaction has been demonstrated throughout the gastrointestinal tract. In the gastric-small intestinal system, 41.0% ± 5.8% (n = 3) of the ingested trans-cinnamic acid was converted into p-coumaric acid after 4 h of digestion, as well as 8.9% ± 1.6% (n = 3) in the stomach, 13.8% ± 3.3% (n = 3) in the duodenum, 11.8% ± 3.4% (n = 3) in the jejunum, and 6.5% ± 1.0% (n = 3) in the ileum. In the large intestinal system, cinnamate 4-hydroxylase activity was detected but was too weak to be quantified. These results suggest that S. cerevisiae may afford a useful host for the development of biodrugs and may provide an innovative system for the prevention or treatment of diseases that escape classical drug action. In particular, yeasts may provide a suitable vector for biodetoxication in the digestive environment.
The development of innovative biodrugs by using recombinant living microorganisms active in the human digestive environment has been recently considered (1). Potential medical applications are numerous and include the correction of gastric or intestinal deficiencies (e.g., by increasing lipase [10, 11], trypsin, or lactase) or organ failure (e.g., by removing urea in cases of kidney failure) (8, 22), biodetoxication (12), or direct production in the digestive tract of therapeutic proteins, such as biological mediators (e.g., insulin, growth factor, or interleukin [24]) or vaccines (21, 28). Recombinant bacteria, particularly lactic acid bacteria, have been mostly suggested as potential hosts (9). However, yeasts can offer advantages, especially when a eukaryotic environment is required for the functional expression of the heterologous gene (4). The common baker's yeast Saccharomyces cerevisiae emerges as a promising candidate owing to its generally recognized as safe status and its easy genetic engineering. For instance, Saccharomyces spp. have been already used in humans as probiotics (6), in enzyme substitution therapy for congenital sucrase-isomaltase deficiency (13), and in the treatment of intestinal functional disorders such as colitis or antibiotic-associated diarrhea (3).
Genetically modified microorganisms have potential as a biodetoxication system in the digestive environment. In particular, recombinant yeasts expressing phase I xenobiotic metabolizing enzymes, mostly represented by cytochrome P450, or phase II xenobiotic metabolizing enzymes, such as glutathione S-transferase or N-acetyltransferase, could be used to increase the defense of the host and/or patient against environmental xenobiotics, mainly those ingested with food (e.g., pesticides, procarcinogens, or chemical additives). The cytochrome P450 enzymes play a major role in the human detoxication system by metabolizing xenobiotics such as drugs, alcohol, procarcinogens [e.g., benzo(a)pyrene or dioxin], dyes, and pesticides (2). In our work, the cinnamate 4-hydroxylase (CA4H) activity of a plant (Helianthus tuberosus) P450 73A1 was chosen as a model for a reaction catalyzed by a P450 (owing to the nontoxicity and easy quantification of both substrate and product). The membranous CA4H catalyzes the 4-hydroxylation of trans-cinnamic acid into p-coumaric acid, reproducing the first oxidative step in the plant phenylpropanoid pathway (29).
In the first stages of the development of biodrugs, one of the challenges is to find the most appropriate model that simulates the human digestive environment, in order to evaluate the viability and activity of recombinant microorganisms. In vitro digestive systems (19, 27) or animal models, such as germfree rodents colonized with human flora (23) can be considered. In the system used for our study, the TNO gastrointestinal tract model (TIM), the overall digestive tract is reproduced by two separate systems: the gastric-small intestinal system (TIM 1) (17) and the large intestinal system (TIM 2) (18). These multicompartmental, dynamic, computer-controlled systems were designed to accept parameters and data obtained from in vivo studies with human volunteers. The main parameters of the digestion, such as pH; temperature; peristaltic movements; gastric, biliary, and pancreatic secretions; absorption of nutrients and water; and colonic microflora are reproduced as accurately as possible. They are useful study tools for the development of biodrugs, offering accuracy, reproducibility, easy manipulation, and the possibility of collecting samples at any level of the digestive tract and at any time during digestion. These systems have been validated by several studies (15, 17), including the evaluation of the microorganism survival rate in the digestive tract (16).
To validate the use of living recombinant yeasts as convenient hosts for the development of biodrugs, the survival rate and bioconversion activity of model recombinant S. cerevisiae expressing the heterologous CA4H were studied in these gastrointestinal tract systems.
MATERIALS AND METHODS
Yeast strain.
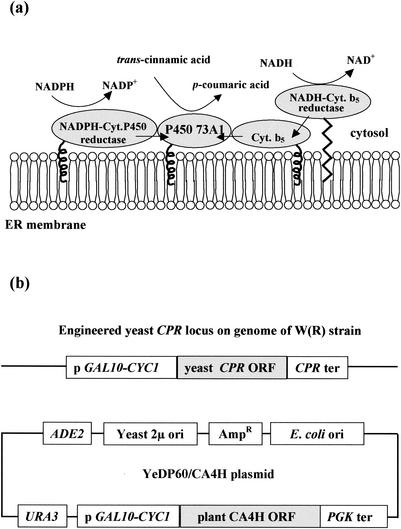
The S. cerevisiae strain (kindly provided by Denis Pompon, CNRS, Gif-sur-Yvette, France) was derived from the haploid strain W303-1B (MATα ade2-1 his3-11,15 leu2-3,112 ura3-1 Canr cyr+) (25). The strain was genetically engineered to overexpress yeast NADP-cytochrome P450 reductase (CPR) and H. tuberosus CA4H when grown in the presence of galactose (Fig. 1). The PCR-amplified CA4H open reading frame was inserted into the yeast expression vector pYeDP60. This plasmid was based on the origin of replication of the yeast 2μ minicircle, URA3 and ADE2 selection markers, and an expression cassette composed of GAL10-CYC1 promoter and phosphoglycerate kinase terminator sequences. The resulting S. cerevisiae catalyzes the second step of the plant phenylpropanoid pathway (29), metabolizing trans-cinnamic acid into p-coumaric acid.
FIG. 1.
The recombinant model of S. cerevisiae expresses the plant cytochrome P450 73A1 and catalyzes the bioconversion of trans-cinnamic acid into p-coumaric acid (CA4H activity). (a) Schematic representation of the P450 73A1 and its yeast-associated proteins, the NADPH CPR, the cytochrome (Cyt.) b5, and the NADH cytochrome b5 reductase bound to the endoplasmic reticulum (ER) membrane. (b) Genetic construction of the recombinant model of S. cerevisiae. The YeDP60/CA4H plasmid was used to transform the S. cerevisiae W303-1B strain overproducing yeast CPR [W(R) strain]. ORF, open reading frame; PGK, phosphoglycerate kinase; ter, terminator; ori, origin of replication.
Yeast culture conditions.
The S. cerevisiae strain was precultured to stationary phase at 28°C in 30 ml of SGI broth (7 g of yeast nitrogen base without amino acids/liter, 1 g of Bacto Casamino Acids/liter, 20 mg of tryptophan/liter, and 20 g of glucose/liter). A 1/10 dilution was made in 250 ml of YPGE (10 g of yeast extract/liter, 10 g of Bacto peptone/liter, 5 g of glucose/liter, and 3% [vol/vol] ethanol), and cells were grown in a shaking incubator (28°C, 200 rpm, 36 h). Induction was started by adding a 10% (vol/vol) aqueous solution of 200 g of galactose/liter and continued for 12 h (28°C, 200 rpm) until the cell density reached 5 × 107 cells/ml. At the end of culture, yeast cells were in the beginning of their stationary growth phase. The cells were then harvested, washed with sterile physiological saline solution, and resuspended in 300 ml of YPL medium (10 g of yeast extract/liter, 10 g of Bacto peptone/liter, 20 g of galactose/liter) when introduced in the gastric-small intestinal system or in 10 ml of YPL in the case of the large intestinal system.
Artificial gastrointestinal digestions.
In our study, the gastric-small intestinal system was not connected to the large intestinal system.
(i) Gastric-small intestinal system.
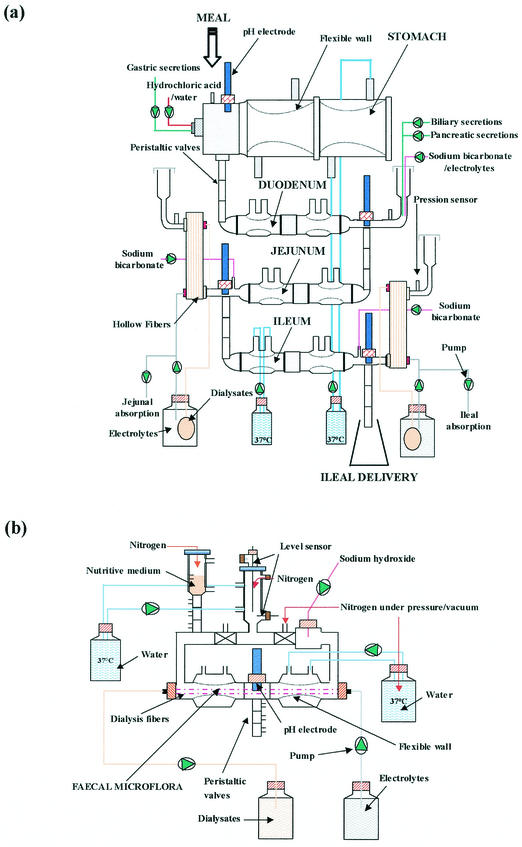
The gastric-small intestinal system (TIM 1) (17) consisted of four successive compartments simulating the stomach, the duodenum, the jejunum, and the ileum (Fig. 2a). Each compartment was composed of glass units with a flexible inside wall. The system was kept at 37°C by pumping water into the space between the glass jacket and the flexible wall. Peristaltic movements were simulated by changing the water pressure. Mathematical modeling of gastric and ileal deliveries with power exponential equations [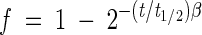 , where f represents the fraction of meal delivered, t represents the time of delivery, t1/2 represents the half-time of delivery, and β represents a coefficient describing the shape of the curve] was used for the computer control of meal transit. In our study, the gastric-small intestinal system was programmed to reproduce the human digestion of liquids, according to in vivo data. The half-time of gastric emptying was 30 min, and the β coefficient of the power exponential equation was 1. The half-time of small intestinal delivery of the meal was 160 min, and the β coefficient was 1.6. Chyme transit was regulated by opening or closing the peristaltic valves that connect the compartments. The volume in each compartment was monitored with a pressure sensor connected to the computer and regulated with jejunal absorption. The pH was computer monitored and continuously controlled in each compartment. In the stomach, the pH followed a preset curve: pH 4.5, 4.2, 2.1, and 1.7 at 5, 20, 60, and 90 min, respectively. In the small intestine, the pH was maintained at 6.5, 6.8, and 7.2 in the duodenum, jejunum, and ileum, respectively. Gastric, biliary, and pancreatic secretions (17) were introduced into the corresponding compartments by computer-controlled pumps. Water and products of digestion were removed from the jejunal and ileal compartments by pumping dialysis liquid (flow rate, 10 ml/min) through the hollow fiber membrane units (HG 400; Hospal Cobe, Lyon, France).
, where f represents the fraction of meal delivered, t represents the time of delivery, t1/2 represents the half-time of delivery, and β represents a coefficient describing the shape of the curve] was used for the computer control of meal transit. In our study, the gastric-small intestinal system was programmed to reproduce the human digestion of liquids, according to in vivo data. The half-time of gastric emptying was 30 min, and the β coefficient of the power exponential equation was 1. The half-time of small intestinal delivery of the meal was 160 min, and the β coefficient was 1.6. Chyme transit was regulated by opening or closing the peristaltic valves that connect the compartments. The volume in each compartment was monitored with a pressure sensor connected to the computer and regulated with jejunal absorption. The pH was computer monitored and continuously controlled in each compartment. In the stomach, the pH followed a preset curve: pH 4.5, 4.2, 2.1, and 1.7 at 5, 20, 60, and 90 min, respectively. In the small intestine, the pH was maintained at 6.5, 6.8, and 7.2 in the duodenum, jejunum, and ileum, respectively. Gastric, biliary, and pancreatic secretions (17) were introduced into the corresponding compartments by computer-controlled pumps. Water and products of digestion were removed from the jejunal and ileal compartments by pumping dialysis liquid (flow rate, 10 ml/min) through the hollow fiber membrane units (HG 400; Hospal Cobe, Lyon, France).
FIG. 2.
Diagram of the TNO Nutrition and Food Research Institute (Zeist, The Netherlands) gastric-small intestinal system TIM 1 (a) (17) and large intestinal system TIM 2 (b) (18).
(ii) Large intestinal system.
The large intestinal system (TIM 2) (18) was based on the same concept as that developed for TIM 1 (Fig. 2b). It was inoculated with fresh feces from healthy human volunteers. The microflora was allowed to stabilize for 48 h before introducing the recombinant yeasts and was shown to contain physiological levels of the most representative human bacterial populations (e.g., Bifidobacterium, Lactobacillus, Enterobacteriaceae, and Clostridium) (18). A feeding medium (18) reproducing the ileal effluent was regularly introduced into TIM 2. The feeding rate was set at 4 ml/h, and the chyme was removed at a flow rate of 2 ml/h, providing a half-time for the renewal of the large intestinal content of 36 h. The system was kept anaerobic by flushing it with nitrogen. The pH was maintained at 5.8. Fermentation products, such as short chain fatty acids, and water were removed by pumping dialysate through hollow fiber membranes inside the system at a flow rate of 2 ml/min.
(iii) Sampling.
To evaluate the survival rate of the recombinant model of S. cerevisiae and quantify the CA4H activity in the digestive environment, 1010 yeast cells and 200 μmol of trans-cinnamic acid were simultaneously introduced either into the stomach of TIM 1 or into TIM 2. Both the stability of trans-cinnamic and p-coumaric acids and the specificity of the enzymatic reaction were checked by control experiments. In the control experiments, 200 μmol of trans-cinnamic acid or 200 μmol of p-coumaric acid was introduced either into the stomach of TIM 1 or into TIM 2, without yeasts or with 1010 yeast cells with no CA4H gene in their plasmid.
The survival rate of the yeasts and the CA4H activity were evaluated in TIM 1, taking one sample in the meal before its introduction in the stomach and regularly collecting samples in each digestive compartment until the amount of meal was considered negligible (less than 5% of the food intake, i.e., after 90, 150, 240, and 240 min in the stomach, duodenum, jejunum, and ileum, respectively). The jejunal absorption and the ileal effluents were collected on ice. The collection vessels were replaced at 30, 45, 60, 90, 120, 180, and 240 min. The volumes were measured and samples were taken for each period. The digestive samples were immediately analyzed. In order to evaluate the yeast survival rate in TIM 1, a nonabsorbable, water-soluble, easy-to-quantify marker, blue dextran (0.08% [wt/vol] in water), was added in the artificial stomach at the beginning of digestion, as previously described (17).
The yeast survival rate was evaluated in TIM 2, taking samples every hour for 12 h following the introduction of the yeasts, then at 24 and 36 h. Additional samples were collected every 30 min during the first 4 h of incubation to monitor the CA4H activity. The digestive samples were immediately analyzed.
Computer simulation.
Owing to the dynamics of both the gastric-small intestinal system and the enzymatic reaction, a computer simulation had to be developed to quantify the CA4H activity in each digestive compartment. The bioconversion reaction was quantified by measuring the production of p-coumaric acid. At each time of digestion, the continuous process control (i.e., gastric and ileal deliveries, secretion, and absorption flows) have afforded precise knowledge of the meal dilution rate in each compartment of TIM 1. A simulation based on an algorithm which used Visual Basic for application 6 with Excel 2000 (Microsoft) was developed to combine these data with the p-coumaric acid measurements. By means of the computer simulation, in each compartment, the amount of p-coumaric acid resulting from the activity of the yeasts could be dissociated from that delivered by the previous compartment. The curves of the cumulative deliveries of p-coumaric acid were determined in each compartment, based on values measured in that compartment. These data have afforded mass balances and the calculation of the trans-cinnamic acid conversion rates in each compartment of TIM 1.
Analysis of the digestive samples. (i) Yeast counts.
The digestive samples were diluted in physiological water and plated onto SGI selective medium supplemented with ampicillin (100 μg/ml) for samples collected in TIM 2. The yeast survival rate was expressed as a percentage of the total number of cells introduced (either into TIM 1 or into TIM 2). The factors of dilution due to digestive secretions (TIM 1) or resulting from the regular input of feeding medium and output of colon medium (TIM 2) were taken into account in the calculations.
(ii) HPLC analysis.
The enzymatic reaction was stopped immediately after sampling by adding a solution of trifluoroacetic acid (2.5% wt/vol). Before high-performance liquid chromatography (HPLC) analysis, the digestive samples were centrifuged to remove the cells and filtered (0.45-μm pore size). Ten microliters of the filtrate was analyzed on a Lichrospher 100 RP-18 (5 μm) column (125- by 4-mm inside diameter). Elution was performed with a flow rate of 1 ml/min and a gradient of two solvents, A and B, composed of water-methanol-acetic acid (95/5/0.1, vol/vol/vol) and acetonitrile-methanol-acetic acid (95/5/0.1, vol/vol/vol), respectively. The HPLC analysis was started with 90% of solvent A and 10% of solvent B. After 16 min, solvent B was added, reaching 20% within 2 min. These conditions were maintained for 14 min, and initial conditions were then recovered within 2 min. trans-cinnamic and p-coumaric acids were detected by UV absorbance at 280 and 314 nm, respectively, and quantified by using standard curves established in the digestive medium (in either ileal effluents or large intestinal medium, both recovered from control experiments without p-coumaric and trans-cinnamic acids).
RESULTS
Survival rate of recombinant yeasts in gastrointestinal tract systems. (i) Gastric-small intestinal system.
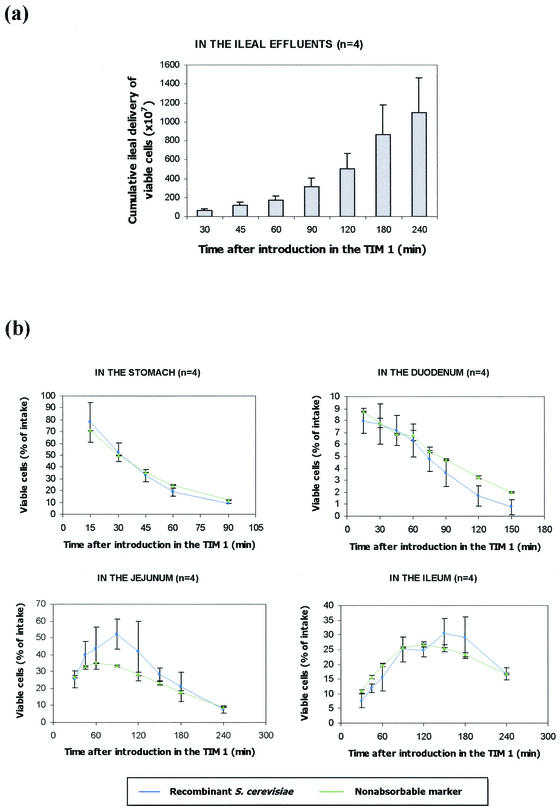
The yeast survival rate in TIM 1 was evaluated in two different ways. First, we compared, at the end of digestion (240 min), the total ingested yeasts with the living yeasts recovered in both the ileal effluents (Fig. 3a) and the residual digestive medium. At the end of the digestion, 75.9% ± 12.1% (n = 4) of the ingested cells were recovered in the ileal effluents, giving a 95.6% ± 10.1% (n = 4) survival rate when the yeasts remaining in the residual digestive medium were added (data not shown). This high survival rate shows the high resistance of the yeasts to gastric (pepsin and lipase) and small intestinal (bile salts and pancreatic enzymes) secretions and low gastric pH. Second, the viability of yeasts was evaluated in each compartment of TIM 1 by comparing the curves obtained for the yeasts and for a nonabsorbable marker, blue dextran. The high survival rate was confirmed, no significant difference being observed in any digestive compartment between the curves for the recombinant yeasts and the nonabsorbable marker (Fig. 3b).
FIG. 3.
Survival rate of the recombinant model of S. cerevisiae in TIM 1. Panel a represents the mean cumulative ileal delivery of viable cells ± standard deviation (n = 4). The curves obtained in each digestive compartment for the recombinant yeasts (blue circles) and for a nonabsorbable marker, blue dextran (green triangles), are shown in panel b. Values are expressed as mean percentages ± standard deviation (n = 4) of the initial intake.
(ii) Large intestinal system.
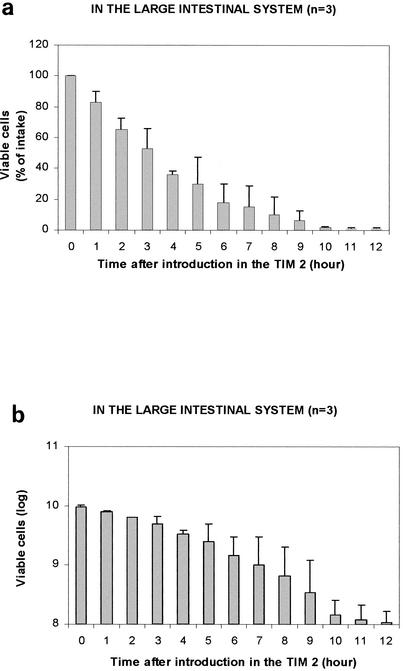
Yeast viability was also evaluated in TIM 2 (Fig. 4). They were more sensitive to the large intestinal conditions, only 35.9% ± 2.7% (n = 3) of the ingested cells remained alive after 4 h of incubation. This survival rate decreased to 1.2% ± 0.4% (n = 3) after 12 h, and no more yeast could be detected in TIM 2 after 24 h of incubation.
FIG. 4.
Survival rate of the recombinant model of S. cerevisiae in TIM 2. Values are expressed as mean percentages ± standard deviations (n = 3) of viable yeasts relative to the total amount introduced in the large intestine (a) or mean number of viable cells ± standard deviation (n = 3) with a logarithmic ladder (b).
Monitoring bioconversion activity in the digestive environment. (i) Gastric-small intestinal system.
Control experiments in TIM 1 showed that both trans-cinnamic and p-coumaric acids were stable under digestive conditions when no yeast was introduced in the artificial stomach. In the presence of yeasts with no CA4H gene in their plasmid, the p-coumaric acid was stable, but the trans-cinnamic acid was not. A loss of about 15% of the ingested trans-cinnamic acid was measured, which was not associated with a production of p-coumaric acid (data not shown), showing the specificity of the enzymatic reaction catalyzed by the recombinant model yeasts. Similar results were obtained in batch cultures (data not shown), suggesting the existence in yeasts of a secondary metabolic pathway which uses trans-cinnamic acid. Consequently, the enzymatic activity of the yeasts was evaluated by measuring the p-coumaric acid production and not the trans-cinnamic acid breakdown.
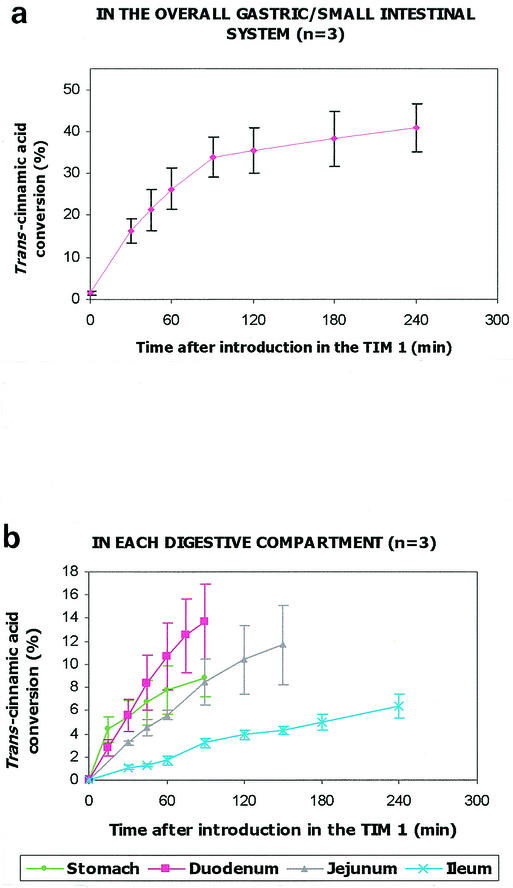
When recombinant yeasts and trans-cinnamic acid were simultaneously introduced in TIM 1, the p-coumaric acid production was regularly measured during digestion to quantify the CA4H activity in the overall in vitro system. For the first time, the ability of recombinant S. cerevisiae to perform a bioconversion reaction is shown in the upper part of the human gastrointestinal tract, as 41.0% ± 5.8% (n = 3) of the ingested trans-cinnamic acid was converted into p-coumaric acid after 240 min of digestion (Fig. 5a). Most of the reaction occurred at the beginning of digestion; a conversion rate of 33.8% ± 4.8% (n = 3) was already reached 90 min after yeast intake.
FIG. 5.
CA4H activity of the recombinant model of S. cerevisiae in TIM 1. The trans-cinnamic acid conversion was evaluated in the overall TIM 1 (a) and, thanks to the computer simulation, in each compartment of the TIM 1 (b). Values are expressed as mean cumulative percentages ± standard deviations (n = 3) of ingested trans-cinnamic acid converted into p-coumaric acid.
By means of the computer simulation, we were able to show that the enzymatic reaction occurred in each digestive compartment and that most of the ingested trans-cinnamic acid was converted into p-coumaric acid in the duodenum and jejunum. Conversion rates of 8.9% ± 1.6%, 13.8% ± 3.3%, 11.8% ± 3.4%, and 6.5% ± 1.0% (n = 3) were found in the stomach, duodenum, jejunum, and ileum, respectively (Fig. 5b). We checked that no trans-cinnamic acid conversion occurred in the ileal effluents.
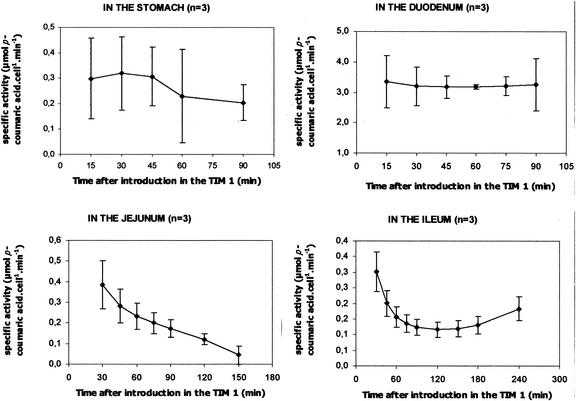
Further calculations were performed to quantify the specific enzymatic activity of the recombinant yeasts in each compartment (Fig. 6). In the stomach and duodenum, the specific activity was stable during digestion (in the stomach, between 0.30 × 10−10 ± 0.16 × 10−10 μmol/cell/min at 15 min and 0.20 × 10−10 ± 0.07 × 10−10 μmol/cell/min at 90 min; in the duodenum, between 3.36 × 10−10 ± 0.86 × 10−10 μmol/cell/min at 15 min and 3.17 × 10−10 ± 0.38 × 10−10 μmol/cell/min at 45 min). In the jejunum, the specific activity regularly decreased from 0.38 × 10−10 ± 0.12 × 10−10 μmol/cell/min at 30 min to 0.05 × 10−10 ± 0.04 × 10−10 μmol/cell/min at 150 min. In the ileum, the activity decreased for the first 90 min from 0.30 × 10−10 ± 0.06 × 10−10 μmol/cell/min to 0.12 × 10−10 ± 0.03 × 10−10 μmol/cell/min and was quite stable from 90 min to the end of digestion. The specific activity was obviously higher in the duodenum than in the other compartments.
FIG. 6.
Specific CA4H activity of the recombinant model of S. cerevisiae in each compartment of TIM 1. Values are expressed as mean percentages ± standard deviations (n = 3) of micromoles of p-coumaric acid produced per yeast cell and per minute (1010).
(ii) Large intestinal system.
CA4H activity was also observed in TIM 2 but was too weak to be quantified. Very small quantities of p-coumaric acid (about 1 μmol) were rapidly detected after the introduction of trans-cinnamic acid only in the presence of recombinant yeasts, showing the specificity of the reaction, and this low level remained until the complete disappearance of the substrate (data not shown). Control experiments showed that both trans-cinnamic and p-coumaric acids were rapidly metabolized by the colonic microflora. Consequently, the p-coumaric acid we measured resulted from a balance between that derived from the 4-hydroxylation of trans-cinnamic acid by the yeasts and that degraded by the colonic microflora. These results suggest that the yeasts were metabolically active in TIM 2 as long as trans-cinnamic acid was available, i.e., for at least 4 h.
DISCUSSION
Our aim was to determine whether recombinant yeasts were able to carry out a reaction of bioconversion directly in the human digestive tract and could serve as hosts for the development of biodrugs, in particular to perform in situ biodetoxication. We therefore studied the viability and enzymatic activity of recombinant S. cerevisiae expressing the model P450 73A1 (CA4H) in artificial digestive systems (TIM) simulating the human digestive environment.
Viability of recombinant yeasts in the digestive environment.
The recombinant model yeasts showed a high survival rate in TIM 1 (95.6% ± 10.1% [n = 4] after 240 min of digestion) in spite of the acid pH of the stomach, the bile salts, and the proteolytic activities of digestive enzymes, whereas in TIM 2, the viability was lower (less than 1% after 12 h of incubation).
Few studies are available on wild S. cerevisiae spp. viability in the human digestive environment. Furthermore, yeast survival rates have only been evaluated in feces (and not throughout the length of the gastrointestinal tract) after oral single or multiple administration of dried yeasts to volunteers. For instance, after a single administration of 1 g of Saccharomyces boulardii (1010.4 CFU) to healthy volunteers, Klein et al. (14) found a fecal recovery of 0.12% ± 0.04% (n = 8). In this study, the mean recovery in stools was found to be less than 5%, independently of the administered dose or whether single or multiple administration was used. This low recovery rate substantiates the results of Blehaut et al. (5), who measured a steady-state fecal recovery of 0.36% ± 0.31% (n = 8) after daily oral administration of 1 g of S. boulardii, and with those of Pecquet et al. (20), who reported a 2% steady-state recovery following daily ingestion of 3 × 108 S. cerevisiae cells. Although the comparison between in vivo data and our in vitro results is hampered by differences in yeast intake and by the fact that TIM 1 and TIM 2 were not directly connected, these in vivo survival rates are very close to those we obtained.
The survival rate of other microorganisms, such as lactic acid bacteria, have also been studied in TIM 1. Marteau et al. (16) observed a bacterial cumulative delivery from the ileal compartment of between 0 and 25% (depending on the bacterial strains) after 240 min of digestion. In our study, under similar experimental conditions, we found a higher survival rate for the recombinant model of S. cerevisiae, 75.9% ± 12.1% (n = 4) of the ingested yeasts being recovered in the ileal effluents after 240 min of digestion. This difference of viability in the digestive environment may favor the choice of yeasts over lactic acid bacteria as hosts for the development of biodrugs (4).
Bioconversion activity of recombinant yeasts in the digestive environment.
In this study, we showed that the recombinant model of S. cerevisiae was able to exert CA4H activity throughout the artificial digestive tract, validating the use of yeasts as potential hosts for the development of biodrugs. In TIM 1, 41.0% ± 5.8% (n = 3) of the ingested trans-cinnamic acid was converted into p-coumaric acid after 240 min of digestion. In TIM 2, the CA4H activity was detected until 4 h after the introduction of the yeasts but could not be quantified, owing to the degradation of both the substrate and the product by colonic microflora (26). A more suitable bioconversion model is consequently being studied for the colon.
In TIM 1, the CA4H activity was quantified by measuring the appearance of p-coumaric acid and not the breakdown of trans-cinnamic acid. A control digestion performed in the presence of the yeasts with no CA4H gene in their plasmid showed a partial loss of the ingested trans-cinnamic acid (not coupled with the production of p-coumaric acid). This result, also observed in classic batch cultures (data not shown), may be explained by an alternative metabolic pathway in S. cerevisiae converting trans-cinnamic acid into the phenylalanine amino acid (D. Pompon, personal communication).
Several hypotheses were advanced to quantify the CA4H activity in TIM 1. First, we hypothesized that the CA4H activity was not substrate limited. This was confirmed because, at the end of digestion, about 30% of the trans-cinnamic acid ingested was recovered. Secondly, we hypothesized that the concentrations of trans-cinnamic and p-coumaric acids in digestive samples could be evaluated by using standard curves plotted in the ileal effluents of a control digestion. We checked the relevance of these standard curves by comparing the signals obtained in HPLC with similar quantities of trans-cinnamic and p-coumaric acids added in samples collected in either the different digestive compartments or the ileal effluents.
CA4H activity was quantified in two independent ways in TIM 1. First, the trans-cinnamic acid conversion was measured in the overall TIM 1 (Fig. 5a). Second, by means of the computer simulation, the trans-cinnamic acid conversion was quantified in each digestive compartment (Fig. 5b). At each sampling time, the sum of the data calculated in each compartment fit with the data measured in the overall TIM 1, validating the computer simulation. This computer simulation should prove useful in future stages of the development of biodrugs, especially if a specific level of the digestive tract has to be targeted for drug administration.
In TIM 1, the bioconversion reaction occurred very fast, with most of the p-coumaric acid produced within the first 90 min of digestion. The same rapidity was observed in batch cultures (pH 5.5), with no more substrate detected after 120 min, when 7 × 106 cells/ml were incubated with 0.2 mM trans-cinnamic acid (25). When the pH of the culture medium was acidified to pH 3, we found that complete substrate conversion was achieved in 60 min (data not shown). We further evaluated in batch cultures the influence of extracellular pH (ranging from pH 1 to 8) on the yeast specific activity, and we showed that pH 3 seems to be the optimal pH of the recombinant CA4H (data not shown). The pH influences the trans-cinnamic acid uptake by yeasts: at a pH of >5, trans-cinnamic acid is essentially in an anionic form, which slowly diffuses through the yeast membrane. Castelli et al. (7) showed in vitro that the trans-cinnamic acid solubility in the lipid membranes of liposomes is 3 times higher at pH 4 than at pH 7.4.
In the digestive environment, numerous factors such as pH, digestive secretions (particularly bile salts), substrate form (complex or simple), and substrate availability could influence the yeast activity. In the artificial stomach, yeasts seemed to be stressed by the acid pH; half of the trans-cinnamic acid conversion in this compartment was achieved within the first 15 min, when the pH was still not too low. Probably owing to the low pH, the specific activity in the stomach was weak in spite of the large number of cells and the elevated concentration of substrate. Most of the trans-cinnamic acid conversion occurred in the duodenum. This data was correlated with a higher specific activity of yeasts in the duodenum than in the other digestive compartments. The high specific activity in the duodenum may firstly be explained by the fact that yeasts were no longer stressed by the acid pH of the stomach and could metabolize the trans-cinnamic acid that had previously easily entered the cells, owing to low gastric pH. Secondly, at this level of the gastrointestinal tract, yeasts can find nutrients in the digestive medium, made available by the proteolytic activity of pancreatic enzymes. Also, some studies have demonstrated that bile salts can favor enzymatic reactions (30). The lower specific activity in the jejunum and ileum might result from a decrease in trans-cinnamic acid availability owing to its previous conversion into p-coumaric acid in the upper digestive compartments.
The recombinant cells were introduced in TIM 1 at the beginning of their stationary growth phase to follow the same experimental protocol of yeast culture and P450 induction as that previously described (25). This enabled us to compare our results (in terms of P450 specific activity) with those previously obtained with classical batch cultures so as to assess the effects of digestive conditions on the CA4H activity of the recombinant S. cerevisiae. We found that yeast specific activity in TIM 1 (ranging from 3.36 × 10−10 ± 0.86 × 10−10 μmol/cell/min and 0.05 × 10−10 ± 0.04 × 10−10 μmol/cell/min, depending on both the compartment and the sampling time) was close to that observed in classic batch cultures, where a bioconversion rate of 8 × 107 p-coumaric acid molecules/cell/min was found, i.e., 1.3 × 10−10 μmol of p-coumaric acid/cell/min (25). These data are very encouraging for the potential use of S. cerevisiae efficiently expressing a P450 as a biodetoxication system in the digestive environment. This high specific activity was particularly remarkable as the expression strategy of CA4H had not yet been adapted to the particular constraints of the digestive environment.
For the first time, we showed the ability of recombinant S. cerevisiae to carry out a reaction of bioconversion throughout the gastrointestinal tract. This work is the first step in the development of recombinant yeasts as innovative vectors for in vivo bioconversion. The main potential application, directly derived from this validation performed with a model P450, is the use of recombinant yeasts as a biodetoxication system in the digestive tract.
Acknowledgments
This work was supported by the French Ministère de l'Education Nationale, de la Recherche, et de la Technologie through a Ph.D. grant.
We thank Denis Pompon for providing the S. cerevisiae strain. We thank J. Cohade and S. Rabot for valuable comments and Virginie Bélard, Joelle Masson, and Séverine Rougeol for technical assistance.
REFERENCES
- 1.Alric, M., S. Blanquet, S. Marol-Bonnin, D. Pompon, and M. Renaud. December 2000. Microorganismes actifs dans l'environnement digestif. International patent WO 01/98461.
- 2.Anzenbacher, P., and E. Azenbacherova. 2001. Cytochrome P450 and metabolism of xenobiotics. Cell. Mol. Life Sci. 58:737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergogne-Berezin, E. 2000. Treatment and prevention of antibiotic associated diarrhoea. Int. J. Antimicrob. Agents 16:521-526. [DOI] [PubMed] [Google Scholar]
- 4.Blanquet, S., S. Marol-Bonnin, E. Beyssac, D. Pompon, M. Renaud, and M. Alric. 2001. The biodrug concept: an innovative approach to therapy. Trends Biotechnol. 19:393-400. [DOI] [PubMed] [Google Scholar]
- 5.Blehaut, H., J. Massot, G. W. Elmer, and R. Levy. 1989. Disposition kinetics of Saccharomyces boulardii in man and rat. Biopharm. Drug Dispos. 10:353-364. [DOI] [PubMed] [Google Scholar]
- 6.Canganella, F., S. Paganini, M. Ovidi, A. M. Vettraino, L. Bevilacqua, S. Massa, and L. D. Trovatelli. 1997. A microbiology investigation on probiotic pharmaceutical products used for human health. Microbiol. Res. 152:171-179. [DOI] [PubMed] [Google Scholar]
- 7.Castelli, F., N. Uccella, D. Trombetta, and A. Saija. 1999. Differences between coumaric and cinnamic acids in membrane permeation as evidenced by time-dependent calorimetry. J. Agric. Food Chem. 47:991-995. [DOI] [PubMed] [Google Scholar]
- 8.Chang, T. M., and S. Prakash. 1998. Therapeutic uses of microencapsulated genetically engineered cells. Mol. Med. Today 4:221-227. [DOI] [PubMed] [Google Scholar]
- 9.Corthier, G., and P. Renault. 1999. Future directions for research on biotherapeutic agents: contribution of genetics approaches on lactic acid bacteria, p. 269-304. In G. W. Elmer, L. McFarland, and C. Surawicz (ed.), Biotherapeutic agents and infections diseases. Humana Press, Inc., Totowa, N.J.
- 10.Drouault, S., G. Corthier, S. D. Ehrlich, and P. Renault. 1999. Survival, physiology, and lysis of Lactococcus lactis in the digestive tract. Appl. Environ. Microbiol. 65:4881-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drouault, S. 1999. Lactococcus lactis, vecteur de lipase dans le tractus digestif, application au traitement de la stéatorrhée. Ph.D. thesis. University of Paris XI, Orsay, France.
- 12.Fahl, W. E., D. Loo, and H. Manoharan. June1999. Chemoprotective bacterial strains. International patent WO 99/27953.
- 13.Harms, H. K., R. M. Bertele-Harms, and D. Bruer-Kleis. 1987. Enzyme-substitution therapy with the yeast Saccharomyces cerevisiae in congenital sucrase-isomaltase deficiency. N. Engl. J. Med. 316:1306-1309. [DOI] [PubMed] [Google Scholar]
- 14.Klein, S. M., G. W. Elmer, L. V. McFarland, C. M. Surawicz, and R. H. Levy. 1993. Recovery and elimination of the biotherapeutic agent, Saccharomyces boulardii, in healthy human volunteers. Pharm. Res. 10:1615-1619. [DOI] [PubMed] [Google Scholar]
- 15.Larsson, M., M. Minekus, and R. Havenaar. 1997. Estimation of the bioavailability of iron and phosphorus in cereals using a dynamic in vitro gastro-intestinal model. J. Sci. Food Agric. 74:99-106. [Google Scholar]
- 16.Marteau, P., M. Minekus, R. Havenaar, and J. H. J. Huis in't Veld. 1997. Survival of lactic acid bacteria in a dynamic model of the stomach and small intestine: validation and the effects of the bile. J. Diary. Sci. 80:1031-1037. [DOI] [PubMed] [Google Scholar]
- 17.Minekus, M., P. Marteau, R. Havenaar, and J. H. J. Huis in't Veld. 1995. A multicompartmental dynamic computer-controlled model simulating the stomach and small intestine. ATLA 23:197-209. [Google Scholar]
- 18.Minekus, M., M. Smeets-Peter, A. Bernalier, S. Marol-Bonnin, R. Havenaar, P. Marteau, M. Alric, G. Fonty, and J. H. J. Huis in't Veld. 1999. A computer-controlled system to simulate conditions of the large intestine with peristaltic mixing, water absorption and absorption of fermentation products. Appl. Microbiol. Biotechnol. 53:108-114. [DOI] [PubMed] [Google Scholar]
- 19.Molly, K., M. Vande Woestyne, and W. Verstraete. 1993. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 39:254-258. [DOI] [PubMed] [Google Scholar]
- 20.Pecquet, S., D. Guillaumin, C. Tancrede, and A. Andremont. 1991. Kinetics of Saccharomyces cerevisiae elimination from the intestines of human volunteers and effect of this yeast on resistance to microbial colonization in gnotobiotic mice. Appl. Environ. Microbiol. 57:3049-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pouwels, P. H., R. J. Leer, M. Shaw, M. J. H. D. Bak-Glashouwer, F. D. Tielen, E. Smit, B. Martinez, J. Jore, and P. L. Conway. 1998. Lactic acid bacteria as antigen delivery vehicles for oral immunization purposes. Int. J. Food Microbiol. 41:155-167. [DOI] [PubMed] [Google Scholar]
- 22.Prakash, S., and T. M. S. Chang. 2000. In vitro and in vivo uric acid lowering by artificial cells containing microencapsulated genetically engineered E. coli DH5 cells. Int. J. Artif. Organs 23:429-435. [PubMed] [Google Scholar]
- 23.Raibaud, P., R. Ducluzeau, F. Dubos, S. Hudault, H. Bewa, and M. C. Muller. 1980. Implantation of bacteria from the digestive tract of man and various animals into gnotobiotic mice. Am. J. Clin. Nutr. 33:2440-2447. [DOI] [PubMed] [Google Scholar]
- 24.Steidler, L., W. Hans, L. Schotte, S. Neirynck, F. Obermeier, W. Falk, W. Fiers, and E. Remaut. 2000. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289:1352-1355. [DOI] [PubMed] [Google Scholar]
- 25.Urban, P., D. Werck-Reichhart, H. G. Teutsch, F. Durst, S. Regnier, M. Kazmaier, and D. Pompon. 1994. Characterization of recombinant plant cinnamate 4-hydroxylase produced in yeast. Kinetic and spectral properties of the major plant P450 of the phenylpropanoid pathway. Eur. J. Biochem. 222:843-850. [DOI] [PubMed] [Google Scholar]
- 26.Van Beek, S., and F. G. Priest. 2000. Decarboxylation of substituted cinnamic acids by lactic acid bacteria isolated during malt whisky fermentation. Appl. Environ. Microbiol. 66:5322-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vatier, J., F. Lionnet, M. T. Vitre, and M. Mignon. 1988. A model of an “artificial stomach” for assessing the characteristics of an antacid. Aliment. Pharmacol. Ther. 2:461-470. [DOI] [PubMed] [Google Scholar]
- 28.Wells, J. M., K. Robinson, L. M. Chamberlain, K. M. Schofield, and R. W. F. Le Page. 1996. Lactic acid bacteria as vaccine delivery vehicles. Antonie Leeuwenhoek 70:317-330. [DOI] [PubMed] [Google Scholar]
- 29.Werck-Reichhart, D. 1995. Cytochromes P450 in phenylpropanoid metabolism. Drug Metab. Drug Interact. 12:221-243. [DOI] [PubMed] [Google Scholar]
- 30.Zarate, G., A. P. Chaia, S. Gonzalez, and G. Oliver. 2000. Viability and beta-galactosidase activity of diary propionibacteria subjected to digestion by artificial gastric and intestinal fluids. J. Food Prot. 63:1214-1221. [DOI] [PubMed] [Google Scholar]