Abstract
Fas is a cell surface death receptor that regulates peripheral tolerance and lymphoid homeostasis. In many pathologic conditions, ectopic Fas activation mediates tissue destruction. Several proteins that can bind to the cytoplasmic death domain of Fas have been implicated in Fas signal transduction. Here we show that FADD, which couples Fas to pro-caspase-8, and, Daxx, which couples Fas to the Jun N-terminal kinase pathway, bind independently to the Fas death domain. We have isolated a death domain mutant, termed FasΔ, that selectively binds Daxx but not FADD. In tranfected tissue culture cells, FasΔ activated Jun N-terminal kinase normally but was impaired in cell death induction. These results suggest that FADD and Daxx activate two independent pathways downstream of Fas and confirm the essential role of FADD binding in apoptosis induction.
Members of the tumor necrosis factor receptor (TNFR) superfamily are cell surface cytokine receptors that control critical cell fate decisions such as proliferation, differentiation, and apoptosis. The TNFR superfamily includes the B cell costimulatory receptor CD40, receptors for the inflammatory cytokine TNF-α, and a growing family of death receptors (1). The prototype of the death receptors is Fas (also named CD95 and APO-1), a ubiquitously expressed receptor for a cytotoxic ligand (FasL) normally expressed on activated T cells and certain immune privileged sites (2, 3). Fas has an essential role in maintaining homeostasis in the immune system, and both deficient or excess Fas signaling can cause human diseases. Following repeated T cell activation, T cells express FasL and commit autocrine suicide, a process termed activation-induced cell death that is important for normal restraint of the immune response (4). Depending on the signal from the B cell antigen receptor, Fas induces either apoptosis or proliferation of B cells in vivo (5). Mice and human deficient in Fas function develop massive lymphadenopathy and are prone to develop severe autoimmune disease (2). On the other hand, ectopic activation of FasL–Fas interaction by cytokines or drugs underlies tissue destruction in fulminant hepatitis, autoimmune Hashimoto’s thyroiditis and type I diabetes, and toxic epidermal necrolysis (6–9). In addition, as a result of Fas-mediated neutrophil activation, FasL causes potent inflammatory responses in many tissues (10).
The signal transduction pathways downstream of Fas have been studied with rapid progress over the last several years. The cytoplasmic domain of Fas has no enzymatic activity but contains a protein-interaction motif termed the “death domain.” An adapter protein, termed FADD or Mort1, binds to the Fas death domain and recruits pro-caspase-8, the zymogen form of an apical cell death protease (11–12). Fas crosslinking by the trimeric FasL or agonisitic antibody induces the proximity of pro-caspase-8 molecules, allowing them to cleave one another and become activated (13–16). Active caspase-8 can then cleave other pro-caspases and activate a caspase cascade, leading to cell death (17). Several viral and cellular pro-caspase-8 like proteins, termed FLIPs (18), contain FADD-interacting DED motifs but inactive protease domains, and they are potent inhibitors of Fas-induced cell death in tissue culture cells and in primary lymphocytes (19–20). An alternate pathway involves the Fas-binding protein Daxx (21). On Fas activation, Daxx interacts with and activates a mitogen-activated protein kinase kinase kinase termed ASK1 (Apoptosis Signal-regulating Kinase 1), leading to the activation of the Jun N-terminal kinase (JNK) and p38 MAP kinase pathways (22). JNK and p38 kinase cascades are also activated by many kinds of stress, and they culminate in the phosphorylation and activation of transcription factors such as AP-1, ATF-2, and other cellular targets (23). JNK has been previously implicated in activating apoptosis in vitro and in vivo (24, 25), but its functional role in death-receptor signaling, as assayed by expressing dominant negative proteins, has been controversial (21, 26–29). JNK activation has also been reported to occur downstream of caspases based on studies with caspase inhibitors (28, 30), but its functional significance and relationship to Daxx-mediated JNK activation are unclear.
The in vivo role of the Fas-FADD-pro-caspase-8 axis has been bolstered by the generation of mice deficient for FADD or overexpressing the FADD death domain (1). FADD-deficient mice are embryonic lethal due to cardiac defects (31). FADD−/− embryonic stem (ES) cells are resistant to Fas, TNF-α, and some (but not all) death receptors (31). Similarly, Casp8-deficient embryos are prenatally lethal, and Casp8−/− fibroblasts are resistant to death induction by Fas, TNF-α, and the death receptor family member DR3 (32). These results strongly suggest that FADD-mediated caspase activation is essential for Fas-induced apoptosis, but possible secondary effects of FADD or caspase-8 deficiency could not be ruled out. For example, FADD-deficient T cells do not proliferate (33), making it impossible to test the role of FADD in the natural context of activation-induced cell death.
In this report, we have used a complementary approach to dissect Fas signaling. We demonstrate that FADD and Daxx bind independently to Fas and identify a Fas mutant that selectively fails to bind FADD. This mutant is substantially impaired in apoptosis induction but is fully capable of activating JNK, illustrating the two distinct pathways mediated by FADD and Daxx downstream of Fas.
MATERIALS AND METHODS
Reagents and Cell Lines.
Anti-murine Fas Jo2 antibody (PharMingen) and anti-Flag mAb M2 (Kodak/IBI) were obtained from the indicated sources. HeLa and 293 cells were obtained originally from American Type Culture Collection.
Plasmid Construction.
Glutathione S-transferase (GST) constructs were made in pGEX vector (Amersham Pharmacia). pEBB-Myc-Fas and pEBB-Myc-FasΔ were each constructed with a Myc epitope tag inserted after the leader sequence by using PCR with Pfu polymerase (Stratagene). Each construct was confirmed by partial DNA sequencing and by immunoblot analysis.
In Vitro Binding Assays.
Glutathione S-transferase (GST) fusions were purified as described (34). 35S-labeled proteins were made with TNT Reticulocyte Lysate System (Promega) from pRK5-FADD(80–205) (35) or pRSET-H6T-DaxxC (21). The in vitro translation (IVT) of FADD(80–205) gave approximately 100-fold more protein than that of DaxxC. 35S-labeled proteins were incubated with 2 μg of each GST fusion protein in 0.1 ml of modified E1A buffer (36) with 50 mM NaCl and 10% glycerol for 1 to 2 hours, washed three times, and analyzed by using SDS/PAGE and autoradiography. Recombinant purified hFADD(95–208) was the gift of B. Huang and S. Fesik (Abbott).
Apoptosis Assay.
Transient transfection and apoptotic morphology assay in HeLa cells were performed as described (21). Specific apoptosis was calculated as the percentage of blue cells with apoptotic morphology in each experimental condition minus the percentage of blue cells with apoptotic morphology in pEBB vector-transfected cells. pEBB vector control transfection was always done in parallel and had ≈5% or less apoptotic cells.
JNK Activation Assay.
Twenty four hours after transient transfection, cells were treated with 0.5 μg/ml Jo2 antibody for 30 min. Flag-JNK1 was immunoprecipitated with anti-Flag antibody, and in vitro kinase assay with 1 μg of GST-cJun(1–79) was performed as described (21).
RESULTS
Daxx and FADD Bind Independently to Fas.
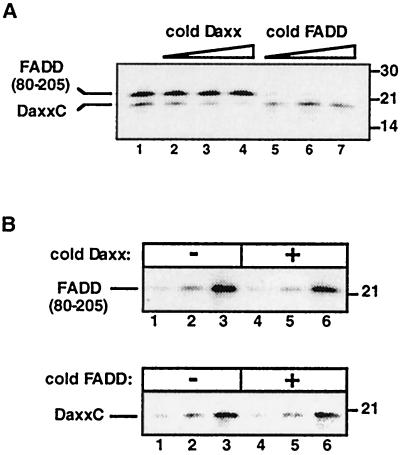
Several proteins, including FADD and Daxx, have been reported to bind to the death domain of Fas (1). FADD binding to Fas occurs via homotypic interactions of their death domains, whereas Daxx binds to Fas death domain via a novel motif, termed DaxxC, that has no sequence similarity to death domains (37, 38, 21). To understand how these interactions affect one another, we carried out binding studies in vitro with these three protein domains. In vitro translated, 35S-labeled DaxxC and the FADD death domain [FADD(80–205)] bound to the immobilized GST fusion protein of Fas intracellular tail (GST-FasIC) (Fig. 1A, lane 1). Addition of cold DaxxC or FADD death domain to the binding reaction competed with binding of the homologous 35S-labeled protein, but no cross competition was observed (Fig. 1A, lanes 2–7). This result suggests that Daxx and FADD do not compete for the same binding site on Fas. Next, to test whether the Daxx and FADD bindings are cooperative, 35S-labeled DaxxC or FADD(80–205) was titrated into the binding reaction in the absence or presence of a saturating amount of the heterologous cold protein. The amount of cold proteins used was sufficient to completely block by competition binding of the homologous material (as determined in Fig. 1A), but they did not affect the binding curve of the heterologous 35S-labeled proteins (Fig. 1B). Because Daxx and FADD binding to Fas is neither competitive nor cooperative, these results indicate that Daxx and FADD bind independently to Fas.
Figure 1.
Independent binding of DaxxC and FADD(80–205) to Fas. (A) Binding of in vitro-translated [35S]DaxxC and FADD(80–205) to GST-FasIC. [35S]DaxxC (5 μl) and [35S]FADD(80–205) (0.1 μl) IVT reactions were added in all lanes. Lanes 2, 3, and 4 contained 5, 10, or 20 μl of IVT nonradioactive DaxxC; lanes 5, 6, and 7 contained 0.5, 2, or 5 μg of recombinant purified hFADD(95–208). (B) Lack of cooperative binding of DaxxC and FADD(80–205) to GST-FasIC. Upper, [35S]FADD(80–205) IVT reaction (0.1 μl in lanes 1 and 4; 0.3 μl in lanes 2 and 5; 1.0 μl in lanes 3 and 6) were incubated with GST-FasIC in the absence (lanes 1–3) or presence (lanes 4–6) of 20 μl of nonradioactive IVT DaxxC. Lower, [35S]DaxxC IVT reaction (2 μl in lanes 1 and 4; 5 μl in lanes 2 and 5; 10 μl in lanes 3 and 6) were incubated with GST-FasIC in the absence (lanes 1–3) or presence (lanes 4–6) of 5 μg of recombinant purified hFADD(95–208). Positions of molecular weight standards (in kDa) are shown at right.
Identification of a Fas Mutant That Selectively Fails to Bind FADD.
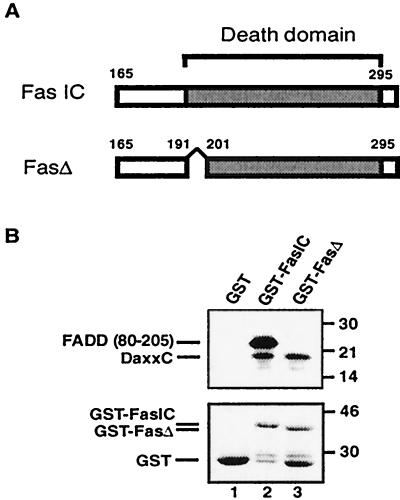
The independent binding of FADD and Daxx to Fas predicts that it should be possible to isolate Fas mutants that selectively abrogate one interaction without affecting the other. The N-terminal boundary of the Fas death domain has been assigned based on sequence similarity to the death domain of TNF receptor 1 (39, 40), and the C-terminal boundary has been determined experimentally by the cytotoxic activities of Fas mutants bearing progressive truncations from the C terminus (39). We noticed that particular N-terminal fusions of the Fas death domain exhibited differential binding properties for Daxx and FADD. These differences were localized to nine amino acids at the N terminus of the Fas death domain and we therefore compared an intact death domain (FasIC) to one lacking the first nine amino acids (192–200; FasΔ) (Fig. 2A). As shown previously, 35S-labeled DaxxC and FADD(80–205) bound specifically to GST-FasIC but not GST (Fig. 2B). GST-FasΔ bound specifically to DaxxC as well as it did to GST-FasIC but did not bind detectably to FADD(80–205). Previous structural studies revealed that the Fas death domain consists of six antiparallel amphipathic helices; charged residues in helix 2 and 3 are thought to mediate electrostatic interactions with FADD (42). Because amino acids 192–200 are positioned far away from this region, the defect of FasΔ in FADD binding may result from allosteric effects. The ability of FasΔ to selectively bind Daxx but not FADD confirms that Daxx and FADD bind independently to Fas and provides a potentially useful tool to dissect Fas signaling.
Figure 2.
Identification of FasΔ. (A) Schematic representation of fusion proteins encompassing the cytoplasmic domain of murine Fas (41). Boundaries of the death domain and FasΔ deletion are indicated. (B) Binding of in vitro translated [35S]DaxxC and FADD(80–205) to GST-fusion proteins. [35S]DaxxC (10 μl) and [35S]FADD(80–205) (0.2 μl) IVT reactions were added to all lanes. Positions of MW standards (in kDa) are shown at right (Upper). Coomassie blue-stained GST fusion proteins from the same gel is shown (Lower).
JNK Activation and Apoptosis by FasΔ.
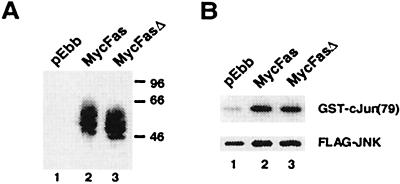
Based on overexpression experiments with wild-type and dominant negative proteins, we previously suggested that Daxx is both sufficient and necessary to mediate JNK activation by Fas in human embryonic kidney 293 cells (21). Fas also activates JNK in HeLa cells by a FADD-independent mechanism (43). To test how the FasΔ mutation may affect Fas signaling, we constructed full-length murine Fas and FasΔ with a Myc-epitope tag on the N-terminal extracellular domain. Myc-Fas and Myc-FasΔ were equivalently expressed after transient transfection into 293 cells; both proteins migrated as a series of bands on SDS/PAGE gels apparently because of glycosylation (Fig. 3A). Fluorescence-activated cell sorting indicated that both Fas proteins were present on the cell surface (data not shown). Importantly, Myc-FasΔ activated JNK as well as Myc-Fas as determined by in vitro kinase assays (Fig. 3B).
Figure 3.
(A) Expression of Myc-Fas and Myc-FasΔ. Human embryonic kidney 293 cells were transfected with the indicated plasmids, immunoprecipitated with anti-Myc antibody conjugated to agarose beads, and immunoblotted with anti-Myc antibody. (B) JNK activation by Myc-Fas and Myc-FasΔ. Flag-tagged JNK1 (Flag-JNK) and the indicated plasmids were cotransfected into 293 cells; 24 hours after transfection, cells were treated with 0.5 μg/ml Jo2 antibody for 30 min and assayed for JNK kinase activity. Upper, Phosphorylation of GST-cJun. Lower, Expression of Flag-JNK. The data shown are representative of three independent assays.
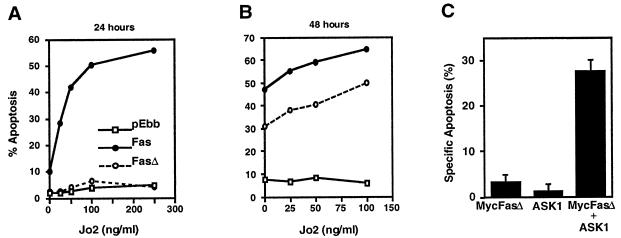
Coexpression of either FADD(80–205) or DaxxC with Fas was previously reported to inhibit Fas killing in HeLa cells (21), implying that both FADD and Daxx pathways were required for apoptosis. Expression of Myc-Fas in HeLa cells potently caused apoptosis in the presence of anti-mouse Fas antibody Jo2 in a dose-dependent manner (Fig. 4A). Strikingly, Myc-FasΔ did not cause appreciable cell death at 24 hours after transfection. At 48 hours after transfection, there was significant apoptosis in cells expressing Myc-FasΔ compared with vector transfected cells, but the level of apoptosis was still less than that caused by Myc-Fas (Fig. 4B). The apoptosis at 48 hours occurred in the absence of Jo2, which is likely the result of the known propensity of Fas death domain at higher concentrations to self-associate and initate signaling (44). The complete loss of cell killing at 24 hours (but efficient killing at 48 hours) by Myc-FasΔ suggests that Fas can activate two pathways for cell killing and that FADD binding is essential for the direct initiation of apoptosis. FADD overexpression can activate caspase-8 and quickly engage the apoptotic caspase cascade (1). Killing by Myc-FasΔ may be relatively slow because the Daxx pathway activates JNK and presumably transcription, which then indirectly activates apoptosis. We previously showed that expression of ASK1, a mitogen-activated protein kinase kinase kinase activated by Daxx, does not induce apoptosis in normal serum conditions, but coexpression with an activated allele of Daxx causes synergistic cell killing (22). We hypothesized that because FasΔ binds Daxx and presumably activates it, FasΔ should be able to activate ASK1-dependent killing. Indeed, coexpression of Myc-FasΔ and ASK1 caused synergistic killing within 24 hours (Fig. 4C). This killing is quantitatively similar to that achieved by ASK1 plus Daxx (22), implying that the Daxx pathway is intact in FasΔ. This result also suggests that the JNK pathway can induce apoptosis quickly if sufficiently activated.
Figure 4.
Apoptosis by Fas and FasΔ. HeLa cells were transfected with pEBB, pEBB-Myc-Fas, or pEBB-Myc-FasΔ plasmids with 0.5 μg of pCMV-lacZ. Jo2 antibody was added 16 hr later. The cells were stained with 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-gal) 24 hours (A) or 48 hours (B) after transfection, and blue cells were analyzed for apoptotic morphology. (C) Synergistic killing of MycFasΔ and ASK1. The indicated plasmids and pCMV-lacZ were cotransfected in HeLa cells; amount of transfected DNA were equalized by adding vector DNA. The cells were stained with X-gal 24 hr after transfection and analyzed for apoptotic morphology as described in Materials and Methods.
DISCUSSION
We have presented evidence that FADD and Daxx bind independently to the death domain of Fas and identified FasΔ, a mutation of the Fas cytoplasmic death domain that selectively fails to bind FADD. These results support previous functional data that FADD and Daxx activate distinct signaling pathways downstream of Fas (21). The independent binding of FADD and Daxx to Fas also suggests that the apoptotic defect in FADD- or casp8-deficient cells and mice reflects the intrinsic signaling role of these molecules instead of possible structural roles in allowing Daxx to bind Fas. Mutations in the Fas gene have been identified in several human lymphoproliferative disorders. Germ-line mutations in Fas were described in kindreds afflicted with Autoimmune Lymphoproliferative Syndrome (ALPS), a disease characterized by peripheral accumulation of CD4−CD8− T cells and autoantibody production, leading to autoimmune symptoms (45, 46). More recently, somatic mutations of Fas have been discovered in a sizable fraction of multiple myelomas and non-Hodgkin’s lymphomas (47, 48). In all of these diseases, heterozygous point mutations in the Fas death domain with intact expression of the wild-type allele are frequently encountered. These findings have suggested that the Fas mutants encode dominant interfering proteins, in particular by interfering with the trimerization of wild-type death domains (45). In most cases, the functional consequences of Fas mutation on downstream signal transduction pathways are not understood. It is now conceivable that in some cases, pathologic Fas mutations may inactivate only FADD or Daxx signaling, and such differences may impact disease presentation and progression. For example, the inhibition of JNK activation downstream of Fas has been shown to interfere with Fas-induced death in multiple myeloma cells (49). Whether such specificity is reflected in the disease-associated Fas mutations should be addressed in the future.
FasΔ activated JNK as effectively as wild-type Fas (Fig. 3), suggesting that JNK activation occurs independently of FADD binding or caspase activation. These results are consistent with the ability of Daxx to directly interact with and activate ASK1 (22). JNK activation also occurred normally in Casp8−/− mouse embryonic fibroblasts (MEFs) and HeLa cells stably overexpressing the FADD death domain (32, 43). Casp8−/− MEFs were resistant to killing by Fas or TNF-α, but JNK activation by either signal was unperturbed compared with wild-type MEFs (32). Together these results provide strong evidence that in some cells, Fas activates JNK by a FADD-independent pathway. Because killing by FasΔ is slow, it suggests that the Daxx pathway is insufficient to compensate completely for the loss of FADD-mediated caspase-8 activation to induce apoptosis. Although it is formally possible that the apoptotic defect in FasΔ is caused by the inability to associate with an unknown Fas-binding protein, the resistance to Fas killing in FADD−/− cells indicates that FADD is most likely the relevant missing partner of FasΔ (31, 33). Sensitivity to FADD-mediated apoptotsis can be modulated by the expression level of FLIP; therefore, a FADD-independent pathway for JNK activation may underlie the ability of Fas to stimulate cell proliferation in vivo (5). However, in Jurkat cells, Fas-mediated JNK activation appears to depend on caspase activation and caspase-8 in particular (28, 50). These differences may reflect tissue-specific regulation of Fas signaling pathways. In this regard, reconstitution of lpr cells with Fas and FasΔ may be an efficient approach to dissect Fas signaling in primary cells. Preliminary results using retroviral transduction of FasΔ into lpr T cells suggest that FADD binding to Fas is required for T cell activation-induced cell death (L. van Parijs, H.Y.C., and D.B., unpublished data).
Many questions remain regarding the role of JNK activation in death receptor signaling. Results from FADD and caspase-8 knockout mice suggest that JNK activation is insufficient to induce apoptosis; unfortunately, these studies mostly compared the killing in the short period of time that is sufficient for wild-type Fas to act. It is well known that the JNK pathway can cause apoptosis if substantially activated (24), and the results in the present study reinforce this idea. However, because FasΔ killing occurred only with high levels of expression (such that the normal dependence on crosslinking antibody is abrogated), it is possible that this does not reflect the physiologic condition. Whether the Daxx pathway is sufficient for apoptosis in vivo in other cell types should be addressed in future studies. The localization of Daxx may be partially or even mostly nuclear, the significance of which is presently unclear (51, 52). In many tissues in vivo, FasL expression predominantly elicits an inflammatory response (8, 10). Recently, Chen et al. (10) demonstrated that the proinflammatory effect of FasL is mediated by Fas-induced p38 activation in neutrophils; activated neutrophils can then kill FasL+ and bystander cells in a nonspecific fashion. These results show a physiologic role for Fas-mediated mitogen-activated protein kinase activation and illustrate an indirect pathway of cell death. Inhibition of the Daxx pathway may block the proinflammatory effect of FasL. Finally, whether the Daxx pathway may be necessary for Fas-mediated apoptosis in vivo is not addressed by the present study. Animals deficient in Daxx or that express a Fas mutant unable to bind Daxx will be helpful in addressing these questions.
Acknowledgments
We thank B. Huang, S. Fesik, and D. V. Goeddel for generous gifts of reagents, and members of the Baltimore lab for discussions and encouragement. This work was supported by the Medical Scientist Training Program at Harvard Medical School (H.Y.C.), Leukemia Society of America (X.Y.), and National Institutes of Health Grant CA51462.
ABBREVIATIONS
- JNK
Jun N-terminal kinase
- TNFR
tumor necrosis factor receptor
- GST
glutathione S-transferase
- IVT
in vitro translation
References
- 1.Ashkenazi A, Dixit V M. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 2.Nagata S, Golstein P. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 3.Abbas A K. Cell. 1996;84:655–658. doi: 10.1016/s0092-8674(00)81042-9. [DOI] [PubMed] [Google Scholar]
- 4.Van Parijs L, Abbas A K. Science. 1998;280:243–248. doi: 10.1126/science.280.5361.243. [DOI] [PubMed] [Google Scholar]
- 5.Rathmell J C, Townsend S E, Xu J C, Flavell R A, Goodnow C C. Cell. 1996;87:319–329. doi: 10.1016/s0092-8674(00)81349-5. [DOI] [PubMed] [Google Scholar]
- 6.Kondo T, Suda T, Fukuyama H, Adachi M, Nagata S. Nat Med. 1997;3:409–413. doi: 10.1038/nm0497-409. [DOI] [PubMed] [Google Scholar]
- 7.Giordano C, Stassi G, DeMaria R, Todaro M, Richiusa P, Papoff G, Ruberti G, Bagnasio M, Testi R, Galluzzo A. Science. 1997;275:960–963. doi: 10.1126/science.275.5302.960. [DOI] [PubMed] [Google Scholar]
- 8.Chernovsky A V, Wang Y, Wong F S, Visintin I, Flavell R A, Janeway C A, Matis L A. Cell. 1997;89:17–24. doi: 10.1016/s0092-8674(00)80178-6. [DOI] [PubMed] [Google Scholar]
- 9.Viard I, Wehrli P, Bullani R, Schneider P, Holler N, Salomon D, Hunziker T, Saurat J H, Tschopp J, French L E. Science. 1998;282:490–493. doi: 10.1126/science.282.5388.490. [DOI] [PubMed] [Google Scholar]
- 10.Chen J-J, Sun Y, Nabel G J. Science. 1998;282:1714–1717. doi: 10.1126/science.282.5394.1714. [DOI] [PubMed] [Google Scholar]
- 11.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 12.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, et al. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 13.Yang X, Chang H Y, Baltimore D. Mol Cell. 1998;1:319–325. doi: 10.1016/s1097-2765(00)80032-5. [DOI] [PubMed] [Google Scholar]
- 14.Muzio M, Stockwell B R, Stennicke H R, Salvesen G S, Dixit V M. J Biol Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 15.Martin D A, Siegel R M, Zheng L, Lenardo M J. J Biol Chem. 1998;273:4345–4349. doi: 10.1074/jbc.273.8.4345. [DOI] [PubMed] [Google Scholar]
- 16.Memon S A, Hou J, Moreno M B, Zacharchuk C M. J Immunol. 1998;160:2046–2049. [PubMed] [Google Scholar]
- 17.Muzio M, Salvesen G S, Dixit V M. J Biol Chem. 1997;272:2952–2956. doi: 10.1074/jbc.272.5.2952. [DOI] [PubMed] [Google Scholar]
- 18.Wallach D, Kovalenko A V, Varfolomeev E E, Boldin M P. Curr Opin Immunol. 1998;10:279–288. doi: 10.1016/s0952-7915(98)80166-0. [DOI] [PubMed] [Google Scholar]
- 19.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer J L, Schroter M, Burns K, Mattmann C, et al. Nature (London) 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 20.Refaeli Y, Van Parijs L, London C A, Tschopp J, Abbas A K. Immunity. 1998;8:615–623. doi: 10.1016/s1074-7613(00)80566-x. [DOI] [PubMed] [Google Scholar]
- 21.Yang X, Khosravi-Far R, Chang H Y, Baltimore D. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang H Y, Nishitoh H, Yang X, Ichijo H, Baltimore D. Science. 1998;281:1860–1863. doi: 10.1126/science.281.5384.1860. [DOI] [PubMed] [Google Scholar]
- 23.Kyriakis J M, Avruch J. BioEssays. 1996;18:567–577. doi: 10.1002/bies.950180708. [DOI] [PubMed] [Google Scholar]
- 24.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 25.Yang D D, Kuan C Y, Whitmarsh A J, Rincon M, Zheng T S, Davis R J, Rakic P, Flavell R A. Nature (London) 1997;389:865–870. doi: 10.1038/39899. [DOI] [PubMed] [Google Scholar]
- 26.Verheij M, Bose R, Lin X H, Yao B, Jarvis W D, Grant S, Birrer M J, Haimovitz-Friedman A, Fuks Z, Kolesnick R N. Nature (London) 1996;380:75–79. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 27.Goillot E, Raingeaud J, Ranger A, Tepper R I, Davis R J, Harlow E, Sanchez I. Proc Natl Acad Sci USA. 1997;94:3302–3307. doi: 10.1073/pnas.94.7.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenczowski J M, Dominguez L, Eder A M, King L B, Zacharchuk C M, Ashwell J D. Mol Cell Biol. 1997;17:170–181. doi: 10.1128/mcb.17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z-g, Hsu H, Goeddel D V, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 30.Cahill M A, Peter M E, Kischkel F C, Chinnaiyan A M, Dixit V M, Krammer P H, Nordheim A. Oncogene. 1996;13:2087–2096. [PubMed] [Google Scholar]
- 31.Yeh W-C, de la Pompa J L, McCurrach M E, Shu H-B, Elia A J, Shahinian A, Ng M, Wakeham A, Khoo W, Mitchell K, et al. Science. 1998;279:1954–1958. doi: 10.1126/science.279.5358.1954. [DOI] [PubMed] [Google Scholar]
- 32.Varfolomeev E E, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann J S, Mett I L, Rebrikov D, Brodianski V M, Kemper O C, Kollet O, et al. Immunity. 1998;9:267–276. doi: 10.1016/s1074-7613(00)80609-3. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Cado D, Chen A, Kabra N H, Winoto A. Nature (London) 1998;392:296–300. doi: 10.1038/32681. [DOI] [PubMed] [Google Scholar]
- 34.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 35.Hsu H, Shu H-B, Pan M-G, Goeddel D V. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 36.Hsu H, Xiong J, Goeddel D V. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 37.Boldin M P, Varfolomev E E, Pancer Z, Mett I L, Camonis J H, Wallach D. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 38.Chinnaiyan A M, O’Rourke K, Tewari M, Dixit V M. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 39.Itoh N, Nagata S. J Biol Chem. 1993;268:10932–10937. [PubMed] [Google Scholar]
- 40.Tartaglia L A, Ayres T M, Wong G H W, Goeddel D V. Cell. 1993;74:845–853. doi: 10.1016/0092-8674(93)90464-2. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe-Fukunaga R, Brannan C I, Itoh N, Yonehara S, Copeland N G, Jenkins N A, Nataga S. J Immunol. 1992;148:1274–1279. [PubMed] [Google Scholar]
- 42.Huang B, Eberstadt M, Olejniczak E T, Meadows R P, Fesik S W. Nature (London) 1996;384:299–308. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- 43.Wajant H, Johannes F J, Haas E, Siemienski K, Schwenzer R, Schubert G, Weiss T, Grell M, Scheurich P. Curr Biol. 1998;8:113–116. doi: 10.1016/s0960-9822(98)70042-9. [DOI] [PubMed] [Google Scholar]
- 44.Boldin M P, Mett I L, Varfolomeev E E, Chumakov I, Shemer-Avni Y, Camonis J H, Wallach D. J Biol Chem. 1995;270:387–391. doi: 10.1074/jbc.270.1.387. [DOI] [PubMed] [Google Scholar]
- 45.Fisher G H, Rosenberg F J, Straus S E, Dale J K, Middleton L A, Lin A Y, Strober W, Lenardo M J, Puck J M. Cell. 1995;81:935–946. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 46.Rieux-Laucat F, Le Deist F, Hivroz C, Roberts I A G, Debatin K M, Fischer A, de Villartay J P. Science. 1995;268:1347–1349. doi: 10.1126/science.7539157. [DOI] [PubMed] [Google Scholar]
- 47.Landowski T H, Qu N, Buyuksal I, Painter J S, Dalton W S. Blood. 1997;90:4266–4270. [PubMed] [Google Scholar]
- 48.Gronbaek K, Stratern P T, Ralfkiaer E, Ahrenkiel V, Andersen M K, Hansen N E, Zeuthen J, Hou-Jensen K, Guldberg P. Blood. 1998;92:3018–3024. [PubMed] [Google Scholar]
- 49.Xu F H, Sharma S, Gardner A, Tu Y, Raitano A, Sawyers C, Lichtenstein A. Blood. 1998;92:241–251. [PubMed] [Google Scholar]
- 50.Juo P, Kuo C J, Yuan J, Blenis J. Curr Biol. 1998;8:1001–1008. doi: 10.1016/s0960-9822(07)00420-4. [DOI] [PubMed] [Google Scholar]
- 51.Kiriakidou M, Driscoll D A, Lopez-Guisa J M, Strauss J F., III DNA Cell Biol. 1997;16:1289–1298. doi: 10.1089/dna.1997.16.1289. [DOI] [PubMed] [Google Scholar]
- 52.Pluta A F, Earnshaw W C, Goldberg I G. J Cell Sci. 1998;111:2029–2041. doi: 10.1242/jcs.111.14.2029. [DOI] [PubMed] [Google Scholar]