Abstract
Tri10, a regulatory gene in trichothecene mycotoxin-producing Fusarium species, is required for trichothecene biosynthesis and the coordinated expression of four trichothecene pathway-specific genes (Tri4, Tri5, Tri6, and Tri101) and the isoprenoid biosynthetic gene for farnesyl pyrophosphate synthetase (FPPS). We showed that six more trichothecene genes (Tri3, Tri7, Tri8, Tri9, Tri11, and Tri12) are regulated by Tri10. We also constructed a cDNA library from a strain of Fusarium sporotrichioides that overexpresses Tri10 (↑Tri10) and used cDNA derived from the ↑Tri10 strain and a non-Tri10-expressing strain (ΔTri10) to differentially screen macroarrays prepared from the cDNA library. This screen identified 15 additional Tri10-regulated transcripts. Four of these transcripts represent Tri1, Tri13, and Tri14 and a gene designated Tri15. Three other sequences are putative orthologs of genes for isoprenoid biosynthesis, the primary metabolic pathway preceding trichothecene biosynthesis. The remaining eight sequences have been designated Ibt (influenced by Tri10) genes. Of the 26 transcripts now known to be positively regulated by Tri10, 22 are positively coregulated by Tri6, a gene that encodes a previously characterized trichothecene pathway-specific transcription factor. These 22 Tri10- and Tri6-coregulated sequences include all of the known Tri genes (except for Tri10), the FPPS gene, and the other three putative isoprenoid biosynthetic genes. Tri6 also regulates a transcript that is not regulated by Tri10. Thus, Tri10 and Tri6 regulate overlapping sets of genes that include a common group of multiple genes for both primary and secondary metabolism.
The trichothecenes are a large group of toxic secondary metabolites produced by a variety of fungi, including Fusarium, Stachybotrys, and Myrothecium (5, 16, 28). These sesquiterpene mycotoxins are strong inhibitors of protein synthesis (29) and can cause toxicoses when humans or animals consume contaminated food or feed (19). They are capable of inducing apoptosis (30) and, in some cases, are influential in plant pathogenesis (4, 11, 24).
The biosynthetic pathway for T-2 toxin, a type A (nonmacrocyclic) trichothecene produced by some Fusarium species, has been well characterized. Most of the chemical intermediates in the pathway are known (10), and most of the genes known to be required for T-2 toxin biosynthesis in Fusarium sporotrichioides NRRL 3299 are organized in a coordinately regulated gene cluster (15); however, not all of the structural genes assumed to be responsible for these reactions have been described (1, 8, 13, 14, 20, 21, 22). Similarly, several genes for pathway regulation and partial self-protection are known (2, 17, 25, 27), but other genes related to toxin production are presumed to exist.
The expression of three representative Tri cluster genes (Tri6, Tri5, and Tri4), the expression of noncluster gene Tri101, and T-2 toxin production are controlled by Tri10, a regulatory gene in the cluster (27). Disruption of the Tri10 coding sequence dramatically reduces the transcription of these four Tri genes and effectively blocks T-2 toxin production. Disruptions in the region upstream of the Tri10 coding sequence result in T-2 toxin hyperproduction and coordinated overexpression of Tri10 and the other four Tri genes.
Tri10 is hypothesized to control all other Tri genes, both inside and outside the Tri gene cluster, in part by positively regulating Tri6 (27). Tri6 encodes a previously characterized Cys2His2 zinc finger DNA-binding protein that functions as a pathway-specific transcription factor and positively regulates the other Tri genes (15, 25). Both Tri10 and Tri6 also control transcript levels for the gene encoding farnesyl pyrophosphate synthetase (FPPS) (27). Since FPPS catalyzes the last step in the isoprenoid biosynthetic pathway, the primary metabolic pathway leading to trichothecene biosynthesis, we further hypothesized that Tri10 and/or Tri6 regulate the expression of additional isoprenoid biosynthetic genes.
Our objective in this study was to identify other genes from F. sporotrichioides involved with or related to T-2 toxin biosynthesis, including those that might be located outside the Tri gene cluster. We used radiolabeled cDNAs prepared from ΔTri10 (Tri10 knocked out) and ↑Tri10 (Tri10 upregulated) strains to screen high-density cDNA macroarrays printed from a ↑Tri10 cDNA library (i.e., a library enriched for genes positively regulated by Tri10). We also further examined the regulatory relationships of Tri10 and Tri6 by determining whether the genes regulated by Tri10 were regulated similarly by Tri6.
MATERIALS AND METHODS
Strains, media, and culture conditions.
Wild-type strains F. sporotrichioides NRRL 3299, Gibberella pulicaris (F. sambucinum) R-6380, and G. zeae (F. graminearum) GZ3639 and transformed strains FsTri10-1-12 (ΔTri10), FsTri10-1-20 (↑Tri10), and NN4 (ΔTri6) were described previously (7, 25, 27). The wild-type strains were maintained on V8 juice agar (200 ml of V8 juice, 3 g of CaCO3, and 20 g of agar per liter), and the transformed strains were maintained on V8 juice agar plus 300 μg of hygromycin B (Calbiochem, La Jolla, Calif.)/ml. Conidia for all strains were stored as frozen (−80°C) glycerol-water (15:85) stocks. Cultures were grown for 23 h at 28°C and 200 rpm in YEPD-5G medium (5% glucose, 0.1% yeast extract, 0.1% peptone) for RNA extraction or YEPD-2G medium (2% glucose, 0.3% yeast extract, 1% peptone) for DNA extraction. Individual bacterial colonies from the cDNA library were maintained on Luria-Bertani agar plates at 4°C and stored at −80°C in 384-well plates (Nalge Nunc International, Naperville, Ill.) containing multiwell plate freezer medium, which is 9 volumes of Luria-Bertani medium combined with 1 volume of 10× freezer medium (containing, per liter, 63 g of K2HPO4, 18 g of KH2PO4, 5 g of sodium citrate, 1 g of MgSO4 · 7H2O, 9 g of (NH4)2SO4, and 440 ml of glycerol). All bacterial culture media were supplemented with ampicillin at 100 μg/ml.
Nucleic acid treatments.
Procedures for the isolation of genomic DNA and mRNA were described previously (12, 27). Plasmid DNA was isolated by using a QIAprep spin miniprep kit (Qiagen, Valencia, Calif.). All RNA samples for cDNA library construction, library screening, and Northern analyses were isolated from cultures grown for 23 h. Both wild-type F. sporotrichioides strain NRRL 3299 and the ↑Tri10 strain show maximum expression of trichothecene genes at about 23 h in liquid cultures, although expression is elevated and persists for an extended period of time in the ↑Tri10 strain (27).
PCR was performed either with Platinum Pfx DNA polymerase (GIBCO BRL, Rockville, Md.) or with Taq DNA polymerase (Promega, Madison, Wis.) according to the manufacturers' instructions. Probes for trichothecene genes were made from three different Fusarium species.
Primer names and sequences for each gene were as follows: Tri3, A-41 (5′-GATGAGCGCTTCATCCTCCTC-3′) and A-42 (5′-CCTAAAGTCGGAAGGCAAGC-3′); Tri4, A-37 (5′-GGAAAGATGGTTGATCAAGACTGG-3′) and A-38 (5′-GCCACTGAAGCTTACAAAGC-3′); Tri5, 60 (5′-CGGATCCATGGAGAACTTTCCCAC-3′) and 59 (5′-CCCATGGTGGATAAGCCCACTC-3′); Tri6, 292 (5′-GCTCTAGATGATTTACATGGCGTCCG-3′) and 293 (5′-GCCTCGAGTCAACACTTGTGTATCCG-3′); Tri7, A-54 (5′-ATGGATATCGCATCGAAAGTGGAAGG-3′) and A-55 (5′-ACTATTGTGGTACAAATACCAGGGCG-3′); Tri8, A-56 (5′-ATGGCTCTTAATCGTTTGGTGTTTTC-3′) and A-57 (5′-TTACCAGGCTGCCGACCAG-3′); Tri9, A-49 (5′-GAATTCAGGCCTTCTACTGACG-3′) and A-50 (5′-CGATTGTAAGCCGCGTCTTATC-3′); Tri10, 320 (5′-CCACCCAGCAATCATCAG-3′) and 356 (5′-GTACCTCGTTTCATGCC-3′); and Tri11, A-43 (5′-GGGCTTGCATATCTTGTGGTAG-3′) and A-44 (5′-AGTTCCTTTAGATTTCAGCCC-3′)—amplified from genomic DNA from F. sporotrichioides NRRL 3299; Tri12, 46MB (5′-GGGTTGCCATAAACCTCG-3′) and 47MB (5′-CAGGCCTTCTCCCTCAAC-3′)—amplified from genomic DNA from G. pulicaris R-6380; and Tri101, B-3 (5′-ATGGCTTTCAAGATACAGCTCG-3′) and B-4 (5′-CTAACCAACGTACTGCGCATAC-3′)—amplified from genomic DNA from G. zeae GZ3639.
Inserts of selected clones from the cDNA library in the pBluescript SK phagemid were excised by using the restriction enzyme pair ApaI and SmaI or SacI and KpnI (New England Biolabs, Beverly, Mass.). Restriction fragments and PCR products were run on 1.2% agarose and 1% agarose (Boehringer Mannheim, Indianapolis, Ind.) gels, respectively, and purified with a QIAquick gel extraction kit (Qiagen). Purified PCR products were cloned into pCR-Script (Stratagene, La Jolla, Calif.). For RNA electrophoresis, 1% agarose gels with 1.1% formaldehyde were used, and 5 μg of total RNA was run for each sample (26). Individual radioactively labeled DNA probes were prepared by using [α-32P]dCTP (Amersham, Piscataway, N.J.) and a nick translation system (GIBCO BRL). All Northern and Southern blots, as well as bacterial colony filters, were made on Hybond-N+ nylon membranes (Amersham) and hybridized, washed, and stripped under high-stringency conditions as specified in the Gene Images nonisotopic nucleic acid detection kit (United States Biochemical Corp., Cleveland, Ohio). Autoradiographic images were produced on BIOMAX MR or X-OMAT LS imaging film (Kodak, Rochester, N.Y.).
cDNA library and macroarray construction.
A cDNA library enriched for genes positively regulated by Tri10 was constructed in Uni-ZAP XR (Stratagene) with mRNA isolated from the ↑Tri10 strain. The library was excised, and 7,680 pBluescript SK phagemid colonies were picked and grown individually in 1 of 20 384-well plates containing freezer medium. High-density cDNA macroarrays were prepared by transferring samples from these plates to nylon membrane filters with a Biomek 2000 workstation (Beckman Coulter, Fullerton, Calif.) such that each clone was represented in duplicate and DNAs from colonies on four plates were combined on a single filter. Thus, each filter contained 1,536 clones in duplicate. Low-density macroarrays of a small sample of the cDNA library also were prepared. Samples from 3 of the 20 plates were transferred to individual filters such that each filter held 384 clones. Candidate genes were identified with both types of arrays, but the majority of the candidate genes came from the high-density arrays.
cDNA library screen.
The arrayed portion of the cDNA library was subjected to several parallel screenings. One set of cDNA macroarrays was differentially screened by first hybridizing the filters with radiolabeled cDNA derived from the non-T-2 toxin-producing ΔTri10 strain and then, after stripping them, hybridizing them with radiolabeled cDNA derived from the T-2 toxin-hyperproducing ↑Tri10 strain. Each cDNA sample was prepared from poly(A)+ mRNA isolated from total RNA samples by using an Oligotex mRNA mini kit (Qiagen). Radiolabeled first-strand cDNA was synthesized from poly(A)+ mRNA by using an adaptation of the Superscript preamplification system (GIBCO BRL). Specifically, the RNA mixture contained 1.5 μg of poly(A)+ mRNA and oligo(dT) as the primer, and [α-32P]dCTP was used in place of dCTP in the deoxynucleoside triphosphate mixture. After incubation with RNase H, the labeled probe was denatured in a boiling water bath for 5 min, chilled on ice for 2 min, and hybridized to the macroarrays as described above. Additional hybridizations with gene inserts from selected cDNA clones and, if necessary, with additional sets of cDNA macroarrays were conducted as needed to identify the entire group of cDNA clones that represented each of the newly identified Tri10-regulated genes. In a confirmatory screen, these gene inserts also were used as probes to hybridize Northern blots containing RNAs from strain NRRL 3299 and the ΔTri10 and ↑Tri10 strains. Northern analyses for the resulting group of genes were repeated at least once, and genes showing any inconsistencies were excluded. Ultimately, only clones representing new sequences clearly regulated by Tri10 were evaluated further.
Additional identical sets of cDNA macroarrays were probed with radiolabeled gene-specific PCR products prepared from available Fusarium trichothecene gene sequences (Tri3, Tri4, Tri5, Tri6, Tri7, Tri8, Tri9, Tri10, Tri11, Tri12, and Tri101).
A final set of Tri genes and Tri10-regulated sequences was used as a probe to hybridize Northern blots containing RNAs from strain NRRL 3299 and the ΔTri10, ↑Tri10, and ΔTri6 strains. All of these Northern analyses were repeated at least once.
DNA sequencing and sequence homology searches.
Inserts isolated from plasmid DNA preparations of cDNA or genomic DNA clones were sequenced by using either an ABI Prism Dye Terminator cycle sequencing core kit or a BigDye Terminator cycle sequencing core kit (Perkin-Elmer, Boston, Mass.). All reactions were run on a model 373 or 377 DNA sequencer (Applied Biosystems, Foster City, Calif.) at the Gene Technologies Laboratory at Texas A&M University. Nucleotide sequences and their putative corresponding amino acid sequences were compared to the National Center for Biotechnology Information nr database (GenBank+EMBL+DDBJ+PDB sequences) by using the blastn and blastx algorithms (3).
RESULTS
Identification of cDNA clones for the first 11 cloned Tri genes.
cDNA clones for Tri101 and 10 Tri cluster genes (Tri3, Tri4, Tri5, Tri6, Tri7, Tri8, Tri9, Tri10, Tri11, and Tri12) were identified by hybridization of the macroarrays with radiolabeled gene-specific PCR products. The arrayed portion of the cDNA library contains clones for all of these genes except Tri6 (Table 1). In general, there is a positive correlation between the percentage of clones for each of the Tri genes on the macroarrays and the corresponding transcript signals seen in Northern hybridizations (see Discussion for exceptions). Collectively, these 11 Tri genes represent 4 to 5% of the clones in the cDNA library.
TABLE 1.
Tri gene cDNA clones identified by hybridization to F. sporotrichioides gene-specific probes
| Gene | % of cDNA librarya | Reference |
|---|---|---|
| Tri4 | 1.61 | 14 |
| Tri5 | 1.07 | 13 |
| Tri101 | 0.79 | 21 |
| Tri8 | 0.53 | 20 |
| Tri3 | 0.23 | 22 |
| Tri11 | 0.13 | 1 |
| Tri12 | 0.08 | 2 |
| Tri7 | 0.07 | 8 |
| Tri9 | 0.07 | 15 |
| Tri10 | 0.01 | 27 |
| Tri6 | 0.00 | 25 |
| Total | 4.59 |
Based on an analysis of 7,680 clones.
Identification of Tri10-regulated genes by the differential screen.
The differential screen of the high-density macroarrays identified 228 clones for genes that appeared to be overexpressed in the toxin-hyperproducing strain relative to the non-toxin-producing strain. Of these 228 clones, 131 represented 5 of the above 11 Tri genes: Tri4 (49 copies), Tri5 (47 copies), Tri101 (28 copies), Tri8 (6 copies), and Tri3 (1 copy). These five genes are among the most highly represented trichothecene genes in the cDNA library in general (Tables 1 and 2). Clones for the other six Tri genes, most of which are expressed at lower levels, were not identified by the differential screen.
TABLE 2.
cDNA clones isolated as positively regulated by Tri10 and/or Tri6
| Genea | % of cDNA library | GenBank accession no. | DNA sequence length (kb) | Similar sequence(s) found by blastx or blastn search |
|---|---|---|---|---|
| Tri1 | 0.31 | AY040587 | 1.86 | Cytochrome P450 monooxygenase |
| Tri13 | 0.29 | AF330109 | 1.88 | Cytochrome P450 monooxygenase |
| Tri14 | 0.48 | AF326571 | 1.18 | No significant match |
| Tri15 | 0.07 | AF327521 | 0.96 | Putative zinc finger protein |
| ACAT | 0.25 | BQ789674, BQ789675 | 0.72 | Acetyl-CoA acetyltransferase |
| HMGS | 0.39 | BQ789676, BQ789677 | 0.82 | Hydroxymethylglutaryl-CoA synthase |
| MK | 0.01 | BQ789678, BQ789679 | 0.80 | Mevalonate kinase |
| Ibt1 | 0.04 | BQ789666, BQ789667 | 0.74 | Aspartyl protease |
| Ibt2 | 0.01 | BQ789668 | 0.28 | No significant match |
| Ibt3 | 0.14 | BQ789669, BQ789670 | 0.80 | Fumarate reductase |
| Ibt4 | 0.01 | BQ789671, BQ789672 | 0.72 | Cellulase, endoglucanase |
| Ibt5 | NAb | BQ789673 | 0.34 | Cytochromeb5 |
| Ibt6 | 0.14 | BQ789680, BQ789681 | 0.89 | Lysophospholipase, phospholipase B |
| Ibt7 | 0.05 | BQ789682 | 0.23 | No significant match |
| Ibt8 | 0.05 | BQ789683, BQ789684 | 0.76 | No significant match |
| Ibs1 | 0.33 | BQ789685, BQ789686 | 0.87 | NADH-dependent flavin oxidoreductase |
Ibt genes are regulated by Tri10; Ibs gene is regulated by Tri6 but not by Tri10.
NA, not available.
The remaining 97 clones, plus a few additional clones from the differentially screened low-density arrays, were characterized further to identify all of the clones for each sequence to permit the selection of one representative clone for each validated Tri10-regulated sequence. This process resulted in the identification of 15 additional sequences under the positive control of Tri10 (Table 2 and Fig. 1). These 15 sequences include three putative orthologs of isoprenoid biosynthetic genes, designated acetyl-coenzyme A (CoA) acetyltransferase (ACAT), hydroxymethylglutaryl-CoA synthase (HMGS), and mevalonate kinase (MK) genes. Two sequences represent Tri13 and Tri14, which reside in the Tri cluster (9, 18); the former encodes a cytochrome P450 monooxygenase that catalyzes C-4 hydroxylation (9, 18), and the function of the latter is unknown (9). Another sequence represents Tri1, a previously identified but uncloned Tri gene (23). TRI1 and TRI13 have been assigned to the families CYP68C1 and CYP526A1, respectively, by the Committee for Standardized Cytochrome P450 Nomenclature (David Nelson, personal communication). We have designated another Tri10-regulated sequence Tri15. The remaining eight new Tri10-regulated sequences (Table 2) have been designated Ibt (influenced by Tri10).
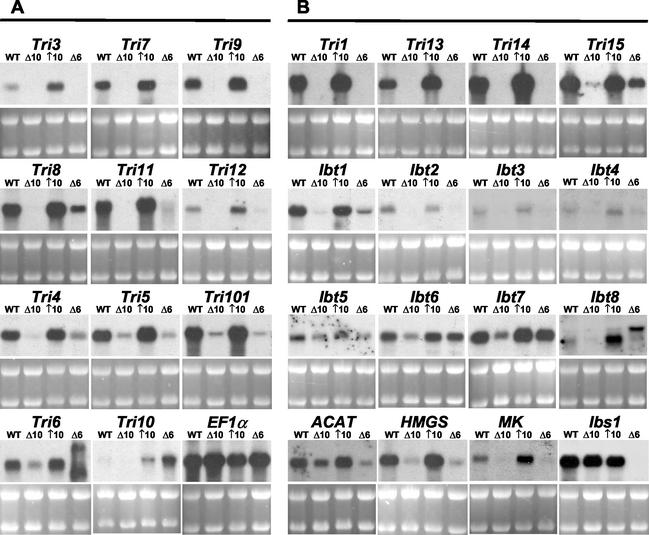
FIG. 1.
Northern analyses of Tri10- and/or Tri6-regulated genes. (A) Genes listed in Table 1. (B) Genes listed in Table 2. Wild-type (WT), ΔTri10 (Δ10), ↑Tri10 (↑10), and ΔTri6 (Δ6) strains of F. sporotrichioides were grown in liquid cultures for 23 h. Five micrograms of total RNA was loaded in each lane. Panels below the Northern analyses depict the corresponding ethidium bromide staining of rRNA in the gels. The transcriptional profile of translation elongation factor 1α (EF1α) is shown as a constitutive control.
Tri10 regulation of known Tri genes.
Northern blot analysis confirmed that, like those for the other identified Tri genes, the transcript levels for Tri3, Tri7, Tri8, Tri9, Tri11, and Tri12 were increased in the ↑Tri10 strain and decreased in the ΔTri10 strain (Fig. 1). However, unlike the transcripts for the 4 previously examined Tri genes (Tri4, Tri5, Tri6, and Tri101), there were no detectable transcripts in the ΔTri10 strain for 9 (Tri1, Tri3, Tri7, Tri8, Tri9, Tri11, Tri12, Tri13, and Tri14) of the 10 other Tri genes. Only Tri15 displayed a low transcript level in the ΔTri10 strain (Fig. 1).
Tri6 regulation of Tri10-regulated sequences.
We compared the expression of the 15 Tri10-regulated sequences and the remaining Tri genes to the expression of Tri4, Tri5, and Tri101 in the ΔTri6 strain. The deletion of Tri6 markedly reduced the transcript levels for 21 of the 24 Tri10-regulated sequences (Fig. 1). There were three broad categories of expression within this group. Six genes (Tri1, Tri3, Tri7, Tri9, Tri13, and Tri14) were under the tight control of both Tri10 and Tri6; that is, the lack of either Tri10 or Tri6 effectively blocked detectable transcript accumulation for these genes. Three genes (Tri8, Tri11, and Tri12) were under the tight control of Tri10 but not Tri6. Ten genes (Tri4, Tri5, Tri101, Tri15, Ibt1, Ibt4, Ibt5, ACAT, HMGS, and MK) were not under the tight control of either Tri10 or Tri6. Although Ibt2 and Ibt3 belonged in one of the two latter categories, we could not assign them to either category with confidence due to their low levels of expression.
Four genes had a pronounced differential pattern of gene expression with regard to Tri10 and Tri6 (Fig. 1B). Transcript levels for three sequences (Ibt6, Ibt7, and Ibt8) whose normal levels of expression at 23 h were equal to or lower than those in the ↑Tri10 strain were reduced in the ΔTri10 strain but not in the ΔTri6 strain. The level of expression of Ibt8 actually increased when Tri6 was knocked out. The size of the Ibt8 transcript also was increased in the ΔTri6 strain. The fourth transcript showed little or no dependence on Tri10 but was strongly dependent on Tri6 and was designated Ibs1 (influenced by Tri6).
DISCUSSION
The results from this study support the hypotheses that Tri10 and Tri6 regulate the expression of (i) all of the Tri genes, both within and outside the core Tri gene cluster, and (ii) additional genes for isoprenoid biosynthesis. Tag et al. previously showed that two Tri cluster genes (Tri4 and Tri5), one noncluster gene (Tri101), and one isoprenoid biosynthetic gene (FPPS) depend on Tri10 and Tri6 for full expression and are overexpressed in a ↑Tri10 strain (27). Here we establish that the other known Tri genes, including Tri1 and the eight remaining Tri cluster genes (Tri3, Tri7, Tri8, Tri9, Tri11, Tri12, Tri13, and Tri14), and three more putative isoprenoid biosynthetic genes (ACAT, HMGS, and MK) also depend on Tri10 and Tri6 for full expression and are overexpressed in the ↑Tri10 strain.
Five additional genes are similarly regulated. One, Ibt2, has no significant similarities to known sequences. Four encode several different putative proteins, including aspartyl protease (Ibt1), reductase (Ibt3), cellulase (Ibt4), and cytochrome b5 (Ibt5). The cellular functions of these genes are currently unknown, although they could have a role in trichothecene production. However, it is unlikely that all of these genes are directly involved in either trichothecene or isoprenoid biosynthesis, suggesting that the regulatory control exerted by Tri10 and Tri6 extends beyond genes for isoprenoid and trichothecene biosynthesis.
Although Tri10 and Tri6 coregulate a fairly large set of transcripts (at least 22), they also regulate some genes (at least four) independently of each other. This regulatory pattern was not unexpected, since the ΔTri6 and ΔTri10 strains have some distinct phenotypic differences (27). One gene, Ibs1, is markedly dependent on Tri6 and has no apparent dependence on Tri10. Three transcripts (Ibt6, Ibt7, and Ibt8) are downregulated in the ΔTri10 strain but not in the ΔTri6 strain and appear to be exceptions to the general hypothesis that Tri10-regulated genes are regulated by Tri6. However, Ibt8 is still influenced by Tri6. Ibt8, like Tri10 (27), is overexpressed with the loss of Tri6. Moreover, the loss of Tri6 also increases the size of the Ibt8 transcript; this result could be due to the use of a different transcriptional start site or different posttranscriptional processing activities. Thus, Tri10 and Tri6 control slightly different sets of genes and, in some instances, may regulate the same gene in different ways.
Meanwhile, Tri6 regulation of most of the Tri10-regulated genes is consistent with the previously reported regulation of some Tri genes (Tri5, Tri4, Tri3, and Tri101) by Tri6 (22, 25, 27) and with the observation that all of the previously known Tri genes (except for Tri10) appear to have TRI6-binding sites in their promoters (15).
One of our main objectives was to identify additional Tri genes both inside and outside the Tri gene cluster by obtaining Tri10-regulated genes. Three of the Tri10-regulated sequences that we identified by using the differential screen represent recognized Tri genes (Tri1, Tri13, and Tri14). Tri1 had been identified on the basis of a UV-induced mutation, but no DNA sequence had previously been available for this gene (6, 23). Both Brown et al. (9) and Lee et al. (18) cloned Tri13 and Tri14. Brown et al. (9) cloned these genes from F. sporotrichioides NRRL 3299 by sequencing a cosmid region downstream of Tri12 in the Tri gene cluster. Lee et al. (18) cloned Tri13 and Tri14 orthologs from G. zeae 88-1 (GenBank accession number AF336365) by PCR with our Tri13 and Tri14 gene sequences and genomic positions from F. sporotrichioides. We named a new Tri10-regulated sequence Tri15 based on preliminary evidence indicating that disruption of this gene alters Tri gene expression and toxin production (A. W. Peplow and M. N. Beremand, unpublished data). Tri15 does not have high sequence similarity with known genes, but it does have two putative Cys2His2 zinc finger DNA-binding domains, consistent with a regulatory function. By using Southern analysis, we detected Tri15, Tri14, and Tri13 orthologs in G. pulicaris R-6380 and G. zeae GZ3639 and a Tri1 ortholog in R-6380 but not GZ3639.
The transcription profiles for the Tri genes and the isoprenoid genes are correlated with the order of the 15 steps that define the T-2 toxin biosynthetic pathway in F. sporotrichioides NRRL 3299 (10). Genes that function before step 9 (Tri6, Tri5, Tri4, Tri101, and the isoprenoid pathway genes ACAT, HMGS, MK, and FPPS) are expressed at low levels in both ΔTri10 and ΔTri6 strains and thus are not under the tight control of either Tri10 or Tri6. The low levels of expression of these early genes may be related to their use in shared pathways. By Southern analysis, it appears that the isoprenoid pathway genes ACAT, HMGS, MK, and FPPS are present in single copies in the genome, and it seems unlikely that trichothecene biosynthesis recruits specific gene paralogs of isoprenoid synthesis that are distinct from those normally used by the cell. Early trichothecene biosynthetic intermediates also are precursors for other metabolites, including apotrichothecenes (14). Genes that most likely function in conjunction with step 9 (Tri11, Tri12, and Tri8) appear to be under the tight control of Tri10 but not Tri6, while genes that function after step 9 (Tri3, Tri13, Tri7, and Tri1) are tightly regulated by both Tri10 and Tri6. These results are consistent with prior observations that neither the ΔTri10 strain nor the ΔTri6 strain makes any T-2 toxin, even though both strains express low levels of Tri5 (25, 27). They also provide experimental evidence supporting the hypothesis that the lack of T-2 toxin production in these strains is due to a lack of transcripts for later pathway-specific genes. Moreover, the loose versus tight division in Tri gene expression mediated by Tri10 and/or Tri6 may define a previously unrecognized important regulatory control point for trichothecene biosynthesis.
The coregulation of FPPS by Tri10 and Tri6 demonstrates a regulatory link between the isoprenoid and trichothecene biosynthetic pathways (27). The coordinated expression of four isoprenoid biosynthetic pathway genes by both Tri10 and Tri6 identifies a regulatory network that links these primary and secondary metabolic pathways. While the regulation of the Tri genes by Tri10 is mediated by Tri6, neither the extent nor the means by which Tri10 and Tri6 control the expression of the isoprenoid genes is known. It will be interesting to determine whether TRI6-binding sites occur in the promoters of the isoprenoid genes and whether Tri10 and Tri6 regulate the expression of additional isoprenoid genes.
It is unlikely that we have identified all of the genes positively controlled by Tri10. While the differential screening of the cDNA macroarrays was fruitful, we did not detect some of the known Tri genes that are expressed at lower levels (e.g., Tri11, Tri12, Tri7, and Tri10) or that have short sequences (e.g., Tri6 and Tri9). Therefore, if other Tri10-regulated genes with a low transcript abundance or with a small transcript size exist, they also might have been missed. Thus, the number of genes regulated by Tri10 identified in this study is probably an underestimate, and it is likely that other Tri10-regulated genes, including additional isoprenoid and trichothecene genes, remain to be identified.
Acknowledgments
We thank Tom Hohn for providing the nucleotide sequences of Tri7, Tri8, and Tri9 prior to their submission to GenBank.
This work was supported by USDA/CSREES NRICGP grant 9503702. A.W.P. and A.G.T. were supported in part by a National Science Foundation grant to the Program for the Biology of Filamentous Fungi at Texas A&M University.
REFERENCES
- 1.Alexander, N. J., T. M. Hohn, and S. P. McCormick. 1998. The TRI11 gene of Fusarium sporotrichioides encodes a cytochrome P-450 monooxygenase required for C-15 hydroxylation in trichothecene biosynthesis. Appl. Environ. Microbiol. 64:221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, N. J., S. P. McCormick, and T. H. Hohn. 1999. TRI12, a trichothecene efflux pump from Fusarium sporotrichioides: gene isolation and expression in yeast. Mol. Gen. Genet. 261:977-984. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai, G. H., A. E. Desjardins, and R. D. Plattner. 2002. Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia 153:91-98. [DOI] [PubMed] [Google Scholar]
- 5.Bean, G. A., T. Fernando, B. B. Jarvis, and B. Bruton. 1984. The isolation and identification of trichothecene metabolites from a plant pathogenic strain of Myrothecium roridum. J. Nat. Prod. 47:727-729. [DOI] [PubMed] [Google Scholar]
- 6.Beremand, M. N. 1987. Isolation and characterization of mutants blocked in T-2 toxin biosynthesis. Appl. Environ. Microbiol. 53:1855-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowden, R. L., and J. F. Leslie. 1999. Sexual recombination in Gibberella zeae. Phytopathology 89:182-188. [DOI] [PubMed] [Google Scholar]
- 8.Brown, D. W., S. P. McCormick, N. J. Alexander, R. H. Proctor, and A. E. Desjardins. 2001. A genetic and biochemical approach to study trichothecene diversity in Fusarium sporotrichioides and Fusarium graminearum. Fungal Genet. Biol. 32:121-133. [DOI] [PubMed] [Google Scholar]
- 9.Brown, D. W., S. P. McCormick, N. J. Alexander, R. H. Proctor, and A. E. Desjardins. 2002. Inactivation of a cytochrome P-450 is a determinant of trichothecene diversity in Fusarium species. Fungal Genet. Biol. 36:224-233. [DOI] [PubMed] [Google Scholar]
- 10.Desjardins, A. E., T. M. Hohn, and S. P. McCormick. 1993. Trichothecene biosynthesis in Fusarium species: chemistry, genetics, and significance. Microbiol. Rev. 57:595-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris, L. J., A. E. Desjardins, R. D. Plattner, P. Nicholson, G. Butler, J. C. Young, G. Weston, R. H. Proctor, and T. M. Hohn. 1999. Possible role of trichothecene mycotoxins in virulence of Fusarium graminearum on maize. Plant Dis. 83:954-960. [DOI] [PubMed] [Google Scholar]
- 12.Hohn, T. M., and A. E. Desjardins. 1992. Isolation and gene disruption of the Tox5 gene encoding trichodiene synthase in Gibberella pulicaris. Mol. Plant-Microbe Interact. 5:249-256. [DOI] [PubMed] [Google Scholar]
- 13.Hohn, T. M., and P. D. Beremand. 1989. Isolation and nucleotide sequence of a sesquiterpene cyclase gene from the trichothecene-producing fungus Fusarium sporotrichioides. Gene 79:131-138. [DOI] [PubMed] [Google Scholar]
- 14.Hohn, T. M., A. E. Desjardins, and S. P. McCormick. 1995. The Tri4 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase involved in trichothecene biosynthesis. Mol. Gen. Genet. 248:95-102. [DOI] [PubMed] [Google Scholar]
- 15.Hohn, T. M., R. Krishna, and R. H. Proctor. 1999. Characterization of a transcriptional activator controlling trichothecene toxin biosynthesis. Fungal Genet. Biol. 26:224-235. [DOI] [PubMed] [Google Scholar]
- 16.Jarvis, B. B., W. G. Sorenson, E. L. Hintikka, M. Nikulin, Y. Zhou, J. Jiang, S. Wang, S. Hinkley, R. A. Etzel, and D. Dearborn. 1998. Study of toxin production by isolates of Stachybotrys chartarum and Memnoniella echinata isolated during a study of pulmonary hemosiderosis in infants. Appl. Environ. Microbiol. 64:3620-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura, M., I. Kaneko, M. Komiyama, A. Takatsuki, H. Koshino, K. Yoneyama, and I. Yamaguchi. 1998. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. Cloning and characterization of Tri101. J. Biol. Chem. 273:1654-1661. [DOI] [PubMed] [Google Scholar]
- 18.Lee, T., Y. K. Han, K. H. Kim, S. H. Yun, and Y. W. Lee. 2002. Tri13 and Tri7 determine deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae. Appl. Environ. Microbiol. 68:2148-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marasas, W. F. O., P. E. Nelson, and T. A. Toussoun. 1984. Toxigenic Fusarium species: identity and mycotoxicology. The Pennsylvania State University Press, University Park.
- 20.McCormick, S. P., and N. J. Alexander. 2002. Fusarium Tri8 encodes a trichothecene C-3 esterase. Appl. Environ. Microbiol. 68:2959-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCormick, S. P., N. J. Alexander, S. E. Trapp, and T. M. Hohn. 1999. Disruption of TRI101, the gene encoding trichothecene 3-O-acetyltransferase, from Fusarium sporotrichioides. Appl. Environ. Microbiol. 65:5252-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick, S. P., T. M. Hohn, and A. E. Desjardins. 1996. Isolation and characterization of Tri3, a gene encoding 15-O-acetyltransferase from Fusarium sporotrichioides. Appl. Environ. Microbiol. 62:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meek, I. B., A. W. Peplow, C. Ake, Jr., T. D. Phillips, and M. N. Beremand. 2003. Tri1 encodes the cytochrome P450 monooxygenase for C-8 hydroxylation during trichothecene biosynthesis in Fusarium sporotrichioides and resides upstream of another new Tri gene. Appl. Environ. Microbiol. 69:1607-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proctor, R. H., T. M. Hohn, and S. P. McCormick. 1995. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant-Microbe Interact. 8:593-601. [DOI] [PubMed] [Google Scholar]
- 25.Proctor, R. H., T. M. Hohn, S. P. McCormick, and A. E. Desjardins. 1995. Tri6 encodes an unusual zinc finger protein involved in regulation of trichothecene biosynthesis in Fusarium sporotrichioides. Appl. Environ. Microbiol. 61:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Tag, A. G., G. F. Garifullina, A. W. Peplow, C. Ake, Jr., T. D. Phillips, T. M. Hohn, and M. N. Beremand. 2001. A novel regulatory gene, Tri10, controls trichothecene toxin production and gene expression. Appl. Environ. Microbiol. 67:5294-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueno, Y., M. Sawano, and K. Ishii. 1975. Production of trichothecene mycotoxins by Fusarium species in shake culture. Appl. Microbiol. 30:4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei, C. M., and C. S. McLaughlin. 1974. Structure-function relationship in the 12,13-epoxytrichothecenes. Novel inhibitors of protein synthesis. Biochem. Biophys. Res. Commun. 57:838-844. [DOI] [PubMed] [Google Scholar]
- 30.Yang, G. H., B. B. Jarvis, Y. J. Chung, and J. J. Pestka. 2000. Apoptosis induction by the satratoxins and other trichothecene mycotoxins: relationship to ERK, p38 MAPK, and SAPK/JNK activation. Toxicol. Appl. Pharmacol. 164:149-160. [DOI] [PubMed] [Google Scholar]