Abstract
Methyl tert-butyl ether (MTBE) is a widespread groundwater contaminant that does not respond well to conventional treatment technologies. Growing evidence indicates that microbial communities indigenous to groundwater can degrade MTBE under aerobic and anaerobic conditions. Although pure cultures of microorganisms able to degrade or cometabolize MTBE have been reported, to date the specific organisms responsible for MTBE degradation in various field studies have not be identified. We report that DNA sequences almost identical (99% homology) to those of strain PM1, originally isolated from a biofilter in southern California, are naturally occurring in an MTBE-polluted aquifer in Vandenberg Air Force Base (VAFB), Lompoc, California. Cell densities of native PM1 (measured by TaqMan quantitative PCR) in VAFB groundwater samples ranged from below the detection limit (in anaerobic sites) to 103 to 104 cells/ml (in oxygen-amended sites). In groundwater from anaerobic or aerobic sites incubated in microcosms spiked with 10 μg of MTBE/liter, densities of native PM1 increased to approximately 105 cells/ml. Native PM1 densities also increased during incubation of VAFB sediments during MTBE degradation. In controlled field plots amended with oxygen, artificially increasing the MTBE concentration was followed by an increase in the in situ native PM1 cell density. This is the first reported relationship between in situ MTBE biodegradation and densities of MTBE-degrading bacteria by quantitative molecular methods.
The fuel oxygenate, methyl tert-butyl ether (MTBE), is a widespread groundwater contaminant in the United States and parts of Europe. MTBE is a water soluble and mobile compound that has the potential to generate long pollution plumes in aquifers impacted by gasoline releases from leaking tanks. In California, at least 10,000 leaking underground storage tank sites are contaminated with MTBE (14).
MTBE biodegradation has been reported in a number of environments, including sewage sludge (29), soils (37), river sediments (3, 4), and biofilters (9, 11). Numerous MTBE-degrading isolates have been discovered, including direct metabolizers and cometabolizers (5, 7, 12, 13, 15, 16, 18, 20, 26, 30, 31, 34). Among these isolates, strain PM 1 is a member of the beta-Proteobacteria and closely related to Aquabacterium, Leptothrix, and Rubrivivax (5). Strain PM1 rapidly degrades MTBE, mineralizes [14C]MTBE to 14CO2, and uses the compound as a sole carbon and energy source (8, 13).
Field evidence indicates that, at many contaminated sites, natural degradation rates are not sufficiently rapid to prevent MTBE plumes from continuing to elongate, except where sufficient concentrations of oxygen are present. In situ technologies for active bioremediation of MTBE-contaminated aquifers have been evaluated at several field sites (3, 4, 6, 23, 31, 36). Oxygen has been added by sparging (31), by oxygen-releasing compounds (23), and via diffusive release across permeable membranes (24, 25, 36). At Vandenberg Air Force Base (VAFB) in Lompoc, California, Mackay et al. (25) and Wilson et al. (36) found that diffusive oxygen release into a portion of the existing MTBE plume can support the in situ aerobic degradation of MTBE by microorganisms indigenous to the VAFB aquifer. In this study, an experimental facility was constructed in situ which allowed for highly controlled tests in which intimate mixing of oxygen with groundwater was ensured, the direction and velocity of the groundwater was well understood, and detailed monitoring in space and time was possible (36).
An enormous challenge in environmental microbiology is to identify and quantify specific members of microbial communities responsible for the degradation of organic pollutants. This knowledge would result in the discovery of novel environmental organisms potentially of use for bioaugmentation as well as guide the development of technologies effective for biostimulation of native populations. Enumerating populations of biodegrading organisms in groundwater, important information for predicting biodegradation rates (33), is particularly challenging. Most previous efforts, such as that of Kao et al. (22), involve most-probable number analyses to measure microbial populations involved in bioremediation at petroleum-hydrocarbon spill sites. Enormous breakthroughs in the development of non-culture-based molecular techniques for characterizing microbial communities are, for the most part, not quantitative and thus provide limited information about population densities. Exceptions include quantitative PCR methods such as TaqMan real-time quantitative PCR (17).
Here we report the discovery of a bacterial strain in MTBE-contaminated groundwater at VAFB that is virtually identical in its 16S ribosomal DNA (rDNA) sequence to strain PM1, an organism demonstrated to rapidly mineralize MTBE as a sole carbon and energy source in pure culture. We demonstrate for the first time, by using quantitative molecular tools, a relationship between the presence of potential MTBE-biodegrading organisms and MTBE biodegradation.
MATERIALS AND METHODS
Field site.
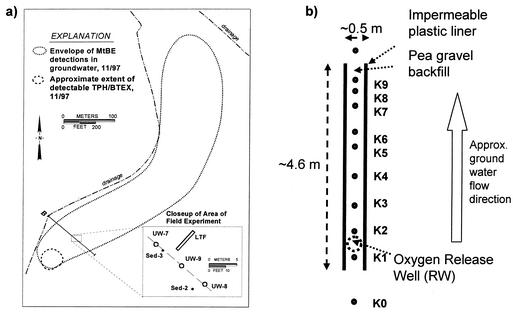
The MTBE plume at VAFB site 60 (Fig. 1a), the focus of our work, is estimated to be at least 520 m long and 80 m wide, weakly anaerobic, and located within a relatively shallow and thin aquifer (36). The contamination originated from a fuel leak at a service station, and despite source removal efforts in 1995, a plume of MTBE and other petroleum hydrocarbons continues to emanate from the source area. The benzene, toluene, ethylbenzene, and xylene plume, however, attenuates to below detection limits within 15 to 30 m of the source. In a controlled field experiment, oxygen was added by diffusive release to contaminated groundwater flowing through a pea-gravel-filled trench (36). Figure 1b depicts the longitudinal test facility (LTF). After 2 months of oxygen release, the concentration of dissolved oxygen (DO) in the oxygen release well (RW) ranged from 15 to 23 mg/liter and an apparent steady-state profile developed along the LTF down gradient of the RW. The groundwater velocity in the LTF was estimated to be 0.88 m/day (36).
FIG. 1.
(a) Map of site 60, VAFB. The dashed line encloses all MTBE detections greater than 2 μg/liter. The close-up view reveals the orientation and scale of the LTF and sample location. (b) Plan view of the LTF. The approximate locations and names of groundwater wells are shown.
Sampling.
Groundwater samples were collected from sampling wells within and adjacent to the LTF by using Cole Parmer Masterflex peristaltic pumps and sterile tubing for each sample. Samples were placed on ice and shipped immediately to the University of California—Davis and filtered within 24 h. Sediment samples were collected at several locations across the plume along transect B (for samples used in this study, locations are shown in Fig. 1). In all cases, a bucket auger was used to collect samples at 2.5 m below the ground surface. Samples were collected sequentially with a sterilized bucket auger, immediately transferred to sterile mason jars, and topped off with groundwater from a well adjacent to the cored location. The sediment and groundwater were shipped on ice for further processing at the University of California—Davis. The sediment was removed from the center of cores after removing the outer layers with a sterile spatula. For DNA extraction, groundwater samples were filtered in the lab through sterile 0.22-μm-pore-size filter (diameter, 47 mm; type GTTP 2500; Millipore, Schawalbach, Germany) units in a laminar flow hood. DNA from the groundwater and sediment samples was extracted as described by Hristova et al. (19).
Laboratory microcosms.
We performed microcosm experiments with groundwater from locations in VAFB site 60 (see Table 2). Microcosms included 40 ml of groundwater in 125-ml amber glass, screw-cap bottles with Teflon-lined septa. We also added 40 g of washed, sterile sand (combusted overnight at 400°C to remove all carbon) to the microcosms after preliminary experiments indicated long lag periods preceding MTBE biodegradation in groundwater alone and to better simulate the groundwater environment. Sterile microcosm controls were prepared by autoclaving sediment and groundwater and adding sodium azide (1 g/liter). MTBE was added as an aqueous stock solution to a final concentration of 5.0 to 10.0 mg/liter, and bottles were incubated at room temperature in the dark. A Shimadzu GC-14A equipped with a photo ionization detector and a 15-m by 0.53-mm DB-1 column (J&W Scientific, Folsom, Calif.) was used for isothermal (150°C) headspace analysis. Helium (99.995% purity) was the carrier gas. Periodically, 50 μl was removed from the gas phase of the microcosms with a glass syringe (Gastight no. 1705RN; Hamilton Company, Reno, Nev.) and analyzed for MTBE degradation. The detection limit for MTBE was approximately 0.01 mg/liter (aqueous concentration).
TABLE 2.
PM1 cell densities measured by TaqMan PCR in groundwater, sediment, and laboratory microcosms
| Locationa in VAFB site 60 | PM1 cell densityb
|
|
|---|---|---|
| Before incubation | After incubation | |
| K3-3 groundwater | 1.3 × 104 ± 0.3 × 104 | 3.3 × 105 ± 0.8 × 105 |
| UW9 groundwater | <DLc | 3.3 × 105 ± 1.0 × 105 |
| Sed.-2 sediment | 2.5 × 103 ± 0.5 × 103 | 2.3 × 105 ± 0.4 × 105 |
| Sed.-3 sediment | 6.4 × 103 ± 0.4 × 103 | 1.2 × 105 ± 0.7 × 105 |
Sample locations are indicated on the map shown in Fig. 1.
PM1 cell densities are given in cells per milliliter of groundwater or cells per gram of sediment. ± standard deviation.
<DL, below detection limit of 30 cells per PCR tube.
ITS analysis.
Internal transcribed spacer (ITS) fingerprints of groundwater microbial communities were generated by using primers 1496F and 23SR, as described by Borneman and Triplett (2). PM1-specific primers 1406F and 1850R (Table 1) were designed based on alignments of sequences of the ITS and the flanking 16S and 23S rRNA genes of the ribosomal operon (Table 1). The reverse primer 1850R is located at a highly variable region of the molecule and is specific for the PM1 sequence. The ITS PM1 primers amplify a 444-bp DNA fragment.
TABLE 1.
Oligonucleotides used as primers and probes
| Namea | Primer | Sequence (5′-3′)b | Reference |
|---|---|---|---|
| 1406F | PM1 ITS forward | TGYACACACCGCCCGT | This study |
| 1850R | PM1 ITS reverse | CGTAAGCCACTGACGCTT | This study |
| 613F | PM1 DGGE forward | GTGACTGCAAGGCTGGAGCG | This study |
| 988R | PM1 DGGE reverse + GC-Clampc | TCTGGTAACTTCTAGACA | This study |
| 963F | TaqMan PCR PM1 forward | CCTTGACATGTCTAGAAGTTACCAGAGA | 19 |
| 1076R | TaqMan PCR PM1 reverse | GCGGGACTTAACCCAACATCT | 19 |
| 1030T | TaqMan PCR probe | ACACGAGCTGACGACGGCCATG | 19 |
| 357F | Bacterial DGGE forward + GC-Clamp | CCTACGGGAGGCAGCAG | 27 |
| 534R | Bacterial DGGE reverse | ATTACCGCGGCTGCTGG | 27 |
| 1406F | Bacterial ITS forward | TGYACACACCGCCCGT | 2 |
| 23SR | Bacterial ITS reverse | GGGTTBCCCCATTCRG | 2 |
Positions are according to Escherichia coli numbering.
The specificity of all listed primers and probes (Table 1) were checked by using the CHECK_PROBE software provided through the Ribosomal Database Project (RDP) and the Basic Local Alignment Search Tool (BLAST) network service in GenBank. Y, C/T; B, G/C/T; R, A/G.
GC-Clamp, CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC G.
Extracted DNA from pure cultures and groundwater samples was amplified by using a Gene Amp 2400 thermal cycler (PE Biosystems). PCR mixtures contained 1 ng of DNA template, 1.5 mM MgCl2, 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 0.1% Triton X-100, 0.25 mM concentrations of each of the deoxynucleoside triphosphates, 2.5 U of Taq polymerase (Promega, Madison, Wis.) and 10 pmol (each) of primers 1406F and 1850R (Table 1) in a final volume of 25 μl. Following initial denaturation at 94°C for 4 min, the PCR program consisted of denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 1 min 30 s (30 cycles) followed by a final extension of 7 min at 72°C. PCR products were applied (5- to 10-μl aliquots) to 5% polyacrylamide gels (0.75 mm thick, 150 by 150 mm) and run on an electrophoresis gel at 150 V for 4 h in 1× TBE (Tris-borate-EDTA, pH 8.0) buffer. Gels were stained with SYBR Green and photographed through a yellow filter with a Kodak EDAS 290 charge-coupled device camera and one-dimensional image analysis software (version LE 3.5.3; Kodak).
DGGE analyses with 16S rDNA universal bacterial and PM1-specific primers.
A denaturing gradient gel electrophoresis (DGGE) fingerprinting approach of 230-bp 16S rDNA PCR products (27) (Table 1) was used to analyze the microbial community at the VAFB site 60 groundwater and sediments.
Based on alignments of 30 bacterial 16S rDNA sequences (from beta-Proteobacteria representatives), PM1-specific DGGE primers (Table 1) were designed by using the CLUSTAL_X (version 1.8) program. The PM1-specific DGGE primers were used to check if PM1 is present in the VAFB site 60 groundwater and sediments. The optimized PCR conditions are initial denaturation at 94°C for 4 min; 30 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 1 min, and extension at 72°C for 1 min 30 s; and final extension at 72°C for 10 min. The PCR mixture was similar to that used for the PM1 ITS PCRs, with the exception that 1.5 U of Taq polymerase (Promega) and 25 pmol of primers were used. PCR products were analyzed by using 6% acrylamide gels and a denaturing gradient of 30 to 60% or 20 to 60% (100% denaturant corresponds to 7 M urea plus 40% deionized formamide). DGGE was performed in 0.5× Tris-acetate-EDTA buffer at 60°C at 150 V for 2 h and then at 200 V for 1 h. After electrophoresis, the gels were stained for 30 min with SYBR Green solution (1:10,000 dilution of the solution from Molecular Probes).
Cloning and sequencing.
DNA bands containing PCR products in DGGE or ITS gels were excised and eluted with 200 μl of Tris-EDTA buffer overnight. One microliter from the elute was used as a PCR template in a second PCR amplification with the corresponding ITS or DGGE PCR primers and conditions. The reaction mixtures were purified by using the Qiaquick PCR purification kit (Qiagen), and 4 μl of the mix was tested for purity by polyacrylamide gel electrophoresis or DGGE. The products were then cloned into TOPO TA cloning vector according to the procedure recommended by the manufacturer (TOPO TA cloning kit; Invitrogen, Carlsbad, Calif.). Clones were randomly selected, grown, and after plasmid extraction with the Qiagen plasmid mini prep kit, used as templates in PCRs to produce products. Selected clones were sequenced with an ABI Prism automatic sequencer (Applied Biosystems, Foster City, Calif.) at Davis Sequencing Inc., Davis, Calif. Sequence identities were determined by BLAST and Ribosomal Database Project analyses.
TaqMan real-time quantitative PCR.
PCR was performed in 25-μl volumes with MicroAmp optical 96-well reaction plates and MicroAmp optical caps (Applied Biosystems). A 113-bp product was amplified by using primers 963F and 1076R and probe 1030T (Table 1). DNA extraction of environmental samples was performed in duplicate, and three PCRs were run for each extraction. PCR conditions and data analyses were performed as described by Hristova et al. (19). The cycle at which a sample crosses the threshold (CT, a PCR cycle where the fluorescence emission exceeds that of nontemplate controls) is called the threshold cycle CT. A high CT value corresponds to a low amount of template DNA, and a low CT corresponds to a large amount of template. We prepared a standard curve relating the threshold cycle CT to 10-fold serial dilutions of DNA or cells of PM1. The corresponding cells per PCR were calculated based on plate counts of PM1 pure culture. The detection limit for the PM1 TaqMan assay is 30 cells per PCR, or 180 cells per ml of groundwater or g of sediment (19).
Nucleotide sequence accession number.
The sequences were submitted to the GenBank nucleotide sequence database and assigned accession numbers AY196467 to AY196475.
RESULTS
In situ remediation of MTBE at VAFB.
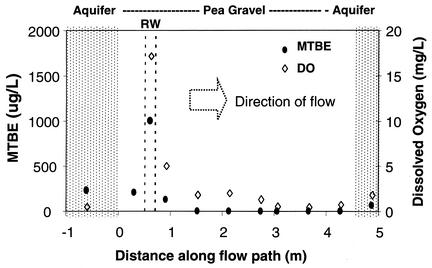
Previous studies indicated that oxygen addition can support MTBE biodegradation in VAFB sediments and groundwater, demonstrated both in laboratory microcosm studies (32, 36) and by field measurements within the biotreatment zone within the LTF (36). The LTF was located, as illustrated in Fig. 1a, near the apparent centerline of the MTBE plume and about 60 m down gradient of the source area. Field experiments within the LTF (Fig. 1b) were initiated in August 1999 when the diffusive release of oxygen began. During the next year, research focused on treatment of plume water flowing under the natural gradient through the LTF; during this period, the influent MTBE concentrations generally ranged from 200 to 500 μg/liter. Starting in the fall of 2000, MTBE concentrations influent to the aerobic treatment zone down gradient of the oxygen RW were artificially increased in a stepwise fashion by adding MTBE into the RW to achieve approximately 800, 1,300, and 2,100 μg of MTBE/liter (36). Figure 2 shows MTBE removal along the groundwater flow path through the LTF 2 days after increasing the MTBE concentrations influent to the treatment zone to 1,300 μg/liter. As illustrated in Fig. 2, MTBE continued to be degraded within the LTF by MTBE-degrading bacteria whose activity had been stimulated and sustained by the release of oxygen. Thus, as the groundwater flowed through the biotreatment zone, both the DO and MTBE decreased, the latter to below 20 μg/liter.
FIG. 2.
Concentrations of MTBE (dots) and DO (diamonds) versus distance along the flow path through the LTF. Samples were collected 3 days after the intentional increase of MTBE addition to 1,300 μg/liter.
Molecular analyses of groundwater microbial communities.
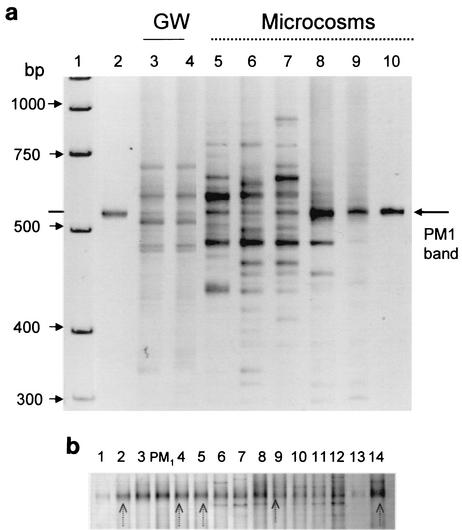
We used universal bacterial PCR primers targeting 16S rRNA or the ITS region to study groundwater microbial communities in the VAFB MTBE-contaminated aquifer. We generated DGGE and ITS PCR fingerprints from community DNA extracted from groundwater samples collected both from within and outside the LTF (Fig. 1). Substantial bacterial diversity in the microbial communities was found in all locations. Both methods indicated the presence of a band on the electrophoresis gels with the same migration distance as the band for strain PM1 when the latter was run in an adjacent lane (Fig. 3a). The ITS PCR fingerprints targeting bacterial DNA also showed the enrichment of a band similar to that of a PM1 pure culture in groundwater microcosms after incubation with 10 μg of MTBE/liter in the lab (Fig. 3a, lanes 5 to 9).
FIG. 3.
(a) Detection of PM1 band in ITS fingerprints of groundwater (GW) or sand samples from the LTF and after incubation in laboratory microcosms. Lane 1, a DNA marker; lanes 2 and 10, PM1 pure culture DNA; lanes 3 and 4, groundwater from well K3 in the LTF; lanes 5 to 7, K3 groundwater from lab microcosms containing sand; lanes 8 and 9, DNA extracted from sand from the same microcosms. (b) Detection of PM1 band in DNA amplified with16S rDNA PM1 DGGE primers extracted from groundwater samples from the LTF and after incubation in laboratory microcosms. Lanes 1 to 4 are groundwater from the microcosms shown in Fig. 5, lanes 5 to 13 are groundwater from the LTF, and lane 14 is a microcosm with a sediment sample collected from the UW8 location outside the LTF. PM1 pure culture DNA has been used as a control. Arrows show bands that were sequenced.
We developed 16S rDNA primers unique to strain PM1 for use in DGGE analysis. We confirmed with DGGE that PM1 sequences were present in all tested wells in the LTF and in the laboratory microcosms (Fig. 3b). DNA sequences of the bands from three microcosm and two field samples indicated that the 16S sequences were ≥99% similar to the laboratory strain of PM1.
We designed another set of PM1-specific primers targeting the ITS region (Table 1) to try to achieve greater resolution in differentiating the native from the laboratory strain of PM1. ITS analysis has been successful in distinguishing bacterial strains and between closely related species (1, 10). PM1 ITS analysis on field and microcosm samples from VAFB site 60 showed the presence of the PM1 ITS band in all microcosm samples and all samples within the trench but not in groundwater from adjacent areas outside the trench (data not shown). Sequences of a total of five ITS bands of amplified DNA from VAFB trench groundwater were 99% similar to one other and ≥98% similar to PM1.
Quantitative PCR of PM1 in VAFB field samples.
We used TaqMan quantitative PCR with primers and a probe specific to the 16S rDNA region of strain PM1 (19) to measure the population density of strain PM1 at different locations in the LTF. We detected native PM1 at densities of 103 to 104 cells per ml in groundwater samples collected at different locations along the LTF down gradient of the oxygen RW (Fig. 1b). PM1 was detected at a density of ≤180 cells/ml (the detection limit) in water samples collected immediately up gradient of the oxygen RW (well K0) and in locations outside of the LTF (UW9 and UW8). Within the LTF (wells K1 to K9), PM1 cells were detected at a density of ≥103 cells/ml of groundwater when oxygen release was occurring. Native PM1 was also detected in sediment samples collected adjacent to the LTF at densities of 103 cells/g of sediment (Table 2).
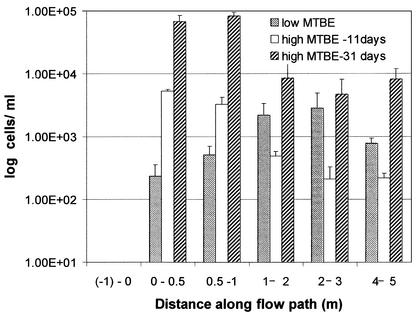
To determine the relationship between PM1 density and MTBE concentration in situ, we sampled the LTF during field experiments in which MTBE was intentionally released at concentrations higher than typically observed in the field. This was part of a larger study measuring the capacity of the LTF to release sufficient oxygen for MTBE treatment (36). A PM1 cell density of 104 cells/ml was observed in close proximity to the oxygen RW 11 days after increasing the MTBE concentration to 1,300 μg/liter (Fig. 4). The PM1 cell density was lower (4.2 × 102 ± 1.0 × 102 cells/ml) in the trench at locations 2 to 4 m down gradient from the oxygen RW. Twenty days after the MTBE was increased to 2,100 μg/liter, PM1 population densities reached 105 cells/ml close to the RW and 104 cells/ml further down gradient (Fig. 4).
FIG. 4.
Relationship between MTBE degradation and PM1 cell densities at different MTBE influent concentrations (all measured in the RW). Low MTBE, 500 μg/liter (samples collected immediately); medium MTBE, 1,300 μg/liter (samples collected 11 days after concentration changed); high MTBE, 2,100 μg/liter (samples collected 20 days after increase).
Linking PM1 population densities to the MTBE disappearance in laboratory microcosms.
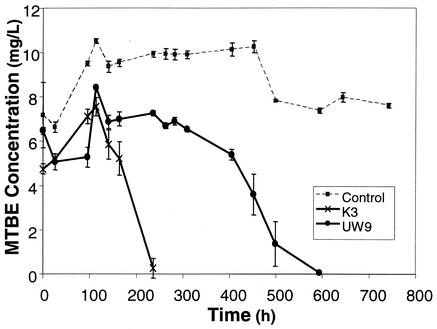
We conducted microcosm studies with sediment and groundwater collected from the LTF and adjacent areas at the VAFB site to test, under controlled conditions, whether MTBE disappearance is associated with increases in PM1 densities. Microcosms with groundwater from within or near the LTF degraded MTBE (Fig. 5). Control microcosms with mineral media and MTBE indicated no contamination by laboratory strain PM1 during incubation.
FIG. 5.
Biodegradation of MTBE in aerobic microcosms containing groundwater and sterile sand as porous media. K3 is within the LTF, and UW9 is outside the LTF.
Preliminary studies were conducted with groundwater and sediment samples from site 60, incubated with or without 10 μg of MTBE/ml, to determine how added MTBE affected PM1 cell density during laboratory incubations. Initial PM1 cell densities of 2.5 × 102 ± 1.8 × 102 cells per ml in groundwater increased during 22 days of incubation to 2.6 × 103 ± 0.5 × 103 cells per ml without MTBE and to 2.9 × 105 ± 1.2 × 105 cells per ml after degradation of the MTBE. Initial densities of 7.6 × 104 ± 0.2 × 104 cells of PM1 per g in sediment samples collected from the field increased during 22 days of incubation to 8.4 × 104 ± 5.6 × 104 cells per g without MTBE and to 1.9 × 106 ± 0.5 × 106 per g of sediment after degradation of MTBE. In conclusion, the presence of MTBE increased initial cell densities far above the small increases possibly associated with growth on indigenous organic compounds.
At the start of the experiments linking MTBE removal to PM1 cell density (Fig. 5), PM1 was not detected in water samples from the UW9 well located outside the LTF but was present at a density of 1.3 × 104 ± 0.3 × 104 PM1 cells/ml in well K3 in the LTF (Table 2). After MTBE was degraded, PM1 reached the same cell density in microcosms regardless of whether the sample was collected from within or outside the LTF: 3.3 × 105 ± 0.8 × 105 cells/ml in K3 and 3.3 × 105 ± 1.0 × 105 cells/ml in UW9 (Table 2). The rate of MTBE biodegradation, however, did reflect differences in initial population density, taking 200 and 600 h to degrade 10 μg of MTBE/ml in K3 and UW9, respectively (Fig. 5). UW9 microcosms showed a substantially higher rate of degradation of a second than first input of 10 μg of MTBE/ml (data not shown). Sediment samples incubated with MTBE showed a 2 log order increase in the initial density of PM1 (Table 2).
PM1 was also detected in all triplicate microcosm bottles by ITS PCR specific to PM1 in DNA extracted from groundwater and sediment samples from all microcosms except the sodium azide-killed controls (data not shown).
DISCUSSION
It was surprising to find DNA sequences virtually identical to the 16S rDNA and ITS regions of the PM1 genome present in MTBE-contaminated groundwater at VAFB. The laboratory strain PM1 was isolated from a compost-filled biofilter treating off-gases from a Los Angeles, Calif., sewage treatment plant that received some portion of its discharges from local oil refineries (13). Biodegradation activity by the microbial consortium from which PM1 was isolated was not detected until approximately a year after MTBE began to be measured in the biofilter influent (9). It is not possible for us to determine whether PM1 was derived from the compost, evolved de novo under selection in the biofilter, or entered the biofilter in association with aerosols from waste streams.
The distribution of native PM1 density was positively associated with oxygen presence and higher MTBE concentrations. Within the LTF, before the MTBE concentration was artificially increased, the highest densities were detected somewhat down gradient from the oxygen RW. After increasing the MTBE concentration, however, the highest densities were immediately adjacent to the oxygen RW, suggesting that higher oxygen concentrations were needed to remove the higher MTBE concentrations. On the other hand, densities of native PM1 were very low or not detectable in anoxic samples collected outside the LTF. The presence of native PM1 in the anoxic groundwater suggests that it is capable of surviving in the absence of oxygen.
The rates of aerobic MTBE biodegradation are strongly influenced by oxygen concentration. In a survey of numerous field and microcosm studies, aerobic MTBE biodegradation half-lives were found to be inversely proportional to DO concentrations (35). In a bioremediation field study at Port Hueneme Naval Facility in Oxnard, Calif., Salanitro et al. (31) found higher MTBE removal rates and higher numbers of MTBE-degrading organisms, estimated by a most-probable number method, in field plots sparged with oxygen than in unamended plots. MTBE biodegradation rates were a strong function of DO concentrations in a study of pilot-scale biotrickling filters treating MTBE-contaminated air streams (11). In the field at VAFB, Wilson et al. (36) found that MTBE degradation by indigenous microorganisms was most rapid in areas that had measurable DO levels of 13 to 23 mg/liter. When the oxygen supply was turned off and DO was confirmed to be absent in the LTF, MTBE concentrations down gradient of the RW rose to values similar to those detected up gradient of the LTF.
The magnitude of increases in densities of native PM1 during MTBE biodegradation was consistent with previous findings that laboratory strain PM1 produces new biomass during MTBE biodegradation (13). Growth of the lab strain PM1 on 25 μg of MTBE/ml was associated with increases in protein concentration with approximately 0.2 μg of biomass formed per 1 μg of MTBE degraded (13). In microcosm experiments in this study, we found approximately 3 × 105 cells of native PM1 produced (from a very low initial population density) when grown on 10 μg of MTBE/ml. Assuming that the dry mass of one cell is 2 pg (28), then this is equivalent to 0.2 μg of cells. This yield for native PM1 is an order of magnitude lower than what we observed for the laboratory strain and may reflect less-than-optimum conditions in the groundwater. We found similar yields for strain PM1 when we incubated groundwater samples from Port Hueneme Naval Facility, which had been inoculated with the lab strain of PM1 in the field 9 months earlier. Initial densities of 102 to 103 cells/ml increased to 3.9 × 105 ± 1.0 × 105 PM1 cells/ml after incubation with 10 μg of MTBE/ml (19).
Strain PM1 has also been reported to be present in other MTBE-contaminated California aquifers. In groundwater samples from Port Hueneme Naval Facility, we found that DNA sequences similar to those of laboratory PM1 were dominant bands on DGGE and ITS gels by using universal bacterial primers (K. Hristova and K. M. Scow, unpublished data). Recently Kane and coworkers reported the presence of dominant DGGE sequences very similar to the laboratory strain of PM1 in aquifer microcosms from two out of four MTBE-contaminated sites in Northern California which were different from those in which we report the presence of PM1 (21). Kane et al. observed a positive correlation between the presence of MTBE biodegradation activity and the PM1 sequence.
Although we demonstrate a relationship between MTBE biodegradation and increases in native PM1 density, we cannot rule out that other microorganisms may also be involved in MTBE biodegradation at VAFB. Defined mixed cultures isolated from VAFB sediments and not including native PM1 showed complete MTBE degradation in mineral media (K. Hristova, unpublished results). Future investigations are exploring the link between the presence of these organisms and MTBE degradation in the field.
In conclusion, a bacterial strain very similar (based on DNA sequences in three different regions of the rDNA) to the well-characterized MTBE-degrading isolate, strain PM1, occurs naturally in MTBE-contaminated groundwater. The densities of the PM1-like organism increase concurrent with MTBE removal in microcosms, and the densities increased when MTBE concentration was artificially increased in situ. Quantitative PCR methods, such as the one used here, can link the densities of specific microbial populations involved in biodegradation to rates of bioremediation, environmental factors, and management practices. Such techniques will be very useful in evaluating the impacts of different bioremediation methods on the actual catalysts (i.e., the organisms carrying out the treatment) of these technologies.
Acknowledgments
This study was supported by the NIEHS Superfund Basic Research Program (2P42 ES04699), EPA Center for Ecological Health Research, UC Toxic Substances Program, Oxygenated Fuels Association, and the Water Resources Center.
Our gratitude goes to Isaac Wood and Ryan Wilson for help in the field study and groundwater sampling. We thank undergraduate students Sarah Adamson and Brian Watanabe for work on the microcosm experiments.
REFERENCES
- 1.Aakra, A., J. B. Utaker, and I. F. Nes. 1999. RFLP of rRNA genes and sequencing of the 16S-23S rDNA intergenic spacer region of ammonia-oxidizing bacteria: a phylogenetic approach. Int. J. Syst. Bacteriol. 49:123-130. [DOI] [PubMed] [Google Scholar]
- 2.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from Eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley, P. M., J. E. Landmeyer, and F. H. Chapelle. 2001. Widespread potential for microbial MTBE degradation in surface-water sediments. Environ. Sci. Technol. 35:658-662. [DOI] [PubMed] [Google Scholar]
- 4.Bradley, P. M., F. H. Chapelle, and J. E. Landmeyer. 2001. Effect of redox conditions on MTBE biodegradation in surface water sediments. Environ. Sci. Technol. 35:4643-4647. [DOI] [PubMed] [Google Scholar]
- 5.Bruns, M. A., J. R. Hanson, J. Mefford, and K. M. Scow. 2001. Isolate PM1 populations are dominant and novel methyl tert-butyl ether degrading bacteria in compost biofilter enrichments. Environ. Microbiol. 3:1-7. [DOI] [PubMed] [Google Scholar]
- 6.Carter, S. R., J. M. Bullock, and W. R. Morse. Enhanced biodegradation of MTBE and BTEX using pure oxygen injection, p. 142-147. In B. C. Alleman and A. Leeson (ed.), In situ and on site bioremediation. Battelle Press, Columbus, Ohio.
- 7.Church, C. D., P. J. Tratnyek, J. F. Pankow, J. E. Landmeyer, A. L. Baehr, M. A. Thomas, and M. Schirmer. 1999. Effects of environmental conditions on MTBE degradation in model column aquifers, p. 93-101. In Proceedings of the Technical Meeting of the USGS Toxic Substances Hydrology Program. Battelle Press, Columbus, Ohio.
- 8.Deeb, R. A., H. Y. Hu, J. R. Hanson, K. M. Scow, and L. Alvarez-Cohen. 2000. Substrate interactions in BTEX and MTBE mixtures by an MTBE-degrading isolate. Environ. Sci. Technol. 35:312-317. [DOI] [PubMed] [Google Scholar]
- 9.Eweis, J. B., E. D. Schroeder, D. P. Chang, and K. M. Scow. 1998. Biodegradation of MTBE in a pilot-scale biofilter, p. 342-346. In G. B. Wickramanayake and R. E. Hinchee (ed.), Natural attenuation of chlorinated and recalcitrant compounds. Batelle Press, Columbus, Ohio.
- 10.Fisher, M. M., and E. W. Triplet. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fortin, N. Y., and M. A. Deshusses. 1999. Treatment of methyl tert-butyl ether vapors in biotrickling filters. Analysis of the rate-limiting step and behavior under transient conditions. Environ. Sci. Technol. 33:2987-2991. [Google Scholar]
- 12.Garnier, P., R. Auria, C. Auger, and S. Revah. 1999. Cometabolic biodegradation of methyl t-butyl ether by Pseudomonas aeruginosa grown on pentane. Appl. Microbiol. Biotechnol. 51:498-503. [DOI] [PubMed] [Google Scholar]
- 13.Hanson, J. R., C. E. Ackerman, and K. M. Scow. 1999. Biodegradation of methyl tert-butyl ether by a bacterial pure culture. Appl. Environ. Microbiol. 65:4788-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Happel, A. M., E. H. Beckenbach, and R. U. Halden. 1998. An evaluation of MTBE impacts to California groundwater resources. UCRL-AR-130897. Lawrence Livermore National Laboratory, Environmental Protection Department, University of California, Davis.
- 15.Hardison, L. K., S. S. Curry, L. M. Ciuffetti, and M. R. Hyman. 1997. Metabolism of diethyl ether and cometabolism of methyl tert-butyl ether by a filamentous fungus, a Graphium sp. Appl. Environ. Microbiol. 63:3059-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatzinger, P. B., K. McClay, S. Vainberg, M. Tugusheva, C. Condee, and R. J. Stefan. 2001. Biodegradation of methyl tert-butyl ether by a pure bacterial culture. Appl. Environ. Microbiol. 67:5601-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez-Perez, G., F. Fayolle, and J.-P. Vandecasteele. 2001. Biodegradation of ethyl t-butyl ether (ETBE), methyl t-butyl ether (MTBE), and t-amyl methyl ether (TAME) by Gordonia terrae. Appl. Microbiol. Biotechnol. 55:117-121. [DOI] [PubMed] [Google Scholar]
- 19.Hristova, K., C. M. Leutenegger, and K. M. Scow. 2001. Detection and quantification of MTBE-degrading strain PM1 by real-time TaqMan PCR. Appl. Environ. Microbiol. 67:5154-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyman, M., P. Kwon, K. Williamson, and K. O'Reilly. 1998. Cometabolism of MTBE by alkane-utilizing microorganisms, p. 321-326. In G. B. Wickramanayake and R. E. Hinchee, (ed.), Natural attenuation of chlorinated and recalcitrant compounds. Battelle Press, Columbus, Ohio.
- 21.Kane, S. R., H. R. Beller, T. C. Legler, C. J. Koester, H. C. Pinkart, R. U. Halden, and A. M. Happel. 2001. Aerobic biodegradation of methyl tert-butyl ether by aquifer bacteria from leaking underground storage tank sites. Appl. Environ. Microbiol. 67:5824-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao, C. M., S. C. Chen, J. K. Liu, and Y. S. Wang. 2001. Application of microbial enumeration technique to evaluate the occurrence of natural bioremediation. Water Res. 35:1951-1960. [DOI] [PubMed] [Google Scholar]
- 23.Koenigsberg, S., C. Sandefur, W. Mahaffey, M. Deshusses, and N. Fortin. 1999. Peroxygen mediated bioremediation of MTBE, p. 13-18. In B. C. Alleman and A. Leeson (ed.), In situ bioremediation of petroleum hydrocarbon and other organic compounds. Battelle Press, Columbus, Ohio.
- 24.Mackay, D. M., R. D. Wilson, M. J. Brown, W. P. Ball, G. Xia, and D. P. Durfee. 2000. A controlled field evaluation of continuous vs. pulsed pump-and-treat remediation of a VOC-contaminated aquifer: site characterization, experimental setup, and overview of results. J. Contam. Hydrol. 41:81-131. [Google Scholar]
- 25.Mackay, D. M., R. D. Wilson, K. M. Scow, M. D. Einarson, B. Fowler, and I. A. Wood. 2001. In situ remediation of MTBE at Vandenberg Air Force Base, California. Contam. Soil Sediment Water Spring special issue p. 43-46.
- 26.Mo, K., C. O. Lora, A. E. Wanken, M. Javanmardian, X. Yang, and C. F. Kulpa. 1997. Biodegradation of methyl t-butyl ether by pure bacterial cultures. Appl. Microbiol. Biotechnol. 47:69-72. [DOI] [PubMed] [Google Scholar]
- 27.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 28.Neidhardt, F. C., J. L. Ingraham, and M. Schaecter. 1990. The physiology of the bacterial cell. Sinauer Associates, Inc., Sunderland, Mass.
- 29.Park, K., and R. M. Cowan. 1997. Biodegradation of gasoline oxygenates, p. 17. In B. Alleman and A. Leeson (ed.), Proceedings of the Fourth International In Situ and On-Site Bioremediation Symposium. Battelle Press, Columbus, Ohio.
- 30.Salanitro, J. P., L. A. Diaz, M. P. Williams, and H. L. Wisniewski. 1994. Isolation of a bacterial culture that degrades methyl t-butyl ether. Appl. Environ. Microbiol. 60:2593-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salanitro, J. P., P. C. Johnson, G. E. Spinnler, P. M. Maner, H. L. Wisniewski, and C. Bruce. 2000. Field-scale demonstration of enhanced MTBE bioremediation through aquifer bioaugmentation and oxygenation. Environ. Sci. Technol. 34:4152-4162. [Google Scholar]
- 32.Scow, K. M., J. Leung, D. Mackay, R. Wilson, and A. Smith. 2000. Oxygen stimulates MTBE biodegradation by indigenous microbial populations in groundwater, p. 552. In Proceedings of the 100th General Meeting of the American Society for Microbiology. ASM Press, Washington, D.C.
- 33.Simkins, S., and M. Alexander. 1984. Models of mineralization kinetics with the variables of substrate concentration and population density. Appl. Environ. Microbiol. 51:1153-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steffan, R. J., K. McClay, S. Vainberg, C. W. Condee, and D. Zhang. 1997. Biodegradation of the gasoline oxygenates methyl tert-butyl ether, ethyl tert-butyl ether, and tert-amyl methyl ether by propane-oxidizing bacteria. Appl. Environ. Microbiol. 63:4216-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stocking, A. J., R. A. Deeb, A. E. Flores, W. Stringfellow, J. Talley, R. Brownell, and M. C. Kavanaugh. 2000. Bioremediation of MTBE: a review from a practical perspective. Biodegradation 11:187-201. [DOI] [PubMed] [Google Scholar]
- 36.Wilson, R., K. M. Scow, and D. Mackay. 2002. In situ MTBE biodegradation supported by diffusive oxygen release. Environ. Sci. Technol. 36:190-199. [DOI] [PubMed] [Google Scholar]
- 37.Yeh, C. K., and J. T. Novak. 1994. Anaerobic biodegradation of gasoline oxygenates in soils. Water Environ. Res. 66:744-752. [Google Scholar]