Abstract
We have constructed mutants of chymotrypsin inhibitor 2 with short glutamine repeats inserted into its inhibitory loop. These mutants oligomerize when expressed in Escherichia coli. The dimer of a mutant with four glutamines now has been crystallized, and its structure has been solved by molecular replacement by using the wild-type monomer as a search model. The structure of each half of the dimer is found to be the same as that of the wild-type monomer, except around the glutamine insertion. It was proposed that the components of the oligomers are held together by hydrogen bonds between the main-chain and side-chain amides of the glutamine repeats. Instead, they appear to form by swapping domains on folding in E. coli, and the glutamine repeats connecting the components of the dimers are disordered.
Over the past 5 years, a growing number of dominantly inherited neurodegenerative diseases, including Huntington’s disease and several different forms of spinocerebellar ataxia, have been linked to abnormally expanded CAG repeats in the coding regions of certain genes. The diseases are collectively known as polyglutamine-expansion diseases because the CAG repeats code for stretches of polyglutamine in the affected proteins. Progressive neurodegeneration begins in late adulthood if a single glutamine repeat exceeds a critical length, usually ≈40 residues, and begins earlier and becomes more severe as the repeat gets even longer in successive generations of affected individuals. The proteins causing the different diseases are unrelated except for the glutamine repeats. It is likely, therefore, that the diseases share a common pathogenic mechanism that depends only on the presence of an abnormally expanded glutamine repeat and not on its protein context. Together with the autosomal dominant genetics of the diseases, this mechanism implies that abnormally expanded glutamine repeats are toxic. Several groups have confirmed this by demonstrating the cytotoxicity of long glutamine repeats in various model systems (1, 2).
Neither the normal function of glutamine repeats in proteins nor the mechanism by which their abnormally expanded counterparts cause progressive neurodegeneration is known. Perutz et al. proposed that glutamine repeats in proteins can associate with each other by forming polar zippers in which β-strands are held together by hydrogen bonds between their main-chain and side-chain amides (3). They went on to suggest that the threshold effect of the polyglutamine-expansion diseases might reflect a delicate balance of two counteracting factors that determine whether a stable polar zipper is formed: the entropy gain or loss of the glutamines versus that of the solvent molecules. Abnormally expanded glutamine repeats form stable polar zippers because the increase in entropy caused by the liberation of many bound water molecules offsets the entropy loss caused by the formation of a rigid secondary structure, and this leads to the proteins’ aggregation, which causes progressive neurodegeneration; by contrast, shorter glutamine repeats do not form stable polar zippers because the entropy loss cannot be offset by the liberation of fewer solvent molecules (4).
To test the initial hypothesis of Perutz et al. (3) that glutamine repeats in proteins can associate with each other by forming polar zippers, we inserted a 10-glutamine repeat into the inhibitory loop of chymotrypsin inhibitor 2 (CI2), a naturally monomeric protein, to see whether this caused the protein to associate into oligomers (5). This mutant, CI2-Q10i, did indeed form dimers and trimers as well as monomers, and the circular dichroism spectra of these oligomers indicated that they had associated by the formation of polar zippers. However, they were so stable that they could be dissociated into monomers only by denaturation of the protein. Therefore, loop-insertion mutants with shorter glutamine repeats were constructed in the hope that a dynamic equilibrium could be established. These mutants also formed extremely stable oligomers, and it became clear that some other mechanism must be responsible for their formation. Oligomerization has now been shown to occur by domain swapping (K.S., P. Scamborova, T. Galvao, and M.F.P., unpublished results), but polar zipper interactions have not been ruled out. Here, we set out to determine the crystal structure of one of the mutant oligomers, to see whether such interactions could be observed. We tried to purify the monomers, dimers, and trimers of all of these loop-insertion mutants for crystallization. The CI2-Q10i monomer gave crystals that diffracted only to low resolution (K.S., unpublished observations), but the CI2-Q4i dimer produced two suitable crystal forms. The structure of the hexagonal form has been solved and is described below.
MATERIALS AND METHODS
Crystallization.
The protein CI2-Q4i consists of the residues Gly-Gln4-Gly-Met (GQQQQGM) inserted into the inhibitory loop of truncated CI2 (first 20 disordered residues removed and replaced by an N-terminal methionine) immediately after the native residue Met59. For residue numbering, we adopt the wild-type CI2 convention for the convenience of structure comparison. We used “N domain” (residues 21 to 57) and “C domain” (residues 61 to 83) to identify the two fragments that constitute a globular unit (pseudomonomer) of the dimer. For discussion purpose, the residues inserted after Met59 are identified as Gly59A, Gln59B, Gln59C, Gln59D, Gln59E, Gly59F, and Met59G, to comply with the Protein Data Bank convention (6, 7). The CI2-Q4i mutant protein was expressed in Escherichia coli strain NM554 by using plasmid pCI2-Q4i (K.S., P. Scamborova, T. Galvao, and M.F.P., unpublished results). The dimeric fraction of this mutant was purified as described for the corresponding 10-glutamine loop insertion mutant CI2-Q10i (5) and was crystallized at 4°C by the hanging drop method. The drops were prepared by mixing 2 μl of the purified dimer at 25 mg/ml with an equal volume of crystallization buffer (30% wt/vol polyethylene glycol 400, 1.0 M lithium sulfate, and 1 mM calcium chloride, in 0.1 M Tris⋅HCl at pH 7.5), which also was used as the well buffer (1 ml volume). Crystal growth typically took 4 weeks. A single hexagonal crystal measuring 0.3 × 0.3 × 0.2 mm was selected by using a 0.4-mm loop and was mounted in a cryostream at 100 K for data collection; the transfer of crystals had to be done extremely quickly to avoid fragmentation of the crystal. No cryo protectant was added because the high concentration of polyethylene glycol 400 in the crystallization medium made it viscous. The dimeric CI2-Q4i also crystallizes in a cubic form (K.S., unpublished observations).
Structure Determination.
The hexagonal crystals belong to the space group P622, with cell dimensions of a = b = 68.27 Å and c = 60.83 Å. This is similar to the wild type, which also crystallizes in P622, with a = b = 69.02 Å but a smaller unit cell dimension c of 52.89 Å (8). The crystallization conditions of CI2-Q4i were similar to those of the wild-type CI2 (9). The crystal parameters suggest that there is one molecule of CI2-Q4i per asymmetric unit, with a solvent content of 53% and Matthew’s coefficient, Vm = 2.6 Å3/dalton. A single crystal was used to collect two continuous segments of data (40 and 72 frames, respectively, with 1° oscillation per frame) at the X11 beamline in Hamburg. Diffraction images were processed with mosflm (10) and then were scaled and reduced with programs from the CCP4 suite (11). The data were significantly anisotropic: while the crystal diffracts strongly beyond 1.8 Å along h and k, the diffraction intensities fall off rapidly beyond 2.4 Å along l. Ten percent of the data were set aside for cross-validation and were omitted from all stages of refinement. The structure was solved by the molecular replacement method (12) with the program amore (11, 13, 14). The search model was constructed from wild-type CI2 (PDB ID code 2CI2) (6, 7), with the first two disordered residues at the N terminus (residues I19 and I20) as well as the residues flanking the insertion site (residues I54–I63) removed. A clear solution was found with an R-factor of 0.42 and a correlation coefficient of 70.3 for data in the range of 8–2.5 Å. The R-factor dropped to 0.39 after rigid-body refinement.
Refinement.
The structure was refined with refmac (11, 15), with alternating cycles of manual rebuilding. The inclusion of anisotropic scaling in subsequent steps proved to be essential. A bulk solvent model also was used. The computer graphics program o (16) was used for rebuilding, aided by σA-weighted maps with 2mFo DFc and mFo − DFc coefficients (17). arp (18) was used to add and refine water molecules automatically. After four cycles, the refinement converged, and the resulting model has an R-factor of 0.24 and Rfree of 0.30, using all data to 1.8 Å. The stereochemistry of the final model was checked with procheck (19) and was found to be good. The relatively high R-factors are probably attributable to the disorder of ≈14% (10 residues of 71) of all protein atoms. The structural model of CI2-Q4i consists of residues 21 to 57 and 61 to 83, which are called the N and C domains. Most residues in the model have clear electron density. A high average real-space correlation coefficient of 0.89 (20) was calculated with o (16), which shows that the model correlates well with the x-ray data. No electron density was observed for residues 20, 58, 59, 59A–59G, and 60. Despite extensive density modification techniques using the programs dm and dmmulti (11, 21), including multiple crystal form averaging with noncrystallographic symmetry (the cubic form has three monomers in the asymmetric unit), the connectivity between residue 57 and 62 could not be found. In case the space group of CI2-Q4i was of a lower symmetry than P622—i.e., trigonal—and the hexameric rings could actually be trimers of domain-swapped adjacent dimers, in the same layer, processing the data in P622 would have averaged out the dimeric linkage electron density. We also processed them in P312 and P321. We then used the final model from the P622 data to generate a suitable dimeric model for each of these two trigonal space groups and calculated 2mFo − DFc and mFo − DFc maps. No electron density was observed for the loops in either space group. We also found that both the R-factor and Rfree were very similar (R = 0.23, Rfree = 0.27 for P312; R = 0.24, Rfree = 0.26 for P321) in both trigonal space groups: the two trigonal space groups are indistinguishable, confirming that it is more accurate to refine the structure in P622.
RESULTS AND DISCUSSION
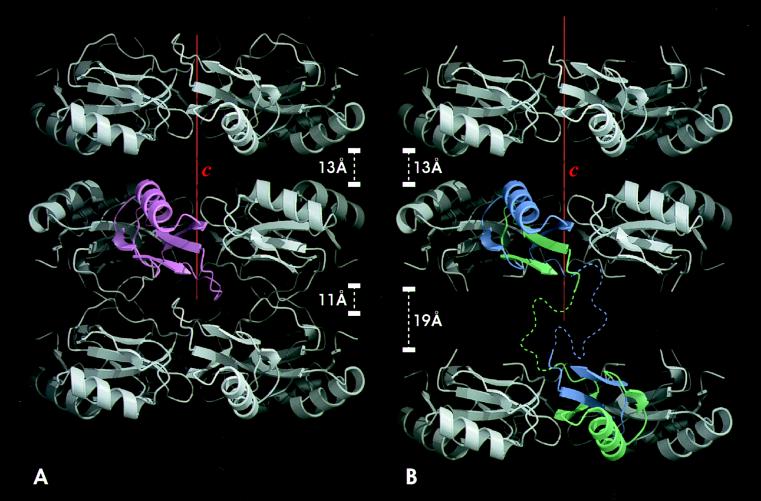
Each member of the dimer is made up of two domains, one from each member; one domain lies N-terminal and the other C-terminal to the inserted residues. We shall call the members pseudomonomers (Fig. 1). Like wild-type CI2, the dimer crystallizes in space group P622. The unit cell contains one pseudomonomer in the asymmetric unit (Fig. 2). The pseudomonomers must therefore be related by a dyad axis of symmetry. All of the secondary structure elements (an α-helix and a four-stranded mixed parallel and antiparallel β-sheet) and the overall fold of the wild type are preserved (8, 22). In both wild-type CI2 and CI2-Q4i, the proteins are arranged in layers of hexameric rings. In the wild type, one of the spaces between the layers contains the inhibitory loop residues packing against symmetry-related loop residues of the next layer. This space is 11 Å wide and contains extensive hydrophobic interactions (Fig. 3A). The corresponding space in the mutant structure is 19 Å wide (Fig. 3B) and contains no distinct peaks of electron density. The alternate spaces are 13 Å wide in both the wild type and the mutant structures (Fig. 3 A and B). Contacts in the 13 Å layer consist of very few van der Waals interactions between the symmetry-related α-helices containing residues Glu33, Glu34, and Lys37; the weakness of these interactions may explain the fragility of the crystals along this plane.
Figure 1.
Diagrammatic sketch of how monomers associate to form a domain-swapped dimer. N and C denote N-terminal and C-terminal domains, respectively.
Figure 2.
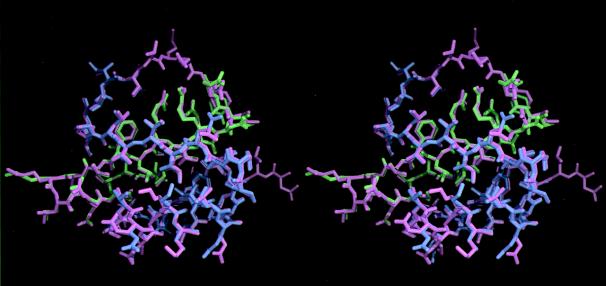
The crystal structure of the pseudomonomer of CI2-Q4i compared with the wild-type CI2 monomer. The structure of CI2-Q4i is colored purple for the N domain (residues 21 to 57) and pale green for the C domain (residues 61 to 83). Wild-type CI2 is colored pink. The rms displacement between all main-chain atoms of the crystal structures of CI2-Q4i and wild-type CI2, when all common residues are superimposed, is 0.20 Å (that for all atoms is 0.88 Å). Figs. 2, 3, and 4 in this work were generated with bobscript (30), which is an enhanced version of molscript (31), and were rendered with raster3d (32).
Figure 3.
The quaternary structure and crystal packing of wild-type CI2 (A) compared with that of dimeric CI2-Q4i (B). Three hexameric ring layers are shown. In B, a CI2-Q4i dimer is colored, and, in A, a CI2 monomer is colored. In the CI2-Q4i structure, one of the spaces between the layers is expanded to 19 Å, compared to 11 Å in the wild-type. The color coding is the same as in Fig. 2. The dotted lines represent the inferred positions of the disordered residues. The red boxes are the unit cells. This view is along the b axis.
The crystal structure of CI2-Q4i does not reveal how the dimer was formed because the loops connecting the N and C domains are disordered. There are two possible mechanisms that would leave the structure of each component of the dimer apparently undisturbed: either domain swapping (for reviews, see refs. 23 and 24) or concatenation of loops on folding. Both mechanisms would account for the pseudomonomers being kept together so firmly that only denaturation can separate them. Biochemical studies showed that CI2-Q4i is domain-swapped rather than concatenated (K.S., P. Scamborova, T. Galvao, and M.F.P., unpublished results). The trace of the polypeptide chain before and after the disordered loop region and the distances between these two residues from two neighboring 2-fold symmetry-related pseudomonomers shows that the only arrangement consistent with the 2-fold symmetry and the distance and volume required to accommodate the bulky inserted residues is that shown in Fig. 3B. This places the symmetry-related pseudomonomers of the dimer in adjacent layers, which are linked covalently by the inserted residues. The space is 19 Å wide, shorter than the 24 Å that would be the length of the seven inserted residues if they formed an extended β-strand. The inserted residues are disordered. Insertion of residues other than glutamines into the CI2 loop also induced oligomerization, which implies that extension of an external loop alone is sufficient to induce misfolding by domain swapping (K.S., P. Scamborova, T. Galvao, and M.F.P., unpublished results). The seven residues introduced into the CI2-Q4i mutant contribute an addition of ≈10% of protein atoms. This was easily tolerated. CI2-Q4i folds like two fragments (residues 20–57 and residues 61–83) docking together to form the pseudomonomer. Similarly, wild-type CI2 (truncated, residues 20–83) can be cleaved into two fragments [CI2 (20–59) and CI2 (60–83)], and these fragments can fold into one structure indistinguishable to the wild type and to our mutant (25, 26).
We found not the results we had hoped for, which was oligomerization by regular hydrogen bonds formed between neighboring β-strands as in a synthetic poly-l-glutamine (3), but instead we found oligomerization by domain swapping. Does this leave anything that we can learn from our results about diseases caused by expansion of glutamine repeat? Insertion of several bulky polar glutamines into a flexible loop does not destroy the structure of our model protein. CI2 loop-insertion mutants have stabilities and folding rates similar to those of the wild type (27). Our mutant CI2-Q4i protein cannot fulfill its normal function of inhibiting serine proteinases because the insertion is at its essential Met59, but if the insertion occurred at some nonessential external loop in a protein, preservation of its structure would allow the protein to function. The puffer fish Huntingtin homologue has a repeat of only four glutamines in the position of the long repeats in human Huntingtin (28), which shows that the ends of the glutamine repeat cannot be farther than 4 × 3.4 Å, or 14 Å, apart. Extension of the glutamine repeat beyond this length may be harmless, as long as its structure remains random. If the repeats form a hairpin, it may loosen the structure either by pulling the two ends too close together or by inserting itself between the β-strands of another molecule as happens in mutant serine proteinase inhibitors (29). So far, there is no evidence for domain-swapping in any of the proteins affected by polyglutamine diseases.
Table 1.
Statistics of data processing and refinement
| Data processing | |
| Resolution, Å | 22-1.8 (1.9-1.8) |
| Number of measurements | 86441 |
| Number of unique reflections | 8153 |
| Multiplicity | 10.6 (10.8) |
| Completeness of data, % | 99.7 (100) |
| Rmerge | 0.074 (0.286) |
| Refinement | |
| Resolution, Å | 22-1.8 (1.9-1.8) |
| Number of reflections in refinement/cross-validation | 7358 (795) |
| Number of protein atoms | 474 |
| Number of water molecules | 55 |
| Number of SO4 atoms | 5 |
| R-factor | 0.24 (0.24) |
| Rfree | 0.30 (0.35) |
| Overall anisotropic B factors, scaling Fo − Fc | |
| B11 | −6.1 |
| B22 | −9.0 |
| B33 | 23.9 |
| B12 | −5.1 |
| Average B factor, Å | 32.2 |
| rms deviations | |
| Bond distance, Å | 0.016 |
| Angle distance, Å | 0.034 |
| Overall error in coordinates from Rfree, Å | 0.15 |
| procheck validation | |
| Ramachandran plot (most favored/additional allowed) | 94.0/6.0 |
| Overall G-factor | −0.2 |
Rmerge is defined as Σ|I − 〈I〉|/ΣI. R-factor = Σ∥Fo| − |Fc∥/Σ|Fo|, where |Fo| and |Fc| are the observed and calculated structure factors, respectively. Rfree is the same as R-factor but calculated with 10% of the data excluded from refinement.
Figure 4.
The 2mFo − DFc electron density near the insertion site of CI2-Q4i. The map is contoured at 1 σ. No connectivity is seen between residues 57 and 61. Color coding is as in Fig. 2. Water molecules are represented by cyan spheres.
Acknowledgments
The authors thank Dr. Ashley Buckle for his help with data collection. This work was supported by a research grant from the Wellcome Trust.
ABBREVIATION
- CI2
chymotrypsin inhibitor 2
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID codes 1CQ4 and R1CQ4SF).
References
- 1.Onodera O, Roses A D, Tsuji S, Vance J M, Strittmatter W J, Burke J R. FEBS Lett. 1996;399:135–139. doi: 10.1016/s0014-5793(96)01301-4. [DOI] [PubMed] [Google Scholar]
- 2.Ordway J M, Tallaksen-Greene S, Gutekunst C A, Bernstein E M, Cearley J A, Wiener H W, Dure L S T, Lindsey R, Hersch S M, Jope R S, et al. Cell. 1997;91:753–763. doi: 10.1016/s0092-8674(00)80464-x. [DOI] [PubMed] [Google Scholar]
- 3.Perutz M F, Johnson T, Suzuki M, Finch J T. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perutz M F. Curr Opin Struct Biol. 1996;6:848–858. doi: 10.1016/s0959-440x(96)80016-9. [DOI] [PubMed] [Google Scholar]
- 5.Stott K, Blackburn J M, Butler P J, Perutz M. Proc Natl Acad Sci USA. 1995;92:6509–6513. doi: 10.1073/pnas.92.14.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abola E E, Bernstein F C, Bryant S H, Koetzle T F, Weng J. In: Crystallographic Databases: Information Content, Software Systems, Scientific Applications. Allen F H, Bergerhoff G, Sievers R, editors. Bonn: Data Commission of the International Union of Crystallography; 1987. pp. 107–132. [Google Scholar]
- 7.Bernstein F C, Koetzle T F, Williams G J B, Meyer E E, Jr, Brice M D, Rodgers J R, Kennard O, Shimanouchi T, Tasumi M. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 8.McPhalen C A, James M N. Biochemistry. 1987;26:261–269. doi: 10.1021/bi00375a036. [DOI] [PubMed] [Google Scholar]
- 9.McPhalen C A, Evans C, Hayakawa K, Jonassen I, Svendsen I, James M N. J Mol Biol. 1983;168:445–447. doi: 10.1016/s0022-2836(83)80028-x. [DOI] [PubMed] [Google Scholar]
- 10.Leslie A G W, Brick P, Wonacott A T. Daresbury Laboratory Information Quarterly for Protein Crystallography. Vol. 18. Warrington, United Kingdom: SERC Daresbury Laboratory; 1986. pp. 33–39. [Google Scholar]
- 11.Collaborative Computational Project, No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 12.Rossmann M G. In: International Science Review Series. Klein L, editor. New York: Gordon and Breach; 1972. [Google Scholar]
- 13.Navaza J, Saludjian P. Methods Enzymol. 1997;276:581–594. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- 14.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 15.Murshudov G N, Vagin A A, Dodson E J. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 16.Jones T A, Zou J-Y, Cewan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 17.Read R J. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- 18.Lamzin V S, Wilson K S. Methods Enzymol. 1997;277:269–305. doi: 10.1016/s0076-6879(97)77016-2. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 20.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.Cowtan K. Jnt CCP 4 ESF-EACBM Newslett Protein Crystallogr. 1994;31:34–38. [Google Scholar]
- 22.Clore G M, Gronenborn A M, Kjaer M, Poulsen F M. Protein Eng. 1987;1:305–311. doi: 10.1093/protein/1.4.305. [DOI] [PubMed] [Google Scholar]
- 23.Xu D, Tsai C-J, Nussinov R. Protein Sci. 1998;7:533–544. doi: 10.1002/pro.5560070301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett M J, Schlunegger M P, Eisenberg D. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neira J L, Davis B, Ladurner A G, Buckle A M, de Prat Gay G, Fersht A R. Fold Des. 1996;1:189–208. doi: 10.1016/s1359-0278(96)00031-4. [DOI] [PubMed] [Google Scholar]
- 26.de Prat Gay G, Fersht A R. Biochemistry. 1994;33:7957–7963. doi: 10.1021/bi00191a024. [DOI] [PubMed] [Google Scholar]
- 27.Ladurner A G, Fersht A R. J Mol Biol. 1997;273:330–337. doi: 10.1006/jmbi.1997.1304. [DOI] [PubMed] [Google Scholar]
- 28.Baxendale S, Abdulla S, Elgar G, Buck D, Berks M, Micklem G, Durbin R, Bates G, Brenner S, Beck S, et al. Nat Genet. 1995;10:67–76. doi: 10.1038/ng0595-67. [DOI] [PubMed] [Google Scholar]
- 29.Carrell R W, Stein P E. Biol Chem Hoppe-Seyler. 1996;377:1–17. doi: 10.1515/bchm3.1996.377.1.1. [DOI] [PubMed] [Google Scholar]
- 30.Esnouf R M. J Mol Graph Model. 1997;15:132–136. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 31.Kraulis P. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 32.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]