Abstract
Escherichia coli isolates were obtained from common host sources of fecal pollution and characterized by using repetitive extragenic palindromic (REP) PCR fingerprinting. The genetic relationship of strains within each host group was assessed as was the relationship of strains among different host groups. Multiple isolates from a single host animal (gull, human, or dog) were found to be identical; however, in some of the animals, additional strains occurred at a lower frequency. REP PCR fingerprint patterns of isolates from sewage (n = 180), gulls (n = 133), and dairy cattle (n = 121) were diverse; within a host group, pairwise comparison similarity indices ranged from 98% to as low as 15%. A composite dendrogram of E. coli fingerprint patterns did not cluster the isolates into distinct host groups but rather produced numerous subclusters (approximately >80% similarity scores calculated with the cosine coefficient) that were nearly exclusive for a host group. Approximately 65% of the isolates analyzed were arranged into host-specific groups. Comparable results were obtained by using enterobacterial repetitive intergenic consensus PCR and pulsed-field gel electrophoresis (PFGE), where PFGE gave a higher differentiation of closely related strains than both PCR techniques. These results demonstrate that environmental studies with genetic comparisons to detect sources of E. coli contamination will require extensive isolation of strains to encompass E. coli strain diversity found in host sources of contamination. These findings will assist in the development of approaches to determine sources of fecal pollution, an effort important for protecting water resources and public health.
Sources of fecal contamination in waterways must be identified in order to adequately address water quality problems and protect public health. Source detection provides direct evidence as to the origin of pollution by identifying indicator organisms by host. Water quality indicator levels are critical parameters that drive management decisions. Studies to investigate fecal pollution sources have focused on either phenotypic or genotypic characteristics of standard water quality indicator bacteria such as fecal streptococci or Escherichia coli (1, 6, 11, 13, 21, 32). Using an indicator to assess water quality is useful because indicator organisms are easier to detect than actual pathogens; not only do the types of pathogenic bacteria, viruses, or protozoans present in contamination vary depending on the host source but pathogens are also often low in numbers and difficult to culture relative to indicator organisms (28).
One approach to source detection is to compare genetic profiles of E. coli strains isolated from contaminated water with strains collected directly from suspected sources. This approach relies on the assumption that host-specific genetic structure exists across the E. coli population (9). Early studies with multilocus enzyme electrophoresis (MLEE) to investigate the genetic structure of natural E. coli populations led to the conclusion that the E. coli population is essentially clonal in nature and experiences infrequent recombination events (12, 19, 24, 31). More recently, recombination has been found to be an important process in E. coli population genetic structure (19). The degree of clonality within the natural E. coli population and the influence of ecological niche on the selection of clonal lines have yet to be clearly defined. Host-specific environments may account for part of the observed diversity within the natural E. coli population; however, the degree to which the host influences the genetic structure of E. coli remains in question (8, 9, 26).
Several studies have employed repetitive element anchored PCR (rep-PCR), which targets repetitive extragenic palindromic (REP), enterobacterial repetitive intergenic consensus (ERIC), or BOX elements, to compare bacterial genome diversity (2, 5, 14, 17, 23). In one study of rep-PCR fingerprinting of human and animal E. coli isolates, 100% of E. coli isolates derived from cows and chickens and between 78 and 90% of human, goose, duck, pig, and sheep E. coli isolates could be correctly assigned to the correct host group by using similarity indices (6). Ribotyping, which targets the conserved 16S and 5S ribosomal DNA regions, has also proven useful for classifying E. coli according to host origin. Discriminant analysis of ribotype patterns of 238 isolates collected from human and nonhuman sources gave an 82% average rate of correct classification (ARCC) (21). Analysis of the banding patterns from these same isolates identified four major groups, all containing both human and nonhuman sources (21). Similar findings were reported in a study where the analysis of ribotype patterns of 287 isolates (collected from 118 individuals) from seven host groups resulted in accurate discrimination of human and nonhuman sources (ARCC of 97.10%), although exclusive grouping of the isolates according to host origin was not reported (1). Amplified fragment length polymorphism fingerprinting coupled with discriminant analysis produced similar results, with rates of correct classification higher than what would be expected if strains were grouped randomly (10). More information is necessary in order to determine the DNA fingerprinting approach that will provide maximum characterization of similarities and differences in the E. coli population. The diversity and relative relatedness of strains within and among different hosts will dictate the numbers of isolates from each host group needed for a representative isolate library and determine to what extent genetic characterizations can be used for E. coli source detection.
This study characterized E. coli from major host sources of fecal pollution: sewage treatment plant influent primarily from residential areas, expected to be predominantly from human sources; gulls from four Lake Michigan beaches; and cattle from four farm sites within a single watershed. Diversity of strains from different hosts was characterized by using DNA fingerprints generated by rep-PCR with REP primers. These DNA fingerprints were compared with those of other DNA fingerprinting approaches in order to evaluate the discriminatory power of each method for the detection of genetic differences in E. coli host strains.
MATERIALS AND METHODS
E. coli isolation and identification.
The host isolates were from animals found within the Milwaukee River basin in Wisconsin (approximately 850 square miles of mixed land use that ultimately drains to Lake Michigan), from two beach sites located on Lake Michigan near the basin discharge point, and two beach sites 30 miles north and south, respectively, along the coastline. Sewage isolates were obtained from raw sewage influent (flow-weighted over 24 h) provided by the Milwaukee Metropolitan Sewage District; for sample isolation, 1 ml of a 1:5 dilution was plated on m-TEC agar (Fisher Scientific, Hanover, Ill.). Gull isolates were obtained by swabbing fresh droppings found on the ground near local roosting gull colonies and inoculating 1 ml sterile saline with the swab in a 15-ml centrifuge tube. Dog fecal samples were collected by dog owners in a similar manner. Dairy cattle isolates were obtained by taking grab samples from manure storage lagoons. For animal fecal sample isolation, approximately 20 μl of fecal matter suspended in sterile water was plated on m-TEC agar. Preliminary DNA fingerprint analyses indicated that a single dominant population was present in an individual animal. Therefore, for the majority of fecal samples from individual animals, only one isolate per sample was used. Additional isolates were obtained from a subset of the fecal samples from individual animals to assess within-animal variation of strains.
E. coli isolates were identified according to the Environmental Protection Agency's original method for E. coli enumeration (29). β-Glucuronidase activity was tested by using EC growth medium containing 4-methylumbelliferyl-β-d-glucuronide (Remel, Lenexa, Kans.); isolated colonies were then confirmed for indole production by using a colorimetric spot test of p-dimethylaminocinnamaldehyde (Remel). The identification rate of E. coli with this combined protocol has been found to be 98% accurate when compared to more-extensive biochemical testing with the API 20E system (bioMerieux, Lyon, France). Approximately 25% of the isolates used in this study were tested by using the API system, and the biochemical characteristics were incorporated into cluster analyses for comparison with DNA fingerprint similarity results. E. coli isolates were grown in Luria broth medium for 18 h at 37°C while shaking at 200 rpm to provide cells for frozen cell stocks and DNA analyses. For PCR analysis, 350 μl of cells (optical density at 600 nm of 0.8 to 1.0) was washed once each with 1 M NaCl and sterile water, resuspended in 100 μl of sterile water, and stored at −20°C (22).
REP and ERIC PCR.
PCR was used to amplify repetitive elements from bacterial isolates to generate DNA fingerprint patterns. The target DNA included two families of noncoding, repetitive sequences found interspersed in bacterial genomes: REP and ERIC elements. Approximately 1 μl of washed cells provided the template for each 25-μl PCR mixture. Primers employed to generate amplified fragments included the REP1R, REP2I, ERIC1R, and ERIC2 primers (30). PCR and cycling parameters were essentially as described by Rademaker and de Bruijn (22), with a modification to the ERIC protocol with 8% dimethyl sulfoxide. Amplifications were performed for 30 cycles with 42 and 52°C annealing temperatures for REP and ERIC reactions, respectively, on a PTC-225 thermocycler (MJ Research, Waltham, Mass.). Separation of amplified genomic fragments was accomplished via gel electrophoresis by using 1% agarose gels made with 1× Tris-acetate-EDTA and run at 70 V for 16 h at 4°C. Gels were stained with 0.6 μg of ethidium bromide/ml in 1× Tris-acetate-EDTA and visualized under UV. Banding patterns were digitally captured by using an EpiChemi II Darkroom bioimaging system (UVP, Inc., Uplands, Calif.).
Validations of reaction conditions were conducted to demonstrate that the fingerprint patterns were reproducible. The initial validation experiments consisted of the comparison of REP and ERIC PCR fingerprinting of 120 isolates analyzed in three separate reactions on different days. The reproducibility of REP and ERIC patterns was further tested by duplicate analysis of >75% of the isolates used for cluster analysis (n = 334) as well as the inclusion of E. coli strain K-12 in every PCR setup as a positive control. No differences in banding patterns from the same template were seen in these analyses, verifying that fragments generated under the standard reaction conditions used in this study were consistent.
Pulsed-field gel electrophoreses (PFGE).
Isolates were analyzed according to the Centers for Disease Control and Prevention method Pulse Net protocol (4, 7). Three standard lanes with E. coli strain G5244 were included on each gel, with duplicate analysis carried out on 25% of the isolates to assure gel normalization and reproducibility. Only fragments within the range of the standard strain (573 to 48 kb) were included in the comparisons of host strains.
Computer analysis of genetic data.
Digital images of gels were entered into the genomic fingerprint analysis program Bionumerics version 2.0 (Applied Maths, Kortrijk, Belgium) and scored for banding patterns by using densitometric curve-based characterization. Band positions were normalized by using a 1-kb molecular size marker (Invitrogen, La Jolla, Calif.) for rep-PCR and E. coli strain G5244 for PFGE to correct for gel irregularities from electrophoresis and allow comparison of multiple gels. For additional standardization, subsets of samples from previous days were included in subsequent gels (5 to 10% of the total sample number). Bands greater than 12,000 bp or less than 300 bp were excluded from REP and ERIC PCR analyses since they either fell outside the coverage of the 1-kb ladder or were consistently indistinct.
Cluster analysis and group statistics.
Cluster analysis, the assignment of similar fingerprints into groups, was performed by using similarity scores that were calculated from cosine coefficients of pairwise comparisons, a curve-based method that accounts for band intensity. For fingerprint pattern comparisons, a 1.0% optimization setting was found to give the highest similarity recognition among multiple samples of strain K-12 for rep-PCR and strain G5244 for PFGE analysis while excluding nonidentical strains. The dendrogram was constructed by using the unweighted pair group method with arithmetic means (UPGMA) tree building method. The most relevant clusters in the dendrograms (within individual host sources and in a global dendrogram of all E. coli isolates in this study) were determined by calculating the similarity cutoff value that produced the highest point-bisectional correlation (Bionumerics manual, version 2.5; Applied Maths). In short, the point-bisectional correlation value is determined at various similarity thresholds by taking the number of clusters defined at that threshold value and creating a new, simplified matrix where all within-cluster values are 100% and all between-cluster values are 0%. The point-bisectional correlation is calculated by comparing values for this new matrix and the original similarity matrix. The most relevant clusters are the similarity cutoff value that offers the highest point-bisectional correlation. Jackknife analysis was used to determine how accurately isolates could be assigned to host groups based on maximum-similarity coefficients. Jackknife analysis entails manual assignment of isolates to a host group, followed by the matching of each isolate to all other isolates in the data set; the percentage of isolates that are correctly identified to the group they were originally assigned is then calculated, as is the percentage of misclassification into other groups. Dog isolates were not included in this analysis due to the small number of strains in the data set. The ARCC was calculated by determining the individual rates of correct classification for each host group and then weighting each value by the number of isolates analyzed from each of the groups.
RESULTS
Computer analysis of DNA fingerprint patterns.
REP primers were used to generate PCR fingerprints for E. coli isolates obtained from major sources of fecal pollution: sewage treatment plant influent from residential areas, samples from cattle feedlot detention systems, and fecal samples from individual humans, gulls, and dogs (Table 1). Reactions with REP primers generated between 13 and 22 amplification products that ranged in size from 300 bp to 6 kb; ERIC primers generated 7 to 13 amplification products in this same size range. PFGE analysis produced restriction fragments ranging from approximately 25 to 600 kb. K-12 fingerprints (n = 47), generated on different days and resolved on separate gels, resulted in similarity scores that ranged from 87.9 to 99.5%. Host isolates with one to two band differences on visual inspection generated similarity indices ranging from 86.7 to 95%, which overlaps the range found for identical isolates; therefore, strains were not designated as identical without manual examination of the fingerprint patterns.
TABLE 1.
E. coli isolates collected from host sources in the Milwaukee River basin
| Host source | No. of isolates used in dendrogram constructiona | No. of samples | Collection time period | Total no. of isolates used to assess within-host variationb (no. of animals) | Total no. of isolates in study |
|---|---|---|---|---|---|
| Sewage treatment plant influent | 180 | 17 (24 hr) flow-weighted samples | 18 months | NA | 180 |
| Human | 3 | 3 individuals | Single collection day | 40 (3) | 40 |
| Dog | 3 | 3 individuals | Single collection day | 40 (3) | 40 |
| Gull | 133 | 4 beach sites | 21 days over 14 mo. | 210 (15) | 328 |
| Cow | 121 | 4 farm sites | 2 collection days per site | NA | 121 |
| Total | 440 | 290 (21) | 709 |
One isolate per individual animal was used for dendrogram construction; sewage and cattle feedlot samples were representative samples of a large number of humans or animals.
Number of individual animals that were sampled is shown in parentheses. NA, not applicable.
Strain diversity within a single animal.
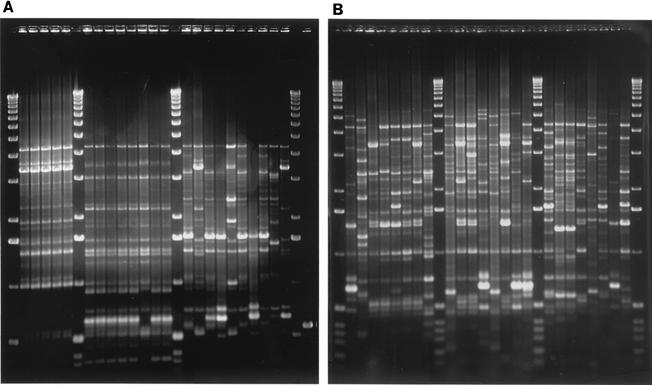
The ability to characterize the E. coli population within a host group requires accurate representation of strain frequency in the population. Multiple strains from a single animal (gull, human, or dog) were analyzed to determine the extent of diversity within a single animal. E. coli isolates that were collected from a single gull fecal sample gave identical REP fingerprint patterns for the majority of isolates that were analyzed (Table 2; Fig. 1A). Additional strains were present in most samples but at a lower frequency. Similar results were obtained with isolates obtained from human and dog samples. In some cases, strains isolated from the same animal were found to have REP patterns similar to the predominate strain, which may indicate the presence of related populations with common parentage. However, most low-frequency isolates produced patterns unrelated to the predominate population (<60% pairwise similarity indices). A subset of isolates from five of the gull samples and one of the human samples was further analyzed with PFGE. The isolates that produced identical REP patterns (gull samples, n = 40; human samples, n = 10) were also genetically indistinguishable by PFGE, with the exception of one gull sample, where two of the eight isolates demonstrated a single band difference. In contrast, a large amount of variation was found among fingerprint patterns of E. coli isolates collected from different gull fecal samples (representative fingerprint patterns are shown in Fig. 1B). Of 133 gull isolates, each collected from individual fecal samples, only nine pairs and one group of three isolates produced identical patterns. Isolates from sewage treatment plant influent, which are expected to provide a broad representation of the E. coli strains in humans, also demonstrated a low occurrence of identical isolates; of 180 isolates, 11 pairs and four groups of 3 to 5 isolates with identical REP PCR patterns were found. However, almost half of the identical isolate groups were obtained from sewage treatment plant influent samples collected on the same day, which may indicate poor mixing of sewage influent samples. The cattle isolates demonstrated the highest amount of homogeneity among strains; six groups of 3 to 10 identical isolates accounted for 35% of the strains that were analyzed. Identical strains occurred across different farm sites as well as within single farms.
TABLE 2.
Distribution of strains isolated from a single animal among the total number of patterns found in that animal
| Host (n)a | Total no. of patterns | Distribution of isolates (no.) among patterns
|
% Similarity between patternsc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MPPb | OP | OP | OP | OP | OP | OP | OP | |||
| Gull (8) | 2 | 7 | 1 | 79.1, 1st and 2nd pattern | ||||||
| Gull (10) | 4 | 5 | 2 | 2 | 1 | |||||
| Gull (11) | 1 | 11 | ||||||||
| Gull (13) | 3 | 9 | 4 | 1 | ||||||
| Gull (14) | 3 | 11 | 2 | 1 | ||||||
| Gull (14) | 7 | 8 | 1 | 1 | 1 | 1 | 1 | 1 | 70.1, 1st, 2nd, and 3rd patterns | |
| Gull (15) | 1 | 15 | ||||||||
| Gull (15) | 1 | 15 | ||||||||
| Gull (15) | 3 | 8 | 3 | 4 | 62.7, 1st and 3rd patterns | |||||
| Gull (15) | 5 | 6 | 5 | 2 | 1 | 1 | 59.0, 2nd and 3rd patterns | |||
| Gull (15) | 6 | 3 | 3 | 3 | 3 | 2 | 1 | |||
| Gull (16) | 3 | 8 | 4 | 4 | ||||||
| Gull (16) | 3 | 11 | 4 | 1 | ||||||
| Gull (16) | 9d | 7 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Gull (17) | 17d | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 86.2, 1st and 2nd 82.7, 3rd and 4th patterns; 81.4, 5th and 6th patterns |
| Human (10) | 1 | 10 | ||||||||
| Human (15) | 1 | 15 | ||||||||
| Human (15) | 1 | 15 | ||||||||
| Dog (10) | 1 | 10 | ||||||||
| Dog (15) | 4 | 8 | 4 | 2 | 1 | 85, 3rd and 4th patterns | ||||
| Dog (15) | 1 | 15 | ||||||||
Number of isolates analyzed from the same fecal sample.
MPP designates the most-predominate pattern found in an animal; the most-predominate pattern was different for every animal that was tested. OP designates other patterns that were found in isolates from a single animal.
Similarity scores were determined by computer comparison of REP PCR fingerprints based on cosine coefficients.
Additional patterns containing a single isolate are not shown in the table.
FIG. 1.
rep-PCR fingerprints generated from REP primers. (A) Isolates collected from three gull fecal samples. Lanes 2 to 6, 8 to 15, and 17 to 26 contain isolates from one of the three samples. Lanes 1, 7, 16, and 27 contain a 1-kb molecular size marker. (B) Single isolates collected from individual gull fecal samples, where one isolate per sample was analyzed. Lanes 2 to 9, 11 to 18, and 20 to 27, gull isolates. Lanes 1, 10, 19, and 28 contain a 1-kb molecular size marker.
Clustering of REP fingerprints in dendrograms.
To evaluate the strain diversity of E. coli within a host group, dendrograms of REP fingerprints were constructed by using the UPGMA method of tree building and significant clusters in each host-specific dendrogram were identified. Pairwise comparison of REP fingerprints of isolates from a single host group generated similarity scores ranging from 15.0 to 98.0% for the group of sewage isolates, 16.4 to 96.8% for gull isolates, and 14.1 to 98.0% for cattle isolates. The E. coli population isolated from sewage samples was divided into five clusters, the population isolated from gulls was divided into five clusters, and the population isolated from cattle was divided into three clusters. Subclusters with high similarity scores (>80% similarity) were found more frequently in the group of sewage isolates than in the group of gull isolates.
A composite dendrogram including all host strains demonstrated subclusters of closely related strains; the majority of these with a similarity index above 80% were exclusive for a host group (Table 3). However, the overall arrangement of the subclusters across the dendrogram was not by host group but was intermixed. Of the 440 isolates analyzed, one-third were <65% similar to other isolates. Clusters defined by >65% similarity contained isolates from more than one source, whereas subclusters defined at approximately 80% similarity produced good differentiation among host sources. Some exceptions were found, such as a single gull isolate that demonstrated high similarity to groups of sewage strains. Crossover (>80% similarity of isolates from different host groups) accounted for less than 10% of the strains analyzed.
TABLE 3.
Groupings of E. coli isolates into clusters by using REP PCR fingerprints
| Major clustera (total no. of isolates) | Subclusterb (>65% similarity) | Host group (no. of isolates) | Results for host-specific groups in each subclusterc
|
||
|---|---|---|---|---|---|
| % Similarity of groups | Major host (no. of isolates) | Minor host (no. of isolates) | |||
| I (15) | I A | Sewd (13), gull (2) | 88.7 | Gull (2) | |
| 90.7 | Sew (7) | ||||
| 74.0 | Sew (8) | ||||
| II (256) | II A | Sew (1), gull (3) | 92.8 | Gull (3) | |
| II B | Sew (9) | 74.3 | Sew (9) | ||
| 80.1 | Cow (9) | Sew (3) | |||
| II C | Sew (13), gull (7), cow (14) | 86.0 | Gull (4) | ||
| 73.6 | Sew (5) | ||||
| II D | Sew (1), gull (4) | 78.5 | Gull (3) | ||
| 95.1 | Sew (6) | ||||
| 94.4 | Cow (3) | ||||
| II E | Sew (27), gull (11), cow (8) | 82.4 | Sew (5) | ||
| 94.8 | Gull (2) | ||||
| 77.3 | Sew (6) | Gull (1) | |||
| II F | Gull (3) | 72.1 | Gull (3) | ||
| 87.9 | Gull (3) | ||||
| II G | Sew (2), gull (5) | 87.9 | Gull (3) | ||
| II H | Sew (3), gull (1) | 88.7 | Sew (2) | ||
| II I | Sew (5), gull (2) | 88.9 | Sew (4) | ||
| II J | Cow (3) | 71.5 | Cow (3) | ||
| 91.0 | Sew (5) | ||||
| II K | Sew (5), gull (7) | 91.6 | Gull (6) | ||
| II L | Sew (4), gull (4), cow (26) | 77.3 | Cow (13) | ||
| 84.2 | Cow (12) | Sew (3), gull (3) | |||
| II M | Sew (5) | 74.2 | Sew (5) | ||
| II N | Cow (3) | 68.7 | Cow (3) | ||
| II O | Cow (3) | 97.5 | Cow (3) | ||
| II P | Sew (1), gull (2), cow (20) | 74.2 | Cow (4) | Gull (1) | |
| 76.8 | Cow (16) | Sew (1), gull (1) | |||
| II Q | Sew (3), gull (5) | 74.0 | Gull (3) | ||
| II R | Sew (9), gull (2), cow (3) | 91.4 | Sew (8) | ||
| 97.2 | Cow (2) | ||||
| II Q | Gull (5), cow (2) | 77.9 | Gull (4) | Cow (1) | |
| III (9) | III A | Gull (4) | 87.6 | Gull (4) | |
| 90.3 | Sew (23) | Gull (6) | |||
| 92.1 | Sew (5) | ||||
| 96.4 | Cow (5) | ||||
| 94.0 | Gull (3) | ||||
| 90.7 | Cow (3) | Gull (1) | |||
| IV A | Sew (47), gull (18), cow (18) | 81.3 | Sew (4) | ||
| 81.3 | Gull (2) | ||||
| 86.3 | Gull (2) | ||||
| 96.8 | Cow (9) | ||||
| IV B | Sew (1), gull (2) | 79.3 | Gull (2) | ||
| IV (157) | IV C | Sew (4), gull (2) | 97.4 | Sew (4) | |
| 87.4 | Gull (4) | ||||
| IV D | Sew (4), gull (10) | 94.4 | Sew (2) | ||
| 72.3 | Gull (3) | ||||
| IV E | Sew (4) | 78.1 | Sew (4) | ||
| IV F | Sew (6), cow (14) | 83.9 | Cow (10) | ||
| 78.1 | Sew (5) | Cow (1) | |||
| IV G | Sew (14), gull (2), cow (2) | 72.9 | Sew (14) | Cow (2) | |
| IV H | Sew (1), gull (3) | 93.3 | Gull (3) | ||
Major clusters were identified by using the cluster cutoff method (see methods); the cutoff values for significant clusters were 33.0, 39.0, 43.7, and 38.9% similarity indices for clusters I, II, III, and IV, respectively. Three isolates did not fall into a significant cluster and are not shown in the table.
Subclusters with three or more isolates with >65% similarity indices are listed. Remaining isolates were either single isolates or pairs of isolates or isolates with <65% similarity to the other isolates in the dendrogram.
The host-specific group was defined as a group of isolates that contained a minimum of 75% of the members from a single host group.
Sew, sewage.
Group statistics.
Jackknife analysis was used to evaluate how accurately similarity coefficients were able to predict host group. The ARCC for the isolates used in dendrogram construction was found to be 79.3%. Gull strains were most often misidentified. The correct classification rate of gull isolates into the gull group was 66.0%, whereas their misclassification as members of the sewage group was 29.0% and their misclassification as members of the cattle group was 5.0%. Sewage isolates were correctly classified at a rate of 83.2%, with misclassification as members of the gull group at 3.3% and misclassification as cattle isolates at 3.5%. Cattle isolates had the highest correct classification rate at 88.2%, with misclassification as belonging to either sewage or gull groups at 5.9%. The high correct classification rate for cattle may be attributed to, in part, the small number of farms sampled.
Comparison of REP PCR with ERIC PCR and PFGE.
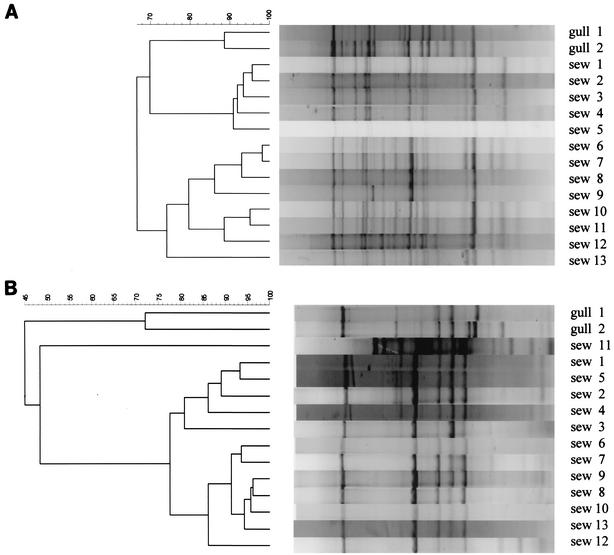
For all of the host groups, REP PCR fingerprinting produced highly similar patterns (65.0 to 99.5% indices) among small groups of isolates. Further analysis was performed on a subset of 101 isolates by using two other fingerprinting techniques, rep-PCR with ERIC primers and PFGE, to compare the relative discriminatory power of each technique. In addition, biochemical profiles were determined. REP and ERIC PCR fingerprints demonstrated various patterns among groups of closely related isolates (Fig. 2). In these instances, cluster analysis did not recover the same arrangement but produced comparable overall cluster similarity indices. Approximately 40% of the strains that were identical by REP PCR were also identical by ERIC PCR and PFGE. In a few instances, strains indistinguishable by REP and ERIC PCR had small genetic differences that were detected by PFGE (e.g., one or two band differences). Notably, API biochemical profiles did not correlate with similarity coefficients; identical API profiles were found in strains with low similarity coefficients, and likewise, strains with identical REP, ERIC, and PFGE fingerprint patterns could be differentiated with API profiles.
FIG. 2.
DNA fingerprints of closely related E. coli strains with REP PCR (A) and ERIC PCR (B). Pairwise similarity scores were obtained by calculating the cosine coefficient, and dendrograms were constructed by using the UPGMA method of tree building. sew, sewage.
DISCUSSION
Accurate representation of the E. coli population from a host group requires collecting isolates that broadly represent the host group of interest. Results from this study indicate that a single animal, e.g., gull or human, generally harbors one predominant strain of E. coli. Although temporal changes within a single animal have been noted, overall temporal changes in strain composition across the greater E. coli population are unexplored (3, 18). In this study, groups of isolates with one or two band differences by REP PCR or PFGE were isolated from an individual human or gull, which suggests that subpopulations may evolve and cooccur in a single animal (20, 27). There is also some evidence of stability in the greater population, as seen by repeated isolation of identical strains from unassociated hosts in this study and past studies (25). For example, sewage and gull isolates with identical fingerprint patterns were recovered from each respective sample type on different days over a 1-year period.
When assessing the genetic composition of an E. coli population by using host strain libraries, including multiple isolates from the same animal in analyses is problematic because these isolates may (i) inflate the estimated frequency of a particular strain in the overall population and (ii) bias assessments of the host strain library (e.g., jackknife analysis) because they will self-identify rather than reflect true genetic similarities across the greater E. coli population. These findings are important to consider when designing sampling strategies that will be representative of strain frequencies in the E. coli population.
Extensive genetic diversity was found within each host group; fingerprint patterns demonstrated fewer than 20% shared bands between two isolates by REP PCR. When all strains were compared, they did not divide into distinct groups according to host source, but rather into multiple closely related subclusters that appeared host-specific, which is consistent with the findings of other studies (1, 6). Jackknife analysis gave correct classification rates for host groups that were higher than what would be expected with random groupings of isolates. While several host-specific subclusters were observed, approximately one-third of strains were <65% similar to any other strain. This suggests that the number of strains analyzed represented only partial coverage of the E. coli population in the hosts that were assessed. The most extensive studies to investigate population structure in the natural E. coli population have been carried out with MLEE; however, these studies have resulted in various conclusions as to the host specificity of E. coli strains (8, 26). Given the large amount of diversity reported in past studies of population structure, discrepant findings among genetic studies with MLEE may be a result of differences in the sampling of the population, where isolate collections represent different aspects of the whole population (V. Souza, M. Rocha, A. Valera, L. E. Eguiarte, Letter, Appl. Environ. Microbiol. 66:5104-5105, 2000). rep-PCR characterizations correlate with MLEE findings (15), and therefore, it can be expected that similar challenges will arise in the development of rep-PCR approaches to characterize E. coli populations for the purpose of identifying host sources of pollution.
Of the fingerprinting approaches used in this study, PFGE gave the highest discrimination among isolates and may be useful for investigating temporal changes in a resident population or confirming the presence of clonal populations. However, since PFGE can detect single base pair changes, PFGE fingerprint patterns were highly diverse and had few common fragments that could be used for pattern comparisons, whereas both REP and ERIC PCR detected similarities as well as differences in the majority of strains that were analyzed. In addition, PCR-based methods may be more useful for large datasets given the capabilities for high-throughput analyses and lower cost relative to other DNA fingerprint methods (15, 22). The REP and ERIC primers produced comparable, but not identical, results in dendrogram groupings. Similar findings were reported with REP and ERIC PCR fingerprint analyses of Bradyrhizobium japonicum, where one set of primers detected differences not found with a second set of primers (16). This suggests that strain characterizations are not dependent on primer selection in rep-PCR with REP or ERIC primers, although BOX primers were not evaluated. Composite fingerprint data sets may produce higher discrimination between closely related strains; however, this resolution may not be necessary since adequate discrimination can be achieved with a single-primer approach.
DNA fingerprinting that targets repetitive elements in PCR may be useful in applications to determine sources of fecal pollution if a representative database of host strains can be achieved. Given the high amount of strain diversity that was found in this study, E. coli characterization may be most feasible within a limited geographic area, e.g., for watershed-specific studies. Application of this approach to the identification of host sources of E. coli in the environment requires more extensive information on the overall genetic structure of the natural populations, particularly the host specificity of strains, as well as net temporal changes in the population and geographical differences in strain occurrence.
Acknowledgments
This work was funded by the Milwaukee Metropolitan Sewage District (contract no. M003002P11) and the Wisconsin Department of Natural Resources.
We thank Magnolia Tulod, Joshua Harris, Emerson Lee, and Jennifer Lee for technical support. We also thank the Racine Health Department for providing E. coli isolates and gull fecal samples and Brian Kinkle for critical review of the manuscript.
Footnotes
†Great Lakes WATER Institute contribution number 436.
REFERENCES
- 1. Carson, C. A., B. L Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvalho de Moura, A. C., K. Irino, and M. C. Vidotto. 2001. Genetic variability of avian Escherichia coli strains evaluated by enterobacterial repetitive intergenic consensus and repetitive extragenic palindromic polymerase chain reaction. Avian Dis. 45:173-181. [PubMed] [Google Scholar]
- 3.Caugant, D. A., B. R. Levin, and R. K. Selander. 1981. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics 98:467-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1999. Division of Bacterial and Mycotic Diseases, Pulse Net: The National Molecular Subtyping Network of Foodborne Disease Surveillance. One-day (24-48 h) standardized laboratory protocol for molecular subtyping Escherichia coli O157:H7 by pulse field gel electrophoresis (PFGE). Centers for Disease Control and Prevention, Atlanta, Ga.
- 5.Dalla-Costa, L. M., K. Irino, J. Rodrigues, I. N. Rivera, and L. R. Trabulsi. 1998. Characterisation of diarrhoeagenic Escherichia coli clones by ribotyping and ERIC-PCR. J. Med. Microbiol. 47:227-234. [DOI] [PubMed] [Google Scholar]
- 6.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the polymerase chain reaction to differentiate Escherichia coli from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon, D. M., and J. Lee. 1999. The genetic structure of enteric bacteria from Australian mammals. Microbiology 145:2673-2682. [DOI] [PubMed] [Google Scholar]
- 9.Gordon, D. M. 2001. Geographic structure and host specificity in bacteria and implications for tracing the source of coliforms contamination. Microbiology 147:1079-1085. [DOI] [PubMed] [Google Scholar]
- 10.Guan, S., R. Xu, S. Chen, J. Odumeru, and C. Gyles. 2002. Development of a procedure for discriminating among Escherichia coli isolates from animal and human sources. Appl. Environ. Microbiol. 68:2690-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagedorn, C., S. L. Robinson, J. R. Filtz, S. M. Grubbs, T. A. Angier, and R. B. Reneau, Jr. 1999. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 65:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartl, D. L., and D. E. Dykhuizen. 1984. The population genetics of Escherichia coli. Annu. Rev. Genet. 18:31-68. [DOI] [PubMed] [Google Scholar]
- 13.Harwood, V. J., J. Whitlock, and V. Withington. 2000. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, J. R., J. J. Brown, U. B. Carlino, and T. A. Russo. 1998. Colonization with and acquisition of uropathogenic Escherichia coli as revealed by polymerase chain reaction-based detection. J. Infect. Dis. 177:1120-1124. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, J. R., and T. T. O'Bryan. 2000. Improved repetitive-element PCR fingerprinting for resolving pathogenic and nonpathogenic phylogenetic groups within Escherichia coli. Clin. Diagn. Lab. Immunol. 7:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Judd, A. K., M. Schneider, M. J. Sadowsky, and F. J. de Bruijn. 1993. Use of repetitive sequences and the polymerase chain reaction technique to classify genetically related Bradyrhizobium japonicum serocluster 123 strains. Appl. Environ. Microbiol. 59:1702-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koeuth, T., J. Versalovic, and J. R. Lupski. 1995. Differential subsequence conservation of interspersed repetitive Streptococcus pneumoniae BOX elements in diverse bacteria. Genome Res. 5:408-418. [DOI] [PubMed] [Google Scholar]
- 18.Mason, T. G., and G. Richardson. 1981. Escherichia coli and the human gut: some ecological considerations. J. Appl. Bacteriol. 51:1-16. [DOI] [PubMed] [Google Scholar]
- 19.Milkman, R. 1997. Recombination and population structure in Escherichia coli. Genetics 146:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olive, D. M., and P. Bean. 1999. Principals and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parveen, S., K. M. Portier, K. Robinson, L. Edmiston, M. L. Tamplin. 1999. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 65:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rademaker, J. L. W., and F. J. de Bruijn. 1997. Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer-assisted pattern analysis, p. 151-171. In G. Gaetano-Anolles and P. M. Gresshoff (ed.), DNA markers: protocols, applications, and overviews. J. Wiley and Sons, New York, N.Y.
- 23.Sadowsky, M. J., and H. Hur. 1998. Use of endogenous repeated sequences to fingerprint bacterial genomic DNA, p.398-413. In F. J. de Bruijn and J. R. Lupski (ed.), Bacterial genomes: physical structure and analysis. Chapman and Hall, New York, N.Y.
- 24.Selander, R. K., and B. R. Levin. 1980. Genetic diversity and structure in Escherichia coli populations. Science 210:545-547. [DOI] [PubMed] [Google Scholar]
- 25.Selander, R. K., D. A. Caught, and T. S. Whittam. 1987. Genetic structure and variation in natural populations of Escherichia coli, p. 1625-1648. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, D.C.
- 26.Souza, V., M. Rocha, A. Valera, and L. E. Eguiarte. 1999. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 65:3373-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toranzos, G. A., and G. A. McFeters. 1997. Detection of indicator microorganisms in environmental fresh waters and drinking waters, p. 184-194. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. American Society for Microbiology, Washington, D.C.
- 29.U.S. Environmental Protection Agency. 2000. Improved enumeration methods for recreational water quality indicators: enterococci and Escherichia coli. EPA/821/R-97/004. Office of Science and Technology, Washington, D.C.
- 30.Versalvoic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whittam, T. S., H. Ochman, and R. K. Selander. 1983. Multilocus genetic structure in natural populations of Escherichia coli. Proc. Natl. Acad. Sci. USA 80:1751-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiggins, B. A., R. W. Andrews, R. A. Conway, C. L. Corr, E. J. Dobratz, D. P. Dougherty, J. R. Eppard, S. R. Knupp, M. C. Limjoco, J. M. Mettenburg, J. M. Rinehardt, J. Sonsino, R. L. Torrijos, and M. E. Zimmerman. 1999. Use of antibiotic resistance analysis to identify nonpoint sources of fecal pollution. Appl. Environ. Microbiol. 65:3483-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]