Abstract
The discrimination between perfect-match and single-base-pair-mismatched nucleic acid duplexes was investigated by using oligonucleotide DNA microarrays and nonequilibrium dissociation rates (melting profiles). DNA and RNA versions of two synthetic targets corresponding to the 16S rRNA sequences of Staphylococcus epidermidis (38 nucleotides) and Nitrosomonas eutropha (39 nucleotides) were hybridized to perfect-match probes (18-mer and 19-mer) and to a set of probes having all possible single-base-pair mismatches. The melting profiles of all probe-target duplexes were determined in parallel by using an imposed temperature step gradient. We derived an optimum wash temperature for each probe and target by using a simple formula to calculate a discrimination index for each temperature of the step gradient. This optimum corresponded to the output of an independent analysis using a customized neural network program. These results together provide an experimental and analytical framework for optimizing mismatch discrimination among all probes on a DNA microarray.
DNA microarray technology provides parallel nucleic acid hybridizations for a large number of immobilized oligonucleotides or larger DNA fragments on a small surface area (21). In clinical and environmental microbiology, this technology has been used for assessing gene expression (19), characterizing whole genomes (5), identifying bacteria (8, 10, 28), and monitoring microbial populations (12, 22). We anticipate that, in the next several years, the application of DNA microarrays to environmental microbiology will greatly improve the understanding of complex microbial communities, which are typically composed of many microbial species.
In general, oligonucleotide DNA microarrays containing 15- to 25-mer oligonucleotide probes provide greater discrimination than microarrays composed of larger PCR-amplified DNA fragments. However, a central challenge to the application of DNA microarrays in environmental microbiology is achieving the specificity needed to resolve complex microbial populations, including discriminating between target and nontarget populations that differ by a single nucleotide (10). This level of specificity is needed to resolve variants of highly conserved genes (e.g., those encoding the rRNAs) and to distinguish between closely related target and nontarget microorganisms.
In conventional hybridization assays, single-base-pair discrimination is achieved by adjusting the hybridization conditions (e.g., temperature, ionic strength, or formamide concentration) or washing conditions (dissociation) of the probe-target duplex (31). In DNA microarray assays, however, this approach is difficult to use since one set of hybridization and wash conditions does not provide optimal target discrimination for all probes on the microarray. We therefore have developed an alternative approach that uses differences in thermal dissociation rates of probe-target duplexes to resolve matched and mismatched probe-target duplexes (13, 25).
The oligonucleotide DNA microarray used in this study is a variant of the more conventional format (15, 22, 29). Rather than being directly attached to glass, the probes are immobilized in three-dimensional polyacrylamide gel pads affixed to the glass (2, 6, 9, 10, 12, 13, 18, 24-26, 30). The gel pads provide a format suitable for the determination of equilibrium and nonequilibrium dissociation kinetics (i.e., melting profiles) of a large number of probe-target duplexes and for determining the dissociation temperature (Td), the temperature at which 50% of the duplexes remain during a specified wash period (13, 25). In this study, we used nonequilibrium dissociation kinetics to derive the optimum washing temperature for each probe, providing for maximum discrimination between target RNA or target DNA and all possible single-nucleotide-mismatch variants.
MATERIALS AND METHODS
Synthetic DNA and RNA targets.
The 16S rDNA gene sequences of Staphylococcus epidermidis (accession no. L37605 and X75943) and Nitrosomonas eutropha (accession no. M96402) were obtained from GenBank in the National Center for Biotechnology Information. Single-strand complements to each probe, containing an additional 10 nucleotides of nontarget flanking sequence at the 5′ and 3′ termini, were synthesized in DNA (Operon Technologies Inc., Alameda, Calif.) and RNA (Dharmacon Research Inc., Lafayette, Colo.) forms to avoid possible biases resulting from sample preparation using native rRNA (e.g., possible variability in the efficiency of fragmentation and labeling) (2). The target molecules were fluorescently labeled with Cy3 at the 5′ terminus. The name of target, sequence, size, location of the sequence in the 16S rRNA gene (Escherichia coli numbering), and probe binding site (underlined) are as follows: Staphylococcus target, 5′-TCTGGTCTGTAACTGACGCTGATGTGCGAAAGCGTGGGG-3′, 39-mer, position 737 to 775; Nitrosomonas target, 5′-ACTACAAAGCTAGAGTGCAGCAGAGGGGAGTGGAATTC-3′, 38-mer, position 643 to 680.
Oligonucleotide probe design and synthesis.
A 19-mer oligonucleotide probe (S-G-Staphy-0747-a-A-19) targeting Staphylococcus 16S rRNA was designed as described previously (25). An 18-mer oligonucleotide probe (S-*-Nsom-0653-a-A-18) targeting halotolerant and obligate halophilic Nitrosomonas (27) was used for the Nitrosomonas target. These probes were complemented by a set of probes having all possible single-mismatch variants at each position (Table 1). Probes having two to five mismatches were also incorporated on the microarray. All probes were synthesized with an amino linker at the 3′ terminus as described previously (26).
TABLE 1.
Oligonucleotide probes used in this study and their corresponding Tds
| Series | Probea | Sequencee | DNA
|
RNA
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Td Mean | SD | ΔTb | CVc | Td Mean | SD | ΔT | CV | |||
| S0 | spm | TCGCACATCAGCGTCAGTT | 45.1 | ±1.2 | 0.0 | 2.7 | 44.8 | ±1.8 | 0.0 | 4.0 |
| S1 | s1aa | ACGCACATCAGCGTCAGTT | 41.4 | ±1.5 | −3.7 | 3.7 | 40.0 | ±1.6 | −4.8 | 4.0 |
| S2 | s1ga | GCGCACATCAGCGTCAGTT | 43.7 | ±1.5 | −1.4 | 3.3 | 42.0 | ±2.5 | −2.8 | 5.9 |
| S3 | s1ca | CCGCACATCAGCGTCAGTT | 40.8 | ±1.0 | −4.3 | 2.3 | 40.4 | ±2.1 | −4.4 | 5.1 |
| S4 | s2ag | TAGCACATCAGCGTCAGTT | 43.1 | ±1.4 | −2.0 | 3.3 | 41.3 | ±2.5 | −3.5 | 5.9 |
| S5 | s2gg | TGGCACATCAGCGTCAGTT | 42.7 | ±0.9 | −2.4 | 2.1 | 40.7 | ±1.8 | −4.1 | 4.3 |
| S6 | s2tg | TTGCACATCAGCGTCAGTT | 41.7 | ±1.0 | −3.4 | 2.5 | 40.7 | ±1.2 | −4.1 | 2.9 |
| S7 | s3cc | TCCCACATCAGCGTCAGTT | 37.8 | ±1.0 | −7.3 | 2.6 | 35.6 | ±0.6 | −9.2 | 1.7 |
| S8 | s3tc | TCTCACATCAGCGTCAGTT | 39.1 | ±0.9 | −6.0 | 2.4 | 37.6 | ±0.8 | −7.2 | 2.2 |
| S9 | s3ac | TCACACATCAGCGTCAGTT | 41.5 | ±0.6 | −3.6 | 1.5 | 38.5 | ±1.1 | −6.3 | 3.0 |
| S10 | s4gg | TCGGACATCAGCGTCAGTT | 41.6 | ±0.6 | −3.5 | 1.3 | 38.4 | ±1.5 | −6.4 | 4.0 |
| S11 | s4tg | TCGTACATCAGCGTCAGTT | 41.0 | ±0.9 | −4.1 | 2.2 | 39.4 | ±1.6 | −5.4 | 4.0 |
| S12 | s4ag | TCGAACATCAGCGTCAGTT | 40.8 | ±0.5 | −4.3 | 1.1 | 36.7 | ±1.2 | −8.1 | 3.2 |
| S13 | s5gt | TCGCGCATCAGCGTCAGTT | 43.2 | ±1.3 | −1.9 | 3.0 | 40.2 | ±1.0 | −4.6 | 2.6 |
| S14 | s5ct | TCGCCCATCAGCGTCAGTT | 39.6 | ±1.0 | −5.5 | 2.5 | 38.0 | ±1.3 | −6.8 | 3.3 |
| S15 | s5tt | TCGCTCATCAGCGTCAGTT | 39.8 | ±2.0 | −5.3 | 4.9 | 39.1 | ±0.9 | −5.7 | 2.2 |
| S16 | s6tg | TCGCATATCAGCGTCAGTT | 41.0 | ±0.6 | −4.1 | 1.5 | 40.1 | ±1.0 | −4.7 | 2.4 |
| S17 | s6ag | TCGCAAATCAGCGTCAGTT | 39.4 | ±0.9 | −5.7 | 2.2 | 37.2 | ±0.8 | −7.6 | 2.1 |
| S18 | s6gg | TCGCAGATCAGCGTCAGTT | 40.2 | ±0.9 | −4.9 | 2.3 | 36.1 | ±1.4 | −8.7 | 3.7 |
| S19 | s7gt | TCGCACGTCAGCGTCAGTT | 35.4 | ±0.4 | −9.7 | 1.1 | 36.0 | ±1.8 | −8.8 | 5.0 |
| S20 | s7ct | TCGCACCTCAGCGTCAGTT | 39.4 | ±0.8 | −5.7 | 2.0 | 36.8 | ±1.3 | −8.0 | 3.5 |
| S21 | s7tt | TCGCACTTCAGCGTCAGTT | 41.3 | ±0.8 | −3.8 | 2.0 | 38.5 | ±1.3 | −6.3 | 3.3 |
| S22 | s8aa | TCGCACAACAGCGTCAGTT | 41.6 | ±1.0 | −3.5 | 2.3 | 37.9 | ±0.8 | −6.9 | 2.0 |
| S23 | s8ga | TCGCACAGCAGCGTCAGTT | 43.7 | ±1.3 | −1.4 | 3.0 | 38.8 | ±1.1 | −6.0 | 2.8 |
| S24 | s8ca | TCGCACACCAGCGTCAGTT | 41.1 | ±0.9 | −4.0 | 2.1 | 38.5 | ±1.1 | −6.3 | 2.8 |
| S25 | s9tg | TCGCACATTAGCGTCAGTT | 41.1 | ±0.9 | −4.0 | 2.2 | 39.4 | ±1.1 | −5.4 | 2.8 |
| S26 | s9ag | TCGCACATAAGCGTCAGTT | 40.2 | ±0.9 | −4.9 | 2.3 | 35.6 | ±2.1 | −9.2 | 5.8 |
| S27 | s9gg | TCGCACATGAGCGTCAGTT | 39.6 | ±0.8 | −5.5 | 2.0 | 36.3 | ±1.5 | −8.5 | 4.2 |
| S28 | s10gt | TCGCACATCGGCGTCAGTT | 43.1 | ±1.2 | −2.0 | 2.7 | 39.9 | ±1.7 | −4.9 | 4.3 |
| S29 | s10ct | TCGCACATCCGCGTCAGTT | 40.6 | ±0.9 | −4.5 | 2.2 | 38.0 | ±1.3 | −6.8 | 3.3 |
| S30 | s10tt | TCGCACATCTGCGTCAGTT | 42.0 | ±1.1 | −3.1 | 2.5 | 38.8 | ±0.9 | −6.0 | 2.2 |
| S31 | s11cc | TCGCACATCACCGTCAGTT | 37.0 | ±0.7 | −8.1 | 1.8 | 35.5 | ±0.2 | −9.3 | 0.6 |
| S32 | s11tc | TCGCACATCATCGTCAGTT | 38.6 | ±0.9 | −6.5 | 2.3 | 36.0 | ±0.6 | −8.8 | 1.6 |
| S33 | s11ac | TCGCACATCAACGTCAGTT | 37.8 | ±0.4 | −7.3 | 1.1 | 34.5 | ±0.5 | −10.3 | 1.5 |
| S34 | s12tg | TCGCACATCAGTGTCAGTT | 41.9 | ±1.1 | −3.2 | 2.5 | 40.1 | ±1.2 | −4.7 | 3.0 |
| S35 | s12ag | TCGCACATCAGAGTCAGTT | 40.6 | ±0.9 | −4.5 | 2.3 | 36.3 | ±1.2 | −8.5 | 3.3 |
| S36 | s12gg | TCGCACATCAGGGTCAGTT | 41.5 | ±0.8 | −3.6 | 2.0 | 38.1 | ±0.8 | −6.7 | 2.1 |
| S37 | s13cc | TCGCACATCAGCCTCAGTT | 37.1 | ±0.7 | −8.0 | 1.9 | 35.8 | ±0.6 | −9.0 | 1.5 |
| S38 | s13tc | TCGCACATCAGCTTCAGTT | 38.5 | ±0.5 | −6.6 | 1.4 | 36.3 | ±0.6 | −8.5 | 1.7 |
| S39 | s13ac | TCGCACATCAGCATCAGTT | 38.9 | ±1.3 | −6.2 | 3.2 | 38.0 | ±0.6 | −6.8 | 1.6 |
| S40 | s14aa | TCGCACATCAGCGACAGTT | 42.1 | ±0.8 | −3.0 | 1.9 | 38.4 | ±1.1 | −6.4 | 2.7 |
| S41 | s14ga | TCGCACATCAGCGGCAGTT | 44.9 | ±0.7 | −0.2 | 1.6 | 40.5 | ±1.5 | −4.3 | 3.6 |
| S42 | s14ca | TCGCACATCAGCGCCAGTT | 42.0 | ±0.6 | −3.1 | 1.3 | 39.8 | ±1.4 | −5.0 | 3.5 |
| S43 | s15tg | TCGCACATCAGCGTTAGTT | 41.2 | ±0.8 | −3.9 | 1.8 | 39.3 | ±1.3 | −5.5 | 3.2 |
| S44 | s15ag | TCGCACATCAGCGTAAGTT | 40.6 | ±0.8 | −4.5 | 2.1 | 36.3 | ±1.5 | −8.5 | 4.1 |
| S45 | s15gg | TCGCACATCAGCGTGAGTT | 40.6 | ±0.8 | −4.5 | 1.8 | 36.8 | ±0.9 | −8.0 | 2.4 |
| S46 | s16gt | TCGCACATCAGCGTCGGTT | 43.9 | ±1.3 | −1.2 | 2.9 | 41.2 | ±0.6 | −3.6 | 1.5 |
| S47 | s16ct | TCGCACATCAGCGTCCGTT | 41.3 | ±0.8 | −3.8 | 1.9 | 38.7 | ±0.6 | −6.1 | 1.6 |
| S48 | s16tt | TCGCACATCAGCGTCTGTT | 42.7 | ±0.7 | −2.4 | 1.6 | 39.2 | ±0.6 | −5.6 | 1.4 |
| S49 | s17cc | TCGCACATCAGCGTCACTT | 42.2 | ±0.8 | −2.9 | 1.8 | 39.8 | ±1.6 | −5.0 | 4.1 |
| S50 | s17tc | TCGCACATCAGCGTCATTT | 41.6 | ±0.7 | −3.5 | 1.8 | 39.0 | ±1.4 | −5.8 | 3.6 |
| S51 | s17ac | TCGCACATCAGCGTCAATT | 41.3 | ±0.8 | −3.8 | 1.9 | 38.7 | ±1.7 | −6.1 | 4.3 |
| S52 | s18aa | TCGCACATCAGCGTCAGAT | 43.8 | ±1.7 | −1.3 | 3.9 | 40.0 | ±1.7 | −4.8 | 4.2 |
| S53 | s18ga | TCGCACATCAGCGTCAGGT | 45.9 | ±1.0 | 0.8 | 2.2 | 43.7 | ±2.2 | −1.1 | 5.1 |
| S54 | s18ca | TCGCACATCAGCGTCAGCT | 41.7 | ±0.6 | −3.4 | 1.4 | 37.2 | ±1.2 | −7.6 | 3.3 |
| S55 | s19aa | TCGCACATCAGCGTCAGTA | 44.6 | ±1.1 | −0.5 | 2.4 | 42.8 | ±1.5 | −2.0 | 3.5 |
| S56 | s19ga | TCGCACATCAGCGTCAGTG | 44.7 | ±1.4 | −0.4 | 3.1 | 41.8 | ±1.0 | −3.0 | 2.3 |
| S57 | s19ca | TCGCACATCAGCGTCAGTC | 44.4 | ±1.4 | −0.7 | 3.2 | 41.5 | ±0.7 | −3.3 | 1.7 |
| S58 | 1aa2gg | AGGCACATCAGCGTCAGTT | 44.2 | ±1.4 | −0.9 | 3.1 | 43.5 | ±1.0 | −1.3 | 2.3 |
| S59 | 3cc4gg | TCCGACATCAGCGTCAGTT | 38.8 | ±1.1 | −6.3 | 2.7 | 36.9 | ±0.8 | −7.9 | 2.2 |
| S60 | 3cc4gg11cc12gg | TCCGACATCACGGTCAGTT | NDd | ND | ND | ND | ND | ND | ND | ND |
| S61 | 11cc12gg | TCGCACATCACGGTCAGTT | 34.2 | ±0.8 | −10.9 | 2.3 | 37.5 | ±2.1 | −7.3 | 5.5 |
| N0 | npm | CCCCTCTGCTGCACTCTA | 43.4 | ±1.3 | 0.0 | 6.5 | 44.3 | ±2.9 | 0.0 | 6.5 |
| N1 | n1gg | GCCCTCTGCTGCACTCTA | 42.5 | ±0.8 | −0.9 | 2.0 | 43.0 | ±2.5 | −1.3 | 5.7 |
| N2 | n1ag | ACCCTCTGCTGCACTCTA | 42.9 | ±2.2 | −0.5 | 5.0 | 43.7 | ±2.7 | −0.6 | 6.1 |
| N3 | n1tg | TCCCTCTGCTGCACTCTA | 42.5 | ±2.2 | −0.9 | 5.3 | 43.5 | ±3.2 | −0.8 | 7.4/PICK> |
| N4 | n2gg | CGCCTCTGCTGCACTCTA | 42.1 | ±2.1 | −1.3 | 5.0 | 43.4 | ±3.0 | −0.9 | 6.8 |
| N5 | n2ag | CACCTCTGCTGCACTCTA | 42.1 | ±2.3 | −1.3 | 5.5 | 43.2 | ±3.0 | −1.1 | 6.9 |
| N6 | n2tg | CTCCTCTGCTGCACTCTA | 41.8 | ±2.3 | −1.6 | 5.4 | 43.5 | ±2.8 | −0.8 | 6.3 |
| N7 | n3gg | CCGCTCTGCTGCACTCTA | 41.1 | ±2.0 | −2.3 | 4.8 | 43.2 | ±2.3 | −1.1 | 5.3 |
| N8 | n3ag | CCACTCTGCTGCACTCTA | 41.4 | ±2.4 | −2.0 | 5.8 | 42.2 | ±1.8 | −2.1 | 4.2 |
| N9 | n3tg | CCTCTCTGCTGCACTCTA | 40.6 | ±1.9 | −2.8 | 4.8 | 42.0 | ±1.3 | −2.3 | 3.1 |
| N10 | n4gg | CCCGTCTGCTGCACTCTA | 41.0 | ±1.4 | −2.4 | 3.4 | 41.9 | ±1.4 | −2.4 | 3.2 |
| N11 | n4ag | CCCATCTGCTGCACTCTA | 40.9 | ±1.2 | −2.5 | 2.9 | 42.0 | ±1.0 | −2.3 | 2.4 |
| N12 | n4tg | CCCTTCTGCTGCACTCTA | 40.6 | ±1.0 | −2.8 | 2.3 | 42.8 | ±2.7 | −1.5 | 6.2 |
| N13 | n5ga | CCCCGCTGCTGCACTCTA | 40.8 | ±1.0 | −2.6 | 2.4 | 42.3 | ±1.7 | −2.0 | 3.9 |
| N14 | n5aa | CCCCACTGCTGCACTCTA | 40.7 | ±1.5 | −2.7 | 3.8 | 42.8 | ±2.8 | −1.5 | 6.6 |
| N15 | n5ca | CCCCCCTGCTGCACTCTA | 39.8 | ±1.8 | −3.6 | 4.4 | 42.5 | ±3.0 | −1.8 | 7.0 |
| N16 | ng6g | CCCCTTTGCTGCACTCTA | 38.6 | ±1.5 | −4.8 | 4.0 | 40.4 | ±2.8 | −3.9 | 7.0 |
| N17 | n6ag | CCCCTATGCTGCACTCTA | 40.1 | ±2.1 | −3.3 | 5.2 | 40.3 | ±2.6 | −4.0 | 6.5 |
| N18 | n6gg | CCCCTGTGCTGCACTCTA | 39.3 | ±1.3 | −4.1 | 3.4 | 40.0 | ±2.3 | −4.3 | 5.8 |
| N19 | n7aa | CCCCTCAGCTGCACTCTA | 40.1 | ±2.0 | −3.3 | 5.0 | 41.1 | ±2.9 | −3.2 | 7.0 |
| N20 | n7ga | CCCCTCGGCTGCACTCTA | 40.9 | ±2.1 | −2.5 | 5.1 | 40.7 | ±2.1 | −3.6 | 5.1 |
| N21 | n7ca | CCCCTCCGCTGCACTCTA | 39.5 | ±1.6 | −3.9 | 4.0 | 40.7 | ±1.9 | −3.6 | 4.7 |
| N22 | n8cc | CCCCTCTCCTGCACTCTA | 34.7 | ±0.6 | −8.7 | 1.6 | 40.1 | ±1.3 | −4.2 | 3.3 |
| N23 | n8tc | CCCCTCTTCTGCACTCTA | 35.6 | ±1.5 | −7.8 | 4.3 | 40.8 | ±1.6 | −3.5 | 3.9 |
| N24 | n8ac | CCCCTCTACTGCACTCTA | 37.6 | ±0.6 | −5.8 | 1.7 | 41.8 | ±1.5 | −2.5 | 3.5 |
| N25 | n9tg | CCCCTCTGTTGCACTCTA | 39.4 | ±0.9 | −4.0 | 2.2 | 44.6 | ±2.3 | 0.3 | 5.1 |
| N26 | n9ag | CCCCTCTGATGCACTCTA | 41.0 | ±1.3 | −2.4 | 3.2 | 42.3 | ±1.7 | −2.0 | 4.1 |
| N27 | n9gg | CCCCTCTGGTGCACTCTA | 39.9 | ±1.3 | −3.5 | 3.3 | 41.4 | ±2.0 | −2.9 | 4.7 |
| N28 | n10aa | CCCCTCTGCAGCACTCTA | 39.8 | ±1.4 | −3.6 | 3.5 | 41.0 | ±1.7 | −3.3 | 4.2 |
| N29 | n10ga | CCCCTCTGCGGCACTCTA | 40.4 | ±1.6 | −3.0 | 4.0 | 40.6 | ±1.2 | −3.7 | 2.9 |
| N30 | n10ca | CCCCTCTGCCGCACTCTA | 39.0 | ±1.3 | −4.4 | 3.4 | 40.1 | ±1.6 | −4.2 | 3.9 |
| N31 | n11cc | CCCCTCTGCTCCACTCTA | 35.2 | ±0.7 | −8.2 | 1.9 | 38.8 | ±1.3 | −5.5 | 3.2 |
| N32 | n11tc | CCCCTCTGCTTCACTCTA | 36.7 | ±0.9 | −6.7 | 2.5 | 38.9 | ±1.6 | −5.4 | 4.1 |
| N33 | n11ac | CCCCTCTGCTACACTCTA | 38.0 | ±1.1 | −5.4 | 2.9 | 40.0 | ±2.0 | −4.3 | 5.0 |
| N34 | n12tg | CCCCTCTGCTGTACTCTA | 39.9 | ±1.3 | −3.5 | 3.2 | 40.7 | ±2.2 | −3.6 | 5.4 |
| N35 | n12ag | CCCCTCTGCTGAACTCTA | 39.2 | ±1.4 | −4.2 | 3.6 | 41.2 | ±1.7 | −3.1 | 4.1 |
| N36 | n12gg | CCCCTCTGCTGGACTCTA | 40.8 | ±0.8 | −2.6 | 2.0 | 42.2 | ±1.6 | −2.1 | 3.8 |
| N37 | n13gt | CCCCTCTGCTGCGCTCTA | 41.9 | ±1.0 | −1.5 | 2.3 | 44.9 | ±3.5 | 0.6 | 7.8 |
| N38 | n13ct | CCCCTCTGCTGCCCTCTA | 38.3 | ±0.5 | −5.1 | 1.2 | 43.4 | ±1.9 | −0.9 | 4.4 |
| N39 | n13tt | CCCCTCTGCTGCTCTCTA | 39.3 | ±0.8 | −4.1 | 2.1 | 41.2 | ±1.6 | −3.1 | 3.8 |
| N40 | n14tg | CCCCTCTGCTGCATTCTA | 38.8 | ±1.2 | −4.6 | 3.1 | 40.9 | ±2.2 | −3.4 | 5.4 |
| N41 | n14ag | CCCCTCTGCTGCAATCTA | 39.0 | ±0.9 | −4.4 | 2.3 | 40.3 | ±1.8 | −4.0 | 4.5 |
| N42 | n14gg | CCCCTCTGCTGCAGTCTA | 39.8 | ±0.9 | −3.6 | 2.2 | 40.2 | ±1.6 | −4.1 | 4.0 |
| N43 | n15aa | CCCCTCTGCTGCACACTA | 39.8 | ±0.4 | −3.6 | 1.0 | 40.1 | ±1.4 | −4.2 | 3.4 |
| N44 | n15ga | CCCCTCTGCTGCACGCTA | 40.8 | ±0.8 | −2.6 | 1.8 | 40.0 | ±1.4 | −4.3 | 3.5 |
| N45 | n15ca | CCCCTCTGCTGCACCCTA | 39.0 | ±1.2 | −4.4 | 3.1 | 40.7 | ±1.5 | −3.6 | 3.6 |
| N46 | n16tg | CCCCTCTGCTGCACTTTA | 40.6 | ±0.9 | −2.8 | 2.3 | 40.7 | ±1.3 | −3.6 | 3.2 |
| N47 | n16ag | CCCCTCTGCTGCACTATA | 41.9 | ±0.6 | −1.5 | 1.4 | 40.9 | ±1.8 | −3.4 | 4.3 |
| N48 | n16gg | CCCCTCTGCTGCACTGTA | 42.6 | ±0.3 | −0.8 | 0.7 | 43.8 | ±1.8 | −0.5 | 4.1 |
| N49 | n17aa | CCCCTCTGCTGCACTCAA | 42.3 | ±1.3 | −1.1 | 3.1 | 43.7 | ±1.8 | −0.6 | 4.1 |
| N50 | n17ga | CCCCTCTGCTGCACTCGA | 44.6 | ±1.5 | 1.2 | 3.4 | 44.5 | ±1.4 | 0.2 | 3.2 |
| N51 | n17ca | CCCCTCTGCTGCACTCCA | 42.0 | ±1.1 | −1.4 | 2.6 | 41.8 | ±1.7 | −2.5 | 4.0 |
| N52 | n18gt | CCCCTCTGCTGCACTCTG | 42.9 | ±1.7 | −0.5 | 3.9 | 43.0 | ±1.3 | −1.3 | 3.0 |
| N53 | n18ct | CCCCTCTGCTGCACTCTC | 42.7 | ±1.7 | −0.7 | 3.9 | 42.9 | ±1.4 | −1.4 | 3.3 |
| N54 | n18tt | CCCCTCTGCTGCACTCTT | 43.5 | ±1.4 | 0.1 | 3.2 | 43.8 | ±1.4 | −0.5 | 3.2 |
| N55 | 1gg2gg | GGCCTCTGCTGCACTCTA | 41.1 | ±1.0 | −2.3 | 2.5 | 43.0 | ±1.8 | −1.3 | 4.1 |
| N56 | 3gg4gg | CCGGTCTGCTGCACTCTA | 37.0 | ±0.9 | −6.4 | 2.4 | 39.3 | ±0.8 | −5.0 | 1.9 |
| N57 | 8cc9gg14gg | CCCCTCTCGTGCACTCTA | 33.6 | ±1.5 | −9.8 | 4.5 | 39.4 | ±1.1 | −4.9 | 2.8 |
| N58 | 8cc9gg11cc12gg | CCCCTCTCGTCGACTCTA | ND | ND | ND | ND | 32.7 | ±2.1 | −11.6 | 6.3 |
| N59 | 6gg8cc9gg11cc12gg | CCCCTGTCGTCGACTCTA | ND | ND | ND | ND | ND | ND | ND | ND |
| N60 | 8cc9gg14gg | CCCCTCTCGTCGAGTCTA | ND | ND | ND | ND | ND | ND | ND | ND |
| N61 | 6gg8cc9gg11cc12gg | CCCCTGTCGTCGAGTCTA | ND | ND | ND | ND | ND | ND | ND | ND |
Probe names incorporate the type of target (s, Staphylococcus; n, Nitrosomonas), position of the mismatch from the 5′ terminus, and the type of mismatch (probe-target). spm and npm, perfect-match probes for Staphylococcus and Nitrosomonas targets, respectively.
ΔT, difference of the Td between the perfect-match probe and each mismatch probe.
CV, coefficient of variation.
ND, not determined due to faint fluorescence signal.
Mismatches are underlined.
Microarray fabrication.
The microarray matrix consisted of 100- by 100- by 20-μm polyacrylamide gel pads at a 100-μm spacing. The gel pads were fixed to a glass slide by photopolymerization (9) and activated as described previously (18), and 1 pmol of probe was applied to each gel pad in one droplet (1 nl) of a 1 mM amino-oligonucleotide solution (24) with a robot arrayer (30). The oligonucleotide probes were immobilized through reductive coupling of a 3′ amino group of the oligonucleotide with the aldehyde group of the activated gel pad on the microarrays (18).
Microarray hybridization.
Hybridizations were conducted at room temperature (20°C) for 12 h in a hybridization chamber affixed to the surface of the glass slide (Grace BioLabs, Bend, Oreg.) containing 40 μl of hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 8.0], 40% formamide) and 1 μl of Cy3-labeled target nucleic acids (each at 25 ng/μl). Following hybridization, the microarray was briefly washed three times at room temperature with 100 μl of wash buffer (20 mM Tris-HCl [pH 8.0], 5 mM EDTA, 4 mM NaCl). After the final wash, 100 μl of wash buffer was added to the wash chamber (Grace BioLabs) for fluorescence monitoring. Image analysis was performed by using a custom-designed fluorescence microscope (State Optical Institute, St. Petersburg, Russia) equipped with a cooled charge-coupled device camera (Princeton Instruments, Trenton, N.J.). Preliminary experiments revealed that there was no cross-hybridization of any probe-target duplexes when both target sequences were used (data not shown). Four microarray slides were used repeatedly in this study.
Generation of melting profiles.
To generate melting profiles, the microarray was fixed to a thermal table mounted on the stage of the microscope. The thermal table was connected to a thermoelectric temperature controller (LFI-3751; Wavelength Electronics, Inc., Bozeman, Mont.) and a water bath (Cole Parmer Instruments Co., Chicago, Ill.). Melting profiles for all gel pads were generated by gradually increasing the temperature (1°C/min) of the thermal table from 20 to 70°C and recording the fluorescence signal intensity of the gel pads at 2°C intervals. Temperature, data acquisition, image processing, and analysis were controlled with custom software written in LabVIEW (version 5.1; National Instruments Co., Austin, Tex.). The signal intensity of each melting profile was normalized, and the Td was calculated by using Td-calculator (http://stahl.ce.washington.edu) as described previously (25). Obtained Tds are listed in Table 1. Hybridization and melting profile analyses were repeated five times for both DNA and RNA targets.
DI.
The optimum wash temperature, defined as that providing maximum discrimination between perfect-match duplexes and those containing mismatches, is generally determined empirically. To refine and systematize the determination of an optimum wash temperature, we introduced a discrimination index (DI). The DI for a specific wash temperature was determined by the following equation: DItemperature = (pmtemperature/mmtemperature)×(pmtemperature − mmtemperature), where pmtemperature is the average signal intensity of perfect-match duplexes at a specific wash temperature and mmtemperature is the average signal intensity of mismatched duplexes, excluding those duplexes which have terminal and next-to-terminal mismatches.
Data for the NN.
The input data set consisted of signal intensity (melting) profiles, with each input record consisting of a single profile of either a perfect-match duplex, a duplex with a mismatch in the ultimate or penultimate position, or a duplex with an internal mismatch. The output data set consisted of one categorical variable that was coded 0 if the corresponding record was a perfect-match duplex, 1 if the duplex had a mismatch in the ultimate or penultimate position, or 2 if the duplex had an internal mismatch. Prior to neural network (NN) analyses, the data were all normalized to have a mean of 0 and a standard deviation (SD) of 1.
NN software and analyses.
The NN software was custom designed by using Java software and was based on the “leave one input out” cross-validation model (3). Rather than leave one input out, we modified the model to use one input (e.g., single intensity values at a specific temperature) to predict a categorical output (e.g., a perfect-match duplex, a duplex with a mismatch in the ultimate or penultimate position, or a duplex with an internal mismatch). We chose this approach because it was difficult to measure the importance of inputs that are statistically dependent (i.e., signal intensities within the same melting profile are highly correlated to one another). The software is available at a World Wide Web-based interface at http://stahl.ce.washington.edu under the heading “Tools for data analyses.”
The network architecture consisted of one input layer, one hidden layer, and one output layer. Neurons in the hidden layer used a hyperbolic tangent activation function, while the neuron in the output layer used a standard purely linear activation function (11). All neurons included a bias term. The Levenberg-Marquardt algorithm was used for training the NN rather than standard back-propagation and conjugate gradient methods because preliminary results showed that the Levenberg-Marquardt algorithm was superior in terms of the number of iterations needed to reach the error minima (11). Since preliminary analysis revealed that the minimum number of hidden neurons needed to produce the highest R2 results was two, only two hidden neurons were used for all NN analyses. A standard least-squares error function was used for training the NN since this function could be easily converted to R2 values.
It should be noted that our method does not produce generalizable NNs since our specific objective was to identify with which inputs the NN learned best. Therefore, no data were used for testing or validation purposes. The NN was deemed to have reached minima (and consequently training was stopped) when the R2 did not increase by more than 0.001 U over a period of 10 s (i.e., approximately 200 megaflops).
For NN analyses, we generated an independent NN for each individual input. If one NN performed better with one input rather than another (i.e., it had a higher R2 value), the input having the better prediction was assumed to be more important. It is essential to recognize that this approach does not provide information on the optimal subsets of inputs but rather identifies which inputs are most important for predicting outputs when presented independently. Since some NNs do not train properly because they reach local minima of their error space, a median of 11 NN runs was conducted for each input. We chose the median rather than the mean since the median minimizes local-minimum effects.
RESULTS AND DISCUSSION
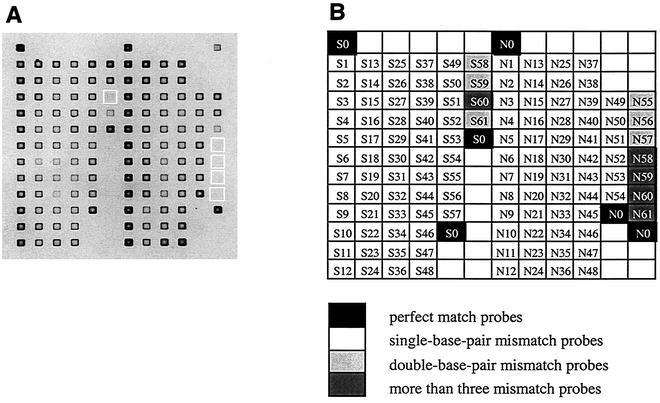
A typical image of the DNA microarray after DNA-DNA hybridization and the corresponding position of each probe on the microarray are shown in Fig. 1. To optimize hybridization and washing conditions, several different types of hybridization buffer containing 0 to 70% formamide and different compositions of wash buffers were tested (12, 25). The optimal conditions were achieved with a hybridization buffer containing 40% formamide and a wash buffer containing 4 mM NaCl. The signal intensities of five probes having more than three mismatches were below detection under these hybridization and wash conditions. However, these conditions did not provide sufficient stringency for discriminating single- or double-base-pair mismatches. For this reason, we examined the melting profiles of probe-target duplexes to determine if adequate discrimination can be attained for single- and double-base-pair mismatches.
FIG. 1.
Typical image of a DNA microarray after hybridization with DNA target sequences (A) and the locations of the oligonucleotide probes (B). Probe labels are as in Table 1. Hybridization and wash conditions are described in the text. Exposure time was 1.0 s. White boxes (A) indicate probes that did not yield detectable fluorescence signals after the wash at 20°C.
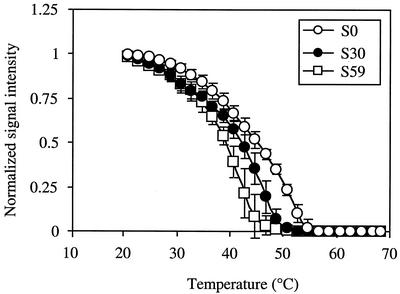
Typical normalized melting profiles of perfect-match probe-target duplexes and those with one or two mismatched base pairs are shown in Fig. 2. For these duplexes, discrimination among perfect-match and mismatched probe-target duplexes was achieved by comparing the Tds. In general, the Td provides an important experimental parameter for distinguishing between probe-target duplexes with and without mismatches (31). For example, a previous study based on melting profiles revealed that Tds provided excellent differentiation among five closely related Bacillus species (13). However, complete resolution is achieved only when the Tds of the perfect-match probe-target and duplexes containing mismatches are sufficiently different.
FIG. 2.
Typical normalized melting profiles of DNA-DNA duplexes of Staphylococcus. S0, perfect-match duplex; S30, single-base-pair-mismatched duplex containing a tt mismatch (probe-target) at position 10 from the 5′ terminus; S59, double-base-pair-mismatched duplex containing cc and gg mismatches at positions 3 and 4, respectively. Error bars, SDs of the data (S0, n = 10; S30, n = 5; S59, n = 4).
The experimentally determined Tds for perfect-match and all single-base-pair-mismatch duplexes are listed in Table 1. The mean Tds for some single-base-pair-mismatch duplexes were slightly higher than the Td for perfect-match duplexes. For example, the Td for probe N50 was 44.6°C (SD, ±1.5°C) in DNA duplexes while the Td for perfect-match probe N0 was 43.4°C (SD, ±1.3°C). In this case, the difference between Tds was not sufficient to adequately resolve perfect-match duplexes and duplexes having a single-base-pair mismatch.
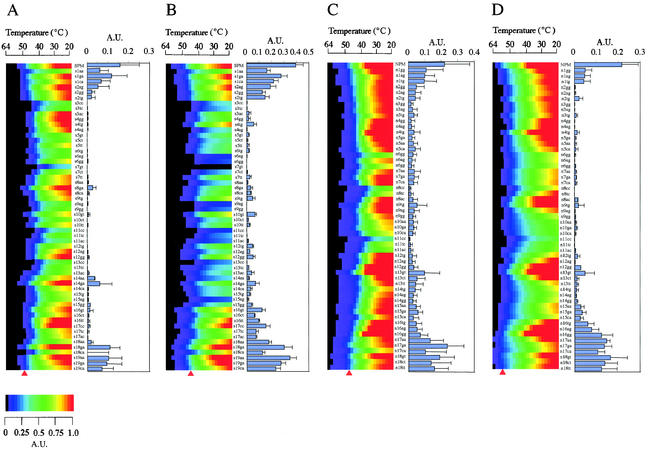
In this study we evaluated the inclusion of signal intensity data to optimize discrimination among perfect-match and mismatched probe-target duplexes. Considering intensity data alone, an optimum corresponds to hybridization and washing conditions at which the signal intensity of mismatches reaches (or approaches) background and the perfect-match duplex maintains a detectable signal. Often these conditions are determined empirically, as represented by Fig. 3. This figure shows the signal intensity for each probe duplex (color) measured at 2°C increments during the thermal dissociation. An empirical estimate of the optimum wash temperature for each probe-target duplex is shown (left section of each panel), and the corresponding intensity data are shown in the adjacent section. For perfect-match duplexes, signal intensities at each empirically defined optimum were approximately 20% of the initial signal intensities. For example, the signal intensity of the perfect-match DNA-DNA duplex of Staphylococcus was 1.11 U at 20°C, while the signal intensity was 0.16 U at the empirically determined optimal wash temperature (52°C) (Fig. 3A, right section). These intensity measurements corresponded to those achieved in a separate experiment in which the microarray was washed at the identified temperature optimum (Fig. 4). However, it was not possible to fully resolve perfect-match probe-target duplexes and those with mismatches at the ultimate or penultimate position. These results were in accordance with the conclusion derived from Td analysis reported previously (25).
FIG. 3.
Signal intensity profile of probe-target duplexes with temperature gradient (color sections; A.U., arbitrary units of fluorescence intensities) and signal intensities at empirically determined optimum wash temperatures (bars). Red triangles, optimum wash temperatures. (A and B) Staphylococcus target DNA (A) and target RNA (B); (C and D) Nitrosomonas target DNA (C) and target RNA (D). Data represent the mean signal intensities of five melting profile analyses and SDs (error bars).
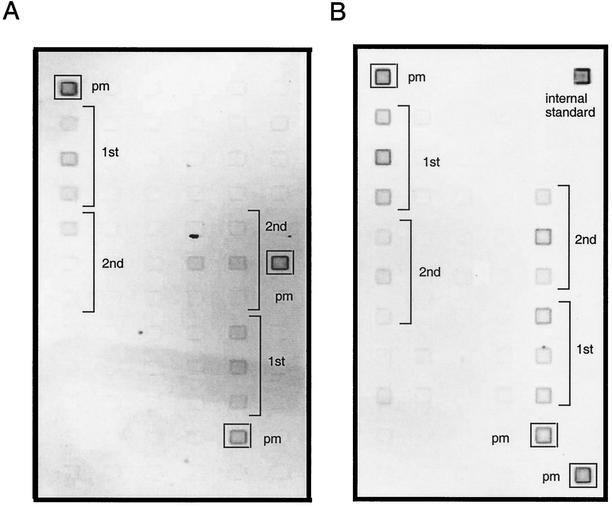
FIG. 4.
Typical images of DNA microarrays washed at optimum temperatures. Melting profiling was terminated at the empirically determined optimum wash temperature. The optimum wash temperatures (shown in Fig. 3) were 52°C for the Staphylococcus DNA probe-target duplexes (A) and 50°C for the Nitrosomonas DNA probe-target duplexes (B). pm, perfect-match probes for Staphylococcus target (S0 in Fig. 1B) or the Nitrosomonas target (N0 in Fig. 1B); 1st, probes define ultimate position; 2nd, probes define penultimate position.
We also observed contrasting relative stabilities of DNA-DNA versus RNA-DNA duplexes for these two probes. For Staphylococcus, the empirically determined optimum wash temperature for DNA-DNA duplexes was higher than that for RNA-DNA duplexes (52 versus 48°C) (Fig. 3A and B). However, for Nitrosomonas, the optimum for the DNA-DNA duplexes was lower than that for RNA-DNA duplexes (50 versus 58°C) (Fig. 3C and D). These contrasting results, also supported by comparing their Tds (P < 0.0001), indicate that the stability of DNA-DNA duplexes and RNA-DNA duplexes is sequence dependent (20, 23) and underscore the difficulty in a priori prediction of duplex stability using currently available models (H. Urakawa et al., unpublished data).
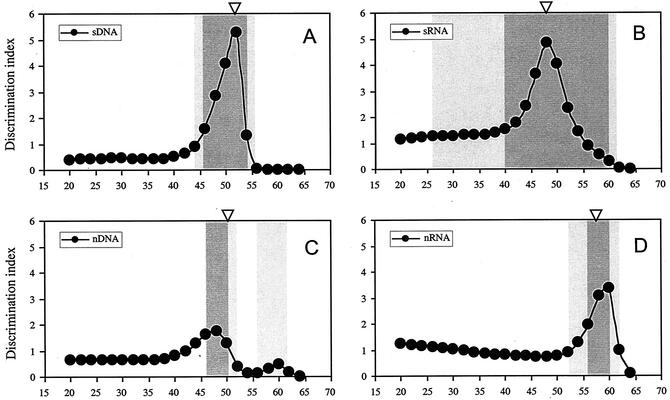
To refine and systematize the above-described empirical approach, we introduced a DI, which is calculated by the formula given in Materials and Methods. This index is defined as the product of difference and ratio of the signal intensities for perfect-match and mismatched duplexes at a given wash temperature. The temperature with the maximum DI is defined as the optimum wash temperature. As shown in Fig. 5, optimum wash temperatures were calculated by using the DI from the signal intensity profiles in the range of 20 to 64°C (Fig. 3). For hybridization with the Staphylococcus target, DI-based and empirically determined optimum wash temperatures for DNA-DNA and RNA-DNA hybridizations were identical (Fig. 3 and 5). For Nitrosomonas, DI-based and empirically determined optimum wash temperatures were within 2°C of each other (i.e., triangle and peak DI values occur at around the same temperature in Fig. 5), suggesting a reasonable match between DI-based prediction and the empirical determination.
FIG. 5.
Inferred optimum wash temperatures for the discrimination of perfect and mismatched duplexes. (A and B) Staphylococcus target DNA (sDNA; A) and target RNA (sRNA; B); (C and D) Nitrosomonas target DNA (nDNA; C) and target RNA (nRNA; D). DI was calculated by using the formula given in Materials and Methods. Triangles, temperatures empirically inferred from melting profiles. Light gray zones, temperature intervals allowing for mismatch discrimination as deduced from NN analysis using all data sets (R2 > 0.7); dark gray zones, temperature intervals deduced from NN analysis using data sets excluding data from ultimate and penultimate positions (R2 > 0.9).
NN analyses were used to further investigate the relationship between terminal and internal mismatches (Fig. 5). The NNs were able to adequately discriminate between perfect-match probe-target duplexes and duplexes with internal mismatches (R2 > 0.90) and between perfect-match probe-target duplexes and duplexes with mismatches at any position (R2 > 0.70) within the temperature intervals indicated. These results are in good agreement with the empirically determined optimum wash temperatures and maxima of DI profiles (Fig. 3 and 5) and support the use of the DI to identify optimum washing temperatures.
The application of NNs to the analysis of complex data in microbiology is relatively new (1). NNs have been used to identify the restriction enzyme profiles for E. coli O156:H7 (4), the pyrolysis mass spectra for Mycobacterium tuberculosis complex species (7), bacterial species from randomly amplified polymorphic DNA patterns (14), fatty acid profiles of microbial communities (17), stable low-molecular-weight rRNA from gel electrophoresis patterns (16), and Td from microarray data (25). However, to our knowledge, no study has used the method outlined in this paper to determine the relative importance of inputs to outputs.
In conclusion, our studies have established an analytical approach to achieving optimum discrimination between target and nontarget duplex structures. Although this objective is important in any application of DNA microarrays to sequence analysis (e.g., identification of point mutations), we note that the application of microarrays to environmental systems must consider a larger and uncharacterized diversity of sequences. Since the character and position of nontarget mismatches in an environmental sample are not known in advance, it is essential that conditions for optimum discrimination be generally defined. Continuing studies are evaluating the number and composition of mismatch probes required to implement the proposed optimization approach in standard applications and possible deviations from model predictions using rRNA derived from natural samples. The melting profiles obtained for this subset of mismatch probes would be used to calculate the maximum DI for each probe. More generally, our results further support the utility of melting profiles for achieving optimum resolution of microarray hybridization data.
Acknowledgments
We thank G. Yershov, A. Kukhtin, and A. Gemmell for their efforts in manufacturing the oligonucleotide microarrays and S. Surzhikov for synthesis of the oligonucleotide probes.
This work was support by grants from the DARPA (DABT63-99-1-0009) and NASA (NAG9-1271) to D.A.S., from the NSF (DEB-0088879) and the EPA (R-82945801) to P.A.N., and from the DARPA to J.J.K.
REFERENCES
- 1.Almeida, J. S., and P. A. Noble. 2000. Neural computing in microbiology. J. Microbiol. Methods 43:1-2. [DOI] [PubMed] [Google Scholar]
- 2.Bavykin, S. G., J. P. Akowski, V. M. Zakhariev, V. E. Barsky, A. N. Perov, and A. D. Mirzabekov. 2001. Portable system for microbial sample preparation and oligonucleotide microarray analysis. Appl. Environ. Microbiol. 67:922-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, C. M. 1995. Neural networks for pattern recognition. Oxford Press, Oxford, United Kingdom.
- 4.Carson, C. A., J. M. Keller, K. K. McAdoo, D. Wang, B. Higgins, C. W. Bailey, J. G. Thorne, B. J. Payne, M. Skala, and A. W. Hahn. 1995. Escherichia coli O157:H7 restriction pattern recognition by artificial neural networks. J. Clin. Microbiol. 33:2894-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong, Y., J. D. Glasner, F. R. Blattner, and E. W. Triplett. 2001. Genomic interspecies microarray hybridization: rapid discovery of three thousand genes in the maize endophyte, Klebsiella pneumoniae 342, by microarray hybridization with Escherichia coli K-12 open reading frames. Appl. Environ. Microbiol. 67:1911-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fotin, A. V., A. L. Drobyshev, D. Y. Proudnikov, A. N. Perov, and A. D. Mirzabekov. 1998. Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res. 26:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman, R., R. Goodacre, P. R. Sisson, J. G. Magee, A. C. Ward, and N. F. Lightfoot. 1994. Rapid identification of species within the Mycobacterium tuberculosis complex by artificial neural network analysis of pyrolysis mass spectra. J. Med. Microbiol. 40:170-173. [DOI] [PubMed] [Google Scholar]
- 8.Gingeras, T. R., G. Ghandour, E. Wang, A. Berno, P. M. Small, F. Drobniewski, D. Alland, E. Desmond, M. Holodniy, and J. Drenkow. 1998. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 8:435-448. [DOI] [PubMed] [Google Scholar]
- 9.Guschin, D., G. Yershov, A. Zaslavsky, A. Gemmell, V. Shick, D. Proudnikov, P. Arenkov, and A. Mirzabekov. 1997. Manual manufacturing of oligonucleotide, DNA, and protein microchips. Anal. Biochem. 250:203-211. [DOI] [PubMed] [Google Scholar]
- 10.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagan, M. T., H. B. Demuth, and M. Beale. 1996. Neural network design. PWS Publishing Co., Boston, Mass.
- 12.Koizumi, Y., J. J. Kelly, T. Nakagawa., H. Urakawa, S. El-Fantroussi, S. Al-Muzaini, M. Fukui, Y. Urushigawa, and D. A. Stahl. 2002. Parallel characterization of anaerobic toluene- and ethylbenzene-degrading microbial consortia by PCR-denaturing gradient gel electrophoresis, RNA-DNA membrane hybridization, and DNA microarray technology. Appl. Environ. Microbiol. 68:3215-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, W. T., A. D. Mirzabekov, and D. A. Stahl. 2001. Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environ. Microbiol. 3:619-629. [DOI] [PubMed] [Google Scholar]
- 14.Moschetti, G., G Blaiotta, F. Villani, S. Coppola, and E. Parente. 2001. Comparison of statistical methods for identification of Streptococcus thermophilus, Enterococcus faecalis, and Enterococcus faecium from randomly amplified polymorphic DNA patterns. Appl. Environ. Microbiol. 67:2156-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray, A. E., D. Lies, G. Li, K. Nealson, J. Zhou, and J. M. Tiedje. 2001. DNA/DNA hybridization to microarrays reveals gene-specific differences between closely related microbial genomes. Proc. Natl. Acad. Sci. USA 98:9853-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noble, P. A., K. D. Bidle, and M. Fletcher. 1997. Natural microbial community compositions compared by a back-propagating neural network and cluster analysis of 5S rRNA. Appl. Environ. Microbiol. 63:1762-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noble, P. A., J. S. Almeida, and C. R. Lovell. 2000. Application of neural computing methods for interpreting phospholipid fatty acid profiles of natural microbial communities. Appl. Environ. Microbiol. 66:694-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Proudnikov, D., E. Timofeev, and A. Mirzabekov. 1998. Immobilization of DNA in polyacrylamide gel for the manufacture of DNA and DNA-oligonucleotide microchips. Anal. Biochem. 259:34-41. [DOI] [PubMed] [Google Scholar]
- 19.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riley, M., and B. Maling. 1966. Physical and chemical characterization of two- and three-stranded adenine-thymine and adenine-uracil homopolymer complexes. J. Mol. Biol. 20:359-389. [DOI] [PubMed] [Google Scholar]
- 21.Service, R. F. 1998. Microchip arrays put DNA on the spot. Science 282:396-399. [DOI] [PubMed] [Google Scholar]
- 22.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugimoto, N., S. Nakano, M. Katoh, A. Matsumura, H. Nakamuta, T. Ohmichi, M. Yoneyama, and M. Sasaki. 1995. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34:11211-11216. [DOI] [PubMed] [Google Scholar]
- 24.Timofeev, E., S. V. Kochetkova, A. D. Mirzabekov, and V. L. Florentiev. 1996. Regioselective immobilization of short oligonucleotides to acrylic copolymer gels. Nucleic Acids Res. 24:3142-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urakawa, H., P. A. Noble, S. El Fantroussi, J. J. Kelly, and D. A. Stahl. 2002. Single-base-pair discrimination of terminal mismatches by using oligonucleotide microarrays and neural network analyses. Appl. Environ. Microbiol. 68:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasiliskov, A. V., E. N. Timofeev, S. A. Surzhikov, A. L. Drobyshev, V. V. Shick, and A. D. Mirzabekov. 1999. Fabrication of microarray of gel-immobilized compounds on a chip by copolymerization. BioTechniques 27:592-594, 596-598, 600. [DOI] [PubMed]
- 27.Wagner, M., G. Rath, H.-P. Koops, and K.-H. Schleifer. 1995. In situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251-264. [Google Scholar]
- 28.Wilson, K. H., W. J. Wilson, J. L. Radosevich, T. Z. DeSantis, V. S. Viswanathan, T. A. Kuczmarski, and G. L. Andersen. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yershov, G., V. Barsky, A. Belgovskiy, E. Kirillov, E. Kreindlin, I. Ivanov, S. Parinov, D. Guschin, A. Drobishev, S. Dubiley, and A. Mirzabekov. 1996. DNA analysis and diagnostics on oligonucleotide microchips. Proc. Natl. Acad. Sci. USA 93:4913-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng, D., E. W. Alm, D. A. Stahl, and L. Raskin. 1996. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl. Environ. Microbiol. 62:4504-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]