Abstract
A protocol was developed to detect bacteria inhabiting microarthropods by means of small-subunit rRNA-targeted fluorescence in situ hybridization and microscopy. The protocol is based on cryosections of whole specimens. In contrast to more commonly applied paraffin-embedding techniques, the protocol is quicker and reduces the number of manipulations which might damage the microscopic material. The method allowed the study of the bacterial colonization of Folsomia candida (Collembola) and the detection of bacteria in both the gut and tissue.
The small-subunit rRNAs and their corresponding genes are highly useful molecules for characterizing microorganisms in and from environmental samples and placing them into a phylogenetic system (20, 26). Small-subunit rRNA genes of unknown organisms can be obtained from noncultivated samples by PCR with nucleic acids directly extracted from environmental samples. For this purpose, primers hybridizing to evolutionarily highly conserved regions are used (18, 25). Sequencing of such amplified genes frequently yields similarities of <100% compared to sequences of cultivated known isolates. It can therefore be concluded that the majority of bacteria from most natural habitats have not yet been cultured and characterized for their phylogenetic affiliations.
The detection of a new rRNA gene by itself will often not be sufficient to understand the activity, and even more, the ecological function of an uncultured organism, because the amplified genes may have been derived from resting or highly active cells. In addition, amplifications of new rRNA genes by PCR do not necessarily reflect the actual abundance of organisms in the original sample (21). A key technique to extend our knowledge about the biology and ecology of uncultivated organisms is in situ hybridization, a technique which allows the PCR-independent detection and localization of bacteria with a selected specificity determined by rRNA-targeted, labeled gene probes (4, 7). Because of the high number of ribosomes and rRNA molecules in most bacterial cells, the hybridized gene probes can be directly visualized, e.g., in bacterial cell suspensions or in microscopic sections. Different labels coupled to a gene probe allow different types of detection, e.g., by staining or epifluorescence microscopy (10, 24). Due to their potential for direct detection and their sensitivity, gene probes coupled with fluorescent dyes are especially useful for the analysis of microscopic structures (1).
Here, we report on a protocol which allows fluorescence in situ hybridization (FISH) analysis to specifically detect bacteria in microarthropods. In our laboratory, we investigate the importance of members of the order Collembola as habitats for microorganisms. The Collembola are microarthropods that have an important function in many soils of enhancing the degradation and restructuring of organic matter. In previous studies, cultivation techniques were used to isolate a number of bacteria from different phylogenetic groups which were able to colonize the collembolan gut (14, 15, 22). In order to further characterize which parts of the collembolan body may serve as microbial habitats, we wanted to use FISH analysis of microscopic sections of the whole specimen.
FISH studies of microscopic sections are commonly done with paraffin- or resin-embedded material (10, 17). For our purposes, however, these techniques were not suitable because the aqueous reagents for dehydration did not efficiently diffuse through the hydrophobic cuticula of the collembolan body. Therefore, we selected the cryosection technique, which is performed on frozen material and is commonly applied without the need for dehydration for histological investigations of tissues from higher organisms. Cryosections combined with FISH have been used to study nitrifying aggregates (19), but to our knowledge, the technique has not been used to study the bacterial colonization of whole organisms. Here, we report how this technique can be modified and optimized for the needs of FISH analysis of microarthropods. In addition, we tested different flourochromes for gene probes and filters for fluorescence microscopy in order to reduce the problems caused by autoflourescence (10) of the investigated material.
MATERIALS AND METHODS
Collembola.
For our analyses, we chose the parthenogenetically reproducing collembole Folsomia candida, which was kept in laboratory stocks in jars with a gypsum-charcoal mixture (plaster of Paris) at the bottom (12). The initial population was obtained from O. Larinck, Technical University, Braunschweig, Germany. The stocks were kept at 18°C in the dark, and the populations were frequently fed with baker's yeast (Saccharomyces cerevisiae). F. candida specimens were taken from these stocks for microscopic analysis.
Preparation of microscopic sections.
In order to preserve the cell and tissue structure, specimens were fixed with formaldehyde (16). A formaldehyde solution (final concentration, 4% [wt/vol] paraformaldehyde) in phosphate-buffered saline (130 mM sodium chloride, 10 mM sodium phosphate buffer, pH 7.2) was prepared as described by Amann (2). To reduce hydrophobic interactions with the cuticula, we added the detergent Triton X-100 (Sigma, Deisenhofen, Germany) to a final concentration of 1% (vol/vol). In order to prevent the formation of air bubbles between the extremities and setae, the solution was first degassed in a vacuum oven (VTR 5022; Heraeus, Hanau, Germany) by boiling it at room temperature. A total of ∼10 adult specimens of F. candida, each with a body length of ∼2 mm, were suspended in 5 ml of the degassed solution. The treatment resulted in the immediate death of the specimens, and the material was then incubated overnight in the refrigerator (4°C) for fixation. After fixation, the material was washed twice in degassed phosphate-buffered saline.
The usual protocol for preparing cryosections of tissue material includes embedding the material in OCT tissue-freezing medium (OCT Tissue-Tex [Miles, Elkhart, Ind.] or Jung [Leica Instruments, Nussloch, Germany]) (6, 8). However, the protocol with the OCT medium could not be applied because the OCT-treated material in our study fragmented the microanatomical structures in the sections of F. candida as a result of adhesion of OCT to the cuticula and formation of drops during the thawing of the material on the microscopic slides. Instead, 5 to 10 specimens were embedded in gelatin-glycerin solution as described by Bancroft (5) (16 g of gelatin, 18.9 g of glycerin, 70 ml of distilled water, and one small crystal of thymole; Merck, Darmstadt, Germany). A volume of ∼0.5 ml of this solidified material was transferred with a spatula from a stock into 2-ml polypropylene reaction tubes. The tubes were incubated in a water bath at 50°C to melt the solution, and then the fixed material was added. The gelatin medium was cooled on ice for 10 min, and 1 ml of chromium(III) potassium sulfate dodecahydrate solution in distilled water (2% [wt/vol]) was then pipetted onto the solidified material and incubated for exactly 1 h on ice to harden the gelatin.
A Frigocut cryostat (Reichert-Jung 2800; Leica Instruments) was used to prepare the cryosection. Still at room temperature, a drop of OCT was added to the tissue holder of the cryostat, and the solidified material was transferred from the reaction tube onto this drop. Subsequently, the mounted tissue holder was incubated at −35°C for 1 h (precooling) in the cryostat.
Sections with a thickness of 0.5 μm were cut in the cryostat with a motor-driven microtome using a type c knife (length, 16 cm; Leica Instruments). Sections were picked up with a needle and transferred onto adhesive, electrostatically charged microscope slides (Superfrost Plus; Menzel, Braunschweig, Germany). This was done immediately after the microscope slides were introduced into the cryostat. It was important that the microscope slides still be warm during this procedure in order to allow adhesion and smoothing out of the thawing microtomic sections. The slides were incubated in dust-free chambers overnight at room temperature. The sections were then dehydrated in an ethanol series, starting with 5% ethanol and increasing to 96% in five steps (2).
The microscope slides used in our study were not coated with an adhesive compound but only electrostatically charged. As an alternative, coating with the adhesive gelatin poly-l-lysine (at 0.1% [wt/vol] [2] or higher [11]) or with silane (9) was tested. However, neither of these products was capable of affixing the cryosections during further treatments.
Gene probe, labeling, and FISH.
In order to detect the general bacterial colonization of the collembolan body, we chose the oligonucleotide probe EUB338 (5′-GCT GCC TCC CGT AGG AGT-3′), which is complementary to a region of the 16S rRNA which is highly conserved in the domain Bacteria (3). The gene probe EUB338 was labeled with the fluorophore Cy3 (extinction wavelength, 555 nm; emission wavelength, 570 nm; Molecular Probes, Eugene, Oreg.). The labeled probe was obtained from MWG Biotech (Ebersberg, Germany).
A total of 150 μl of hybridization buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% sodium dodecyl sulfate, and 30% formamide) containing 50 pmol of the labeled probe ml−1 was carefully pipetted onto the microscopic sections. A large coverslip with small amounts of plasticine at its edges was placed on the microscope slides and carefully pressed until the hybridization solution was evenly distributed over the respective section. A space of ∼0.5 mm was allowed between the slide and the surface of the section. The microscope slides were then transferred horizontally into the hybridization chambers, which consisted of 50-ml test tubes with a sheet of blotting paper that had been moistened with hybridization buffer. After overnight incubation at room temperature in the dark, the coverslips were carefully removed, and the slides were placed in a wash bath with 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature (10). After being washed, the slides were air dried at 37°C and subsequently embedded in Entellan (Merck) and sealed with coverslips.
Fluorescence microscopy.
Fluorescence microscopic analyses were conducted with an epifluorescence microscope (Axioplan; Zeiss, Jena, Germany). In our study, we used two different filter systems: (i) filter 15 (Zeiss) with an excitation of 546 nm and an emission of 590 nm and (ii) the triple-band-pass filter set 25 (Zeiss) with excitation wavelengths of 400, 495, and 570 nm and emission wavelengths of 460, 530, and 610 nm, respectively. Video recordings were taken with a charge-coupled device video camera (Optronics Engineering, Goleta, Calif.) using Lucia G software version 3.52a for digital image analysis (Lim; Laboratory Imaging, Prague, Czech Republic).
RESULTS AND DISCUSSION
In situ detection of bacteria in F. candida.
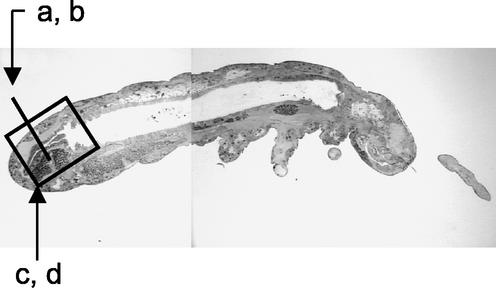
Ultrathin sections were generated from whole specimens of F. candida with resin-embedded material (20) and by the cryosection technique. Both techniques were suitable to preserve the anatomy of the specimens. Resin-embedded material was useful to visualize the structure of the gut, as shown in Fig. 1 for sagittal sections, stained with fuchsin and crystal violet (20). The tubelike midgut and the folded hindgut region are clearly visible (Fig. 1).
FIG. 1.
Sagittal section of a resin-embedded F. candida specimen. The total length of the specimen was ∼2.5 mm. The letters correspond to the sections shown in Fig. 2.
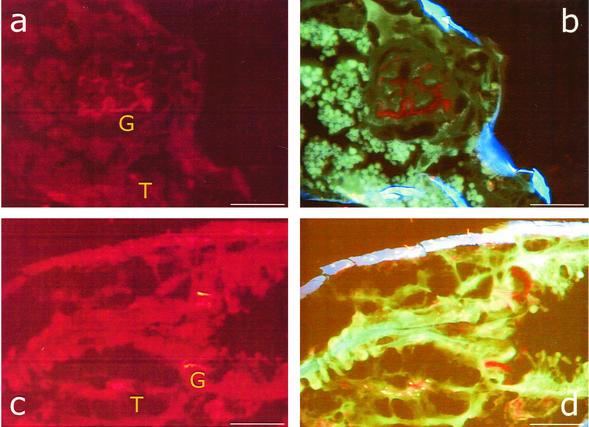
The bacterial colonization of the hindgut region was studied in cryosections by FISH using the probe EUB 388 and two different filter sets (Fig. 2). The positions of both sections shown in Fig. 2 are indicated in Fig. 1. In both sections, transverse (Fig. 2a and b) and sagittal (Fig. 2c and d), in situ hybridization signals caused by bacterial cells could be detected. With filter set 15 (Fig. 2a and c), the tissue showed high nonspecific red autofluorescence. However, the hybridized bacteria increased the signal intensity and appeared significantly brighter in the microscopic image. Much better resolution was obtained with the triple-band filter set 25. Here, the tissue appeared green, and the cuticula, due to the chitin, appeared blue. Bacterial cells could easily be identified by their red-light emission (Fig. 2b and d).
FIG. 2.
FISH with microscopic transverse (a and b) and sagittal (c and d) sections of F. candida as detected with the Cy3-labeled universal bacterial probe EUB388. Shown are microscopic images with filter set 15 (a and c) and with filter set 25 (b and d). T and G indicate tissue and gut regions colonized with bacteria. In panels c and d, the proximal region of the specimen is on the right and the distal region is on the left. Bars, 50 μm.
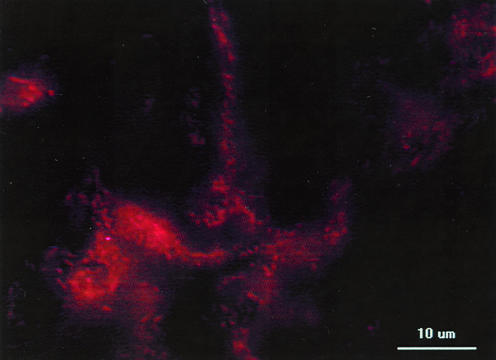
In the food bolus of the midgut region, a large number of rod-shaped cells were detectable with filter set 15 (Fig. 3). Filter set 25 (triple band) was less suitable for studying the bacterial cell morphologies because the filters absorbed some of the light that was necessary for both excitation and emission of Cy3 (not shown).
FIG. 3.
Bacterial colonization of food bolus in the midgut, as detected by FISH and the EUB 338 Cy3-labeled gene probe.
The specificity of the gene probe hybridization for bacterial cells was proven because no tissue cells with eukaryotic nuclear or mitochondrial DNA yielded hybridization signals. Group-specific probes for the Alpha-, Beta-, and Gammaproteobacteria were able to detect different bacterial cells (data not shown). In addition, genus-specific probes were used to detect specific alphaproteobacteria which were localized in different body regions (A. B. Czarnetzki and C. C. Tebbe, unpublished data).
Our FISH analysis clearly showed bacterial colonization of the hindgut (Fig. 2a and b) and the pylorus region; the latter is located at the end of the midgut and the beginning of the hindgut (Fig. 2c and d). Previous cultivation-based studies and scanning electron microscopy done in our laboratory had indicated that the gut of F. candida can be densely colonized with bacteria, especially in the region of the peritrophic membrane (22). In addition, scanning electron microscopy also indicated the presence of intracellular bacteria (unpublished results), and recently, the intracellularly reproducing parasite Wolbachia was identified as an inhabitant of F. candida (23). In our study, FISH showed several red spots in the tissue, and it is likely that they were caused by tissue-colonizing and possibly intracellular bacteria. Due to the specificity of the probe, however, we can indicate only the domain bacteria. With more specific probes, it will be possible in future studies to give a more accurate phylogenetic affiliation of these bacteria that may not be accessible by cultivation techniques.
The selection of the chromophore for labeling the gene probe was a critical step in our study. It is known that in fluorescence light microscopy, autofluorescence of the tissue is a problem, and other authors have therefore used the digoxigenin label instead of a fluorescence label for studying the bacterial colonization of insects by in situ hybridization (10). However, digoxigenin labeling requires additional washing procedures which prolong the protocol and potentially damage the microscopic material. In our study, we used Cy3, because the emission of the yellow-to-red light did not interfere with autofluorescence, as was found with chromophores emitting green or blue light. As an alternative to Cy3, we also found Texas red to be applicable for our purposes (data not shown). Our study demonstrates that cryosections can be an alternative which allows FISH to be performed on microarthropods.
Microscopic sections of larger arthropods (insects) for FISH analyses have been done with paraffin-embedded specimens (13). However, we found this technique unsuitable for the analysis of the smaller microarthropod F. candida, with a body length of 2 mm or less. Prior to paraffin embedding, the investigated materials need to be fixed and dehydrated. Fixation requires the addition of aqueous solutions, such as glutaraldehyde, formaldehyde, or both (M. J. Karnovsky, Abstr. 5th Meet. Am. Soc. Cell Biol., in J. Cell Biol. 27:137a-138a, 1965). However, these solutions did not penetrate efficiently through the chitin cuticula of our material, and chemical fixation was only used to support the main fixation, which was achieved by freezing. In addition, dehydration, which is necessary for paraffin embedding, requires an increasing gradient from 5 to 96% ethanol, and again, the lower ethanol concentrations did not efficiently diffuse through the cuticula into the body cavity.
Our protocol combines two advantages: (i) it is rapid, since there is no necessity for time-consuming fixation and hydration steps and incubation for paraffin hardening, and (ii) it reduces the risk of damaging the microscopic material, e.g., by smearing targeted bacteria into different regions of the body.
Arthropods, especially insects, and bacteria have developed sophisticated relationships over evolutionary periods. Outstanding examples of these interactions are members of the genus Buchnera, the primary endosymbionts of aphids, secondary endosymbionts of weevils or white flies, and bacteria summarized under the name Wolbachia, which inhabit a large number of arthropods from different phylogenetic groups. The majority of arthropods, especially smaller species, are not yet discovered and described, and it can be suspected that microorganisms inhabiting these hosts have a high biotechnological potential which awaits unraveling. FISH with gene probes of different specificities will be an important indicator of whether new organisms have been discovered, and the localization of such organisms in the bodies of their hosts can give clues about their function. In this context, our protocol may be helpful for future studies.
Acknowledgments
We thank Evelin Schummer for excellent technical assistance and Alice B. Czarnetzki for discussions.
We thank the German Ministry for Research and Education for financial support (grant no. 0311769).
REFERENCES
- 1.Amann, R., J. Snaidr, M. Wagner, W. Ludwig, and K. H. Schleifer. 1996. In situ visualization of high genetic diversity in a natural microbial community. J. Bacteriol. 178:3496-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I. 1995. In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, vol. 3.3.6. Kluwer Academic Publishers, Dodrecht, The Netherlands.
- 3.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bancroft, J. D. 1990. Frozen and related sections, p. 81-92. In J. D. Bancroft and A. Stevens (ed.), Theory and practice of histological techniques. Churchill Livingston, London, United Kingdom.
- 6.Bratthauer, G. L. 1999. Preparation of frozen sections for analysis. Methods Mol. Biol. 115:57-62. [DOI] [PubMed] [Google Scholar]
- 7.Cary, S. C., and S. J. Giovannoni. 1993. Transovarial inheritance of endosymbiotic bacteria in clams inhabiting deep-sea hydrothermal vents and cold seeps. Proc. Natl. Acad. Sci. USA 90:5695-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crescenzi, A., C. Panunzi, S. Taccogna, E. Papini, C. M. Pacella, and F. Nardi. 1999. Cytospray fixation of frozen intraoperative sections. Mod. Pathol. 12:92-94. [PubMed] [Google Scholar]
- 9.Fink, S. 1978. Some new methods for affixing sections on glass slides. I. Aqueous adhesives. Stain Technol. 62:27-33. [DOI] [PubMed] [Google Scholar]
- 10.Fukatsu, T., K. Watanabe, and Y. Sekiguchi. 1998. Specific detection of intracellular symbiotic bacteria of aphids by oligonucleotide-probed in situ hybridization. Appl. Entomol. Zool. 33:461-472. [Google Scholar]
- 11.Gerlach, D. 1969. Botanische Mikrotechnik. Georg Thieme Verlag, Stuttgart, Germany.
- 12.Goto, H. E. 1960. Simple techniques for the rearing of collembola and a note on the use of a fungistatic substance in the cultures. Entomol. Mon. Mag. 46:138-140. [Google Scholar]
- 13.Heddi, A., A. M. Grenier, C. Khatchadourian, H. Charles, and P. Nardon. 1999. Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc. Natl. Acad. Sci. USA 96:6814-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann, A., T. Thimm, M. Dröge, E. R. B. Moore, J. C. Munch, and C. C. Tebbe. 1998. Intergeneric transfer of conjugative and mobilizable plasmids harbored by Escherichia coli in the gut of the soil microarthropod Folsomia candida (Collembola). Appl. Environ. Microbiol. 64:2652-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann, A., T. Thimm, and C. C. Tebbe. 1999. Fate of plasmid-bearing, luciferase marker gene tagged bacteria after feeding to the soil microarthropod Onychiurus fimatus (Collembola). FEMS Microbiol. Ecol. 30:125-135. [DOI] [PubMed] [Google Scholar]
- 16.Le Gullec, D. 1998. Ultrastructural in situ hybridization: a review of technical aspects. Biol. Cell 90:297-306. [PubMed] [Google Scholar]
- 17.Moter, A., G. Leist, R. Rudolph, K. Schrank, B.-K. Choi, M. Wagner, and U. B. Goebel. 1998. Fluorescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology 144:2459-2467. [DOI] [PubMed] [Google Scholar]
- 18.Pace, N., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by rRNA sequences. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 19.Schramm, A., D. de Beer, M. Wagner, and R. Amann. 1998. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl. Environ. Microbiol. 64:3480-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stackebrandt, E., and F. A. Rainey. 1995. Partial and complete 16S rDNA sequences, their use in generation of 16S rDNA phylogenetic trees and their implications in molecular ecology studies, p. 1-17. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, vol. 3.1.1. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 21.Stahl, D. A. 1997. Molecular approaches for the measurement of density, diversity, and phylogeny, p. 102-114. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 22.Thimm, T., A. Hoffmann, H. Borkott, J. C. Munch, and C. C. Tebbe. 1998. The gut of the soil microarthropod Folsomia candida (Collembola) is a frequently changeable but selective habitat and a vector for microorganisms. Appl. Environ. Microbiol. 64:2660-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanderkerckhove, T. T. M., S. Watteyne, A. Willems, J. G. Swings, J. Mertens, and M. Gillis. 1999. Phylogenetic analysis of the 16S rDNA of the cytoplasmic bacterium Wolbachia from the novel host Folsomia candida (Hexapoda, Collembola) and its implications for wolbachial taxonomy. FEMS Microbiol. Lett. 180:279-286. [DOI] [PubMed] [Google Scholar]
- 24.Wagner, M., R. Amann, H. Lemmer, and K. H. Schleifer. 1993. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl. Environ. Microbiol. 59:1520-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 26.Woese, C. R., R. O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eukarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]