Abstract
Several bacterial strains that can use organophosphate pesticides as a source of carbon have been isolated from soil samples collected from diverse geographical regions. All these organisms synthesize an enzyme called parathion hydrolase, and in each case the enzyme is encoded by a gene (opd) located on a large indigenous plasmid. These plasmids show considerable genetic diversity, but the region containing the opd gene is highly conserved. Two opd plasmids, pPDL2 from Flavobacterium sp. and pCMS1 from Pseudomonas diminuta, are well characterized, and in each of them a region of about 5.1 kb containing the opd gene shows an identical restriction pattern. We now report the complete sequence of the conserved region of plasmid pPDL2. The opd gene is flanked upstream by an insertion sequence, ISFlsp1, that is a member of the IS21 family, and downstream by a Tn3-like element encoding a transposase and a resolvase. Adjacent to opd but transcribed in the opposite direction is an open reading frame (orf243) with the potential to encode an aromatic hydrolase somewhat similar to Pseudomonas putida TodF. We have shown that orf243 encodes a polypeptide of 27 kDa, which plays a role in the degradation of p-nitrophenol and is likely to act in concert with opd in the degradation of parathion. The linkage of opd and orf243, the organization of the genes flanking opd, and the wide geographical distribution of these genes suggest that this DNA sequence may constitute a complex catabolic transposon.
Organophosphorus pesticides are used worldwide to control major insect pests. These pesticides inhibit acetylcholinesterase in an irreversible manner and cause insect death. Since acetylcholinesterase is also present in all vertebrates, the potential for damage by this class of insecticides to nontarget organisms is extremely high, and the use of many such pesticides is now banned in most developed countries. However, in third-world countries they remain the major insecticides in agricultural pest management, and consequently there is a need for economical and reliable methods of detoxifying organophosphate wastes. A number of soil bacteria have been found to be capable of degrading parathion. They synthesize parathion hydrolase, which is encoded by the opd (for “organophosphate degradation”) gene and which carries out the first step in parathion degradation, leading to the production of p-nitrophenol and diethylphosphoric acid (21). Microbial enzymes such as parathion hydrolase are thought to play a significant role in the degradation of parathion and related organophosphate insecticides in the soil (28), and this has stimulated studies of the organisms that are capable of synthesizing such enzymes (12, 15, 16, 18).
A parathion hydrolase-producing Flavobacterium sp. was isolated in 1972 from a rice field in the Philippines (27). In 1982 a strain of Pseudomonas diminuta that could utilize parathion as a carbon source was isolated in Texas (25), and in 1995 an organism identified as Flavobacterium balustinum, which grew on methylparathion as the sole carbon source, was isolated from agricultural soils close to Anantapur in Andhra Pradesh, India (30). In each of these organisms the opd gene was found to be located on a large plasmid (9, 19, 30). The plasmids were all unrelated as judged by Southern hybridization, but sequencing of the opd genes showed that the nucleotide sequences of the Philippines and Texas isolates were 100% identical and that of the Indian isolate was 98% identical (17, 26, 30a). It should be noted that the P. diminuta opd sequence in the DNA sequence databases (accession number M20392) has been reported to be incorrect (17). The corrected sequence, determined by Serdar et al. (26), is identical to that for the Flavobacterium pPDL2 sequence, but it does not appear to have been lodged in the databases.
Studies of two of the plasmids, pPDL2 from Flavobacterium sp. strain ATCC 27551 and pCMS1 from P. diminuta strain MG, by restriction analysis and hybridization experiments indicated that the opd genes were located in a highly conserved region that was estimated to extend approximately 2.6 kb upstream and 1.7 kb downstream of opd (19, 20). No homology was evident between the two plasmids outside this region, and the possibility that the opd gene was carried on a transposon was discussed (20). However, this hypothesis has not been investigated further, and to date there have been no studies of the sequences flanking the opd genes. We now report the complete sequence of the conserved region of Flavobacterium plasmid pPDL2 and the identification of a number of features suggesting that opd could be carried on a complex transposon. We also describe a novel gene, linked to opd, that is involved in the degradation of p-nitrophenol.
MATERIALS AND METHODS
Bacterial strains and plasmids.
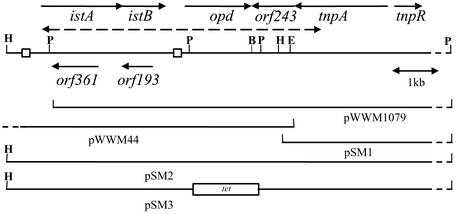
The strains and plasmids used in this study are listed in Table 1. A library of PstI and EcoRI fragments from Flavobacterium plasmid pPDL2 was created by Mulbry et al. (20), and the subclones pWWM1079 and pWWM44, which had been shown to encompass and flank the opd gene, were used as the starting material for sequencing. The relationship to the opd gene cluster of each of the plasmids used in this study is shown in Fig. 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| E. coli JM109(DE3) | recA1 endA1 gyrA96 thi hsrdR17 supE44 relA1 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | 37 |
| E. coli DH5α | F−recA1 endA1 gyrA96 thi-1 hsrdR17 supE44 relA1 deoR Δ(lacZYA-argF)U169 | 35 |
| Plasmids | ||
| pET-15b | Apr, expression vector | Novagen |
| pMMB206 | Cmr, broad-host-range low-copy-number expression vector | 14 |
| pSG335 | Cmr, sacB lacZ | 8 |
| pSM1 | Apr, 5.2-kb fragment (HindIII-PstI) in pUC19 | This work |
| pSM2 | Apr, complete opd gene cluster cloned in pUC19 | This work |
| pSM3 | Apr Tcr, tet gene in the opd coding region of pSM2 | This work |
| pSM4 | orf243 cloned in pET-15b as an NcoI-BamHI fragment | This work |
| pSM5 | opd gene cloned in pMMB206 under the control of the lacZ promoter | This work |
| pSU18 | Cmr, pACYC184-derived cloning vector | 4 |
| pWWM44 | Apr, 9.5-kb EcoRI fragment cloned in pUC19 | 17 |
| pWWM1079 | Apr, 9.3-kb PstI fragment cloned in pUC19 | 17 |
FIG. 1.
Map of the sequenced region from pPDL2. The eight ORFs identified are shown, together with the locations of the IR sequences that flank ISFlsp1 (□). The extent of the plasmid subclones (pWWM1079, pWWM44, and pSM1) used as the starting material for sequencing is indicated, and the region identified by Mulbry et al. (20) as homologous to plasmid pCMS1 is indicated by a dashed arrow. The complete opd gene cluster (on pSM2) and insertion of tet gene in the coding region of the opd gene (pSM3) are indicated. BamHI, EcoRI, HindIII, and PstI sites in the sequence are indicated by B, E, H, and P, respectively.
DNA sequencing and analysis.
Automatic sequencing was performed using BigDye polymerase (Applied Biosystems) followed by analysis on an ABI sequencer. All the pWWM plasmids were cloned in pUC19 (20), and these were sequenced initially using universal and reverse primers. Where necessary, further sequence was derived by subcloning internal fragments or designing specific primers from the known sequence. Computer analysis of the nucleotide sequences was performed with the GCG suite, and open reading frames (ORFs) were identified using FramePlot (11). Analysis of the polypeptides predicted to be encoded by each ORF was performed using BLAST and PSI-BLAST (1, 2), and alignments were carried out with Clustal W (34).
Growth media.
Bacteria were grown at 37°C either in Luria-Bertani LB medium or in M9 medium (24). When necessary, ampicillin (100 μg/ml), tetracycline (10 μg/ml), or chloramphenicol (15 μg/ml) was added to the growth medium. Filter-sterilized sucrose (50%) stock solution was added to Luria agar to prepare Luria agar sucrose (5%) plates.
Cloning and overexpression of orf243.
The product of orf243 was overexpressed by cloning the gene in expression vector pET-15b. Since there are no suitable restriction sites to facilitate cloning, the gene was amplified by PCR using a forward primer (5′ GCACCATGGCGCATGCCCGCGTTC 3′) containing an NcoI site (shown in bold type) overlapping the initiating methionine codon and a reverse primer (5′ CTGTGACTAACCCGGCGCGGTTC 3′) corresponding to the 3′ end of the gene. Since there is a BamHI site downstream of the stop codon of orf243, the PCR-amplified fragment was digested with NcoI and BamHI and cloned in pET-15b digested with similar enzymes. The resulting recombinant plasmid was designated pSM4.
To examine the expression of orf243 in both the presence and absence of opd, the opd gene was cloned under the control of the lacZ promoter in the broad-host-range plasmid vector pMMB206. The opd gene was amplified by PCR from pSM12, which carries opd as an NdeI-XhoI fragment cloned in pET23b, using vector-specific primers. The forward primer (GCCAGAATTCAGGGAGACCACAACGGTTTCACT) contained an EcoRI site (shown in bold type) upstream of the opd ribosomal binding site and the reverse primer (GCCAGGATCCCAAAAAACCCCTCAAGACCC) contained a BamHI site (shown in bold type) downstream of the region specifying the C-terminal His tag encoded in pET23b. The resultant 1.2-kb PCR product was cloned in pMMB206 as an EcoRI-BamHI fragment to give pSM5.
Overnight cultures of Escherichia coli strains JM109(DE3)(pSM5) and JM109(DE3)(pSM5)(pSM4) were subcultured into 10 ml of LB medium and allowed to grow until the culture density, measured by the absorbance at 600 nm, reached 0.5. Gene expression was induced by adding 100 μl of 0.1 M isopropyl-β-d-thiogalactopyranoside (IPTG), and 2 h later protein extracts prepared from these cultures were analyzed by sodium dodecyl sulfate-polyacrylamid gel electrophoresis (12.5% polyacrylamide).
Cloning of the opd gene cluster.
In the plasmid library constructed from the indigenous plasmid pPDL2, the recombinant plasmids pWWM1079 and pWWM44 contained the entire conserved region of the opd gene cluster in an overlapping manner (20). pWWM1079 contains the DNA region located downstream of the opd gene, whereas pWWM44 contains the DNA region upstream of the opd gene; in order to perform a transposition assay, it was necessary to have the entire conserved region on a single plasmid (Fig. 1). There is a unique HindIII site downstream of the opd gene on plasmid pWWM1079 in addition to the HindIII site found in the multiple-cloning site of the vector. Therefore, isolation and religation of the larger (8-kb) HindIII fragment from pWWM1079 gave a plasmid (pSM1) carrying part of orf243 with tnpA and tnpR. The conserved region upstream of opd was isolated on a 5.2-kb HindIII fragment from plasmid pWWM44 and cloned into pSM1 in the correct orientation to reconstitute the entire region, thereby giving pSM2.
Introduction of a tetracycline resistance marker into the opd gene.
To assess the transposition ability of the opd gene cluster, we introduced an antibiotic resistance marker into the cluster. Since there are no known unique restriction sites in pSM2, a tetracycline resistance gene was introduced into the coding region of opd by site-specific recombination. Two PCR primers were synthesized. In both the forward (5′ CGC CGC GCC AGA GCG GCT GGC GTG CGA ACG ATT GTC GAT AGG CCT CGC CGG CTT CCA TTC AGG 3′) and reverse (5′ CCG TGT TTG CCA CGA ACG GAT GCC CAG GAG GGC TGA TGC AGG CCT AAT AGG CGT ATC ACG AGG 3′) primers, the first 39 bases (shown in bold type) represent opd-homologous sequences and the remaining 24 bases are specific to the tetracycline resistance gene. The tetracycline (tet) gene was then amplified using pBR322 as template to give a 1.3-kb PCR fragment containing the tet gene flanked by coding sequences of the opd gene. The PCR fragment was then electroporated into E. coli strain BW25113 containing pSM2, and in vivo mutagenesis was performed as described elsewhere (7). The plasmid containing the tet gene were isolated, and the position of tet gene was confirmed by digesting the plasmid with BamHI. The resultant plasmid, having the tet gene in the correct location, was designated pSM3.
Transposition assay.
E. coli DH5α cells carrying plasmids pSM3 and pSU18 were allowed to grow overnight in LB medium in the absence of any antibiotics. Plasmid DNA was then isolated from this culture and retransformed into E. coli cells; transformants were selected on LB plates containing tetracycline and chloramphenicol. The resultant colonies were replica plated on LB plates containing ampicillin and screened for ampicillin-sensitive colonies that potentially contained the transposed element, marked by tetracycline resistance in pSU18. Alternatively, E. coli DH5α cells containing plasmids pSM3 and pSG335 were allowed to grow for 24 h before being plated on LB plates containing 5% sucrose. Plasmids were isolated from sucrose-resistant colonies and analyzed to investigate the presence of the opd gene cluster in the sacB gene of pSG335.
Degradation of PNP.
E. coli JM109 cells with and without the expression plasmid pSM4 were harvested from 100 μl of overnight culture and washed twice with saline before inoculation into 10 ml of M9 medium. The cultures were allowed to grow until the culture density, measured by the absorbance at 600 nm, reached 0.5. At this stage, the JM109(pSM4) culture was divided into two, and one portion was induced by adding 50 μl of 0.1 M IPTG. Simultaneously, p-nitrophenol (PNP) was added to all the cultures to a final concentration of 20 μg/ml, and the cultures were incubated at 37°C for a further 8 h. After this time, the cells were separated from the medium by a brief centrifugation, the supernatant was collected into a clean conical flask, and the PNP concentration was measured spectrophotometrically as described elsewhere (31).
RESULTS AND DISCUSSION
Previous studies comparing parathion hydrolase plasmids from Flavobacterium sp. (pPDL2) and P. diminuta (pCMS1) suggested, on the basis of hybridization data and restriction mapping, that the opd genes were located within a conserved region of 5 to 6 kb (20). Using the restriction map of pPDL2 (20) as a guide, we used the previously cloned PstI fragments that flank the opd gene and sequenced them upstream and downstream to give a contiguous sequence of just over 7.6 kb (EMBL accession number AJ421424). Every restriction site mapped by Mulbry et al. (20) within this region of pPDL2 was identified at an appropriate location within the sequence, which contains a total of eight ORFs (Fig. 1).
The opd gene of pPDL2.
The sequence of the pPDL2 opd gene was published previously and extends from a BamHI site 420 bp upstream of the opd coding sequence to a PstI site 180 bp downstream of opd (17). There is an untranslated region of 266 bp upstream of opd which contains a number of repeat sequences (see below) and the opd promoter (Siddavatam, unpublished) for which two candidate sequences have previously been proposed. A −35, −10 motif was identified by Mulbry and Karns (17), and a possible σ54-dependent promoter motif was suggested by Harper et al. (9). The latter proposal is untenable because the sequence proposed by these authors has a GG-N11-GC motif and it is now firmly established that all σ54-dependent promoters conform to a GG-N10-GC motif (3). The parathion hydrolase encoded by the opd gene on pPDL2 is 96% identical in its primary amino acid sequence to the enzyme encoded by an indigenous plasmid in F. balustinum (EMBL accession number AJ426431) (30a). A further sequence of a closely related opd gene has recently been reported from an Agrobacterium radiobacter strain (10). The DNA sequence of this gene (designated opdA) is 87% identical to the Flavobacterium gene, but the genomic location of the Agrobacterium gene, i.e., chromosomal or plasmid borne, has not been reported.
The ORF downstream of opd encodes a protein that is involved in the degradation of p-nitrophenol.
The 750 bp downstream of opd contains an ORF transcribed towards opd and potentially encoding a polypeptide of 243 amino acids. BLAST searches found no very strong homologies. The most statistically significant matches were to dipeptidyl aminopeptidases and esterases, but the similarities extended over only part of the polypeptide. However, similarity was detected, over the full length of the polypeptide, to a large protein family (pfam00561) that is characterized by a common alpha/beta-hydrolase fold. An independent analysis used a web-based program for protein fold recognition, 3D-PSSM (http://www.sbg.bio.ic.ac.uk/∼3dpssm/). This program uses a fold prediction algorithm followed by a search of a protein fold library, and all the significant matches identified by this method were also members of the alpha/beta-hydrolase family. The best structural match, with a 95% confidence prediction, was to Pseudomonas fluorescens chloroperoxidase F (Protein Data Bank PDB 1A8S).
Within the alpha/beta-hydrolase family, BLAST analysis indicated that the ORF243 protein showed similarity to a group of proteins that includes Pseudomonas putida DmpD, TodF, and XylF, Pseudomonas azelaica HbpD, Rhodococcus sp. EtbD and BphD, and P. fluorescens CumD. All these proteins are hydrolases that are involved in the degradation of various aromatic compounds including xylene and toluene. A phylogenetic analysis showed ORF243 to be the most distantly related within this group of proteins, being somewhat more closely related to HbpD and BphD, to which it shows 12% identity and 20% similarity, respectively. The proteins in the alpha/beta-hydrolase family have a wide variety of catalytic functions, but they all contain a catalytic triad composed of a serine, an aspartate, and a histidine residue. These residues are borne on a series of loops that are the best-conserved structural features of the fold. Candidate residues for the catalytic triad in ORF243 are Ser58, Asp183, and His215, and it is notable that most of the residues that are conserved between ORF243 and the XylF family are located around these amino acids.
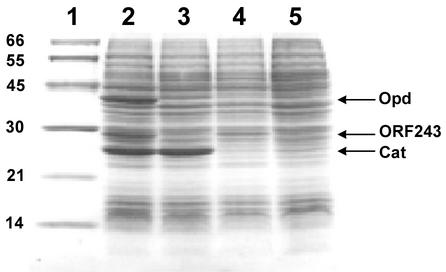
To demonstrate that orf243 encodes the predicted polypeptide, the gene was cloned into the vector pET-15b to give plasmid pSM4 and expression was induced in E. coli strains JM109(DE3) and JM109(DE3)(pSM5). A 27-kDa protein band was present only in the induced cultures containing pSM4, and the polypeptide was present at a notably higher level when Opd was expressed (from pSM5) in the same cells (Fig. 2). This stabilization and the close linkage of the two genes suggested that these proteins may function as a complex and may have a related function. Since orf243 potentially encodes a protein with homology to a family of aromatic compound hydrolases, this raised the question whether the protein might play a role in parathion degradation. Parathion hydrolase (Opd) degrades parathion to diethylphosphoric acid and PNP, which is itself a toxic pollutant. However, a number of organisms, including strains of Flavobacterium, are known to be able to utilize PNP for growth (6).
FIG. 2.
Expression of orf243 in E. coli. Lanes: 1, molecular mass markers (in kilodaltons); 2 and 3, protein extracts prepared from induced (lane 2) and uninduced (lane 3) cultures of E. coli JM109(DE3) cells containing both the expression plasmids pSM4 (orf243) and pSM5 (opd); 4 and 5, protein extracts prepared from the induced (lane 4) and uninduced (lane 5) cultures of E. coli JM109(DE3) cells carrying only pSM4 (orf243). Expression of parathion hydrolase (Opd), ORF243, and vector-encoded chloramphenicol acetyltransferase (Cat) is indicated by arrows.
We therefore assessed the ability of ORF243 to degrade PNP by adding the compound to cultures of JM109(DE3) and JM109(DE3)(pSM4) and measuring the PNP concentration after 8 h of incubation with or without induction of ORF243. In induced cultures of JM109(DE3)(pSM4), the concentration of PNP was reduced from 20 to 3.1 μM, whereas no significant reduction was observed in uninduced JM109(DE3)(pSML) and control JM109(DE3) cultures. Hence, the ORF243 protein does indeed appear to be able to degrade PNP, and the nature of the degradation products is currently under investigation. No other proteins with strong homology to ORF243 have been reported to date, and the catalytic activity of the protein may therefore be novel.
Is opd carried on a transposable element?
The occurrence of a nearly identical gene sequence at three widely separated geographical locations, together with the apparent homology of flanking DNA sequences, suggested that the genes may be carried on a transposable element, but no evidence for transposition has so far been reported (20).
Sequence analysis revealed that opd and orf243 are flanked by an insertion sequence encoding a complete istAB operon and by tnpA and tnpR genes characteristic of Tn3 family transposons (Fig. 1). A 2.5-kb region upstream of opd contains two ORFs transcribed in the same direction as opd and potentially encoding proteins of 507 and 279 amino acids, respectively. A BLAST search showed these proteins to have significant homology to the IstA and IstB proteins encoded by members of the IS21 family of transposons (13, 23). As is frequently observed in IS21-like sequences, the two genes are translationally coupled, indicating that their products are functionally related. Alignment of the predicted amino acid sequences with other IS21 family IstA and IstB proteins showed them to be most closely related to the Ist proteins from two other IS21-like transposons, IS1326 from Pseudomonas aeruginosa (5) and an insertion sequence found in the chromosome of Agrobacterium tumefaciens C58 (EMBL accession number AE008199). The A. tumefaciens proteins IstA and IstB proteins (AAK88585 and AAK88586) are the most closely related to those from Flavobacterium, showing 52 and 63% identity, respectively. The insertion sequence from Flavobacterium pPDL2 was designated ISFlsp1, and the sequence has been deposited in the IS database at the Laboratoire de Microbiologie et Genetique Moleculaire, Centre National de la Resercha Scientifique, Toulouse, France (http://www-is.biotoul.fr).
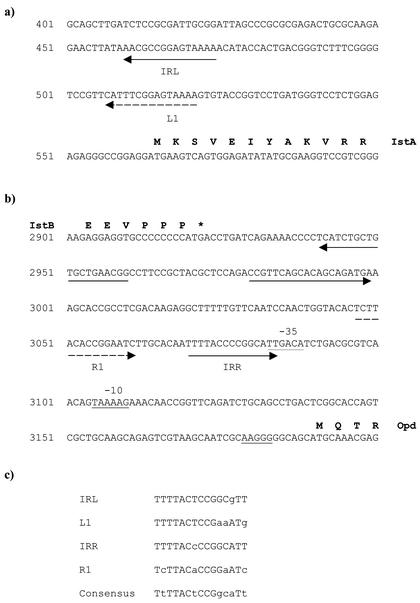
Members of the IS21 family typically have flanking inverted-repeat (IR) sequences of between 11 and 50 bp (13). In ISFIsp1, an IR sequence of 15 bp is found 100 bp upstream of istA (IRL) and 150 bp downstream of istB (IRR) (Fig. 3a and b). Many IS21 members, although not IS21, also contain direct-repeat sequences at each end (13), and in ISFlsp1 imperfect direct repeats of IRL and IRR are present 30 bp downstream of IRL and 10 bp upstream of IRR (Fig. 3c). Searches for repeat sequences in these regions also revealed an extended IR of 20 bp between the 3′ end of istB and IRR, a feature that is not a characteristic of IS21-like sequences (Fig. 3b). Outwardly directed −35 promoter hexamers are located within the terminal IRs of a number of IS elements including IS21 (13, 23). A potential −35 hexamer was originally identified upstream of opd, 102 bp upstream of the initiation codon (17). This −35 sequence is not located wholly within IRR but, rather, spans the IR and the adjacent sequence (Fig. 3b).
FIG. 3.
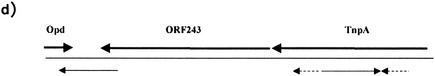
IR sequences of ISFlsp1 and repeat sequences flanking orf243. (a and b) The DNA sequences containing IRL and L1 (a) and IRR and R1 (b) are shown. The coding sequences of IstA, IstB, and Opd are indicated above the DNA sequences. Proposed ribosome binding sites and −35, −10 promoter motifs are underlined. Direct imperfect repeats of IRL and IRR are shown as dashed arrows, and an extended IR sequence downstream of istB is shown by two divergent arrows. (c) Alignment of all four repeat sequences and the derived consensus. (d) Map of the relative positions and orientations of the 87-bp IRs (solid arrows) upstream and downstream of orf243 and the 32-bp direct repeats (broken arrows) within tnpA.
Two ORFs potentially encoding proteins of 193 and 361 amino acids were found on the complementary strand to istA and istB (Fig. 1). BLAST searches of protein databases found no significant matches to either ORF193 or ORF361, but a TBLASTN search identified just two potential homologues, one for each ORF, encoded on the complementary strand of the region encoding IstA and IstB in A. tumefaciens C58 (described above). However, in each case the potential homologues showed only around 30% identity to the ISFlsp1 ORFs, and each was interrupted by a stop codon. The occurrence of ORFs on the complementary strand to istAB has been reported previously in IS1162 and IS1474, which are also members of the IS21 family (29, 38). This led us to investigate the possibility that such ORFs were a characteristic of all IS21-like elements, which could suggest that their products play a role in the transposition process. However, the potential proteins encoded in ISFlsp1 show no homology to those encoded in IS1162 and IS1474. We also examined the sequences of a number of other IS21 family members including IS21, IS232a, IS408, IS1491, and IS5376 but found no evidence for such ORFs being a conserved feature. It therefore remains to be established whether these ORFs encode expressed polypeptides, although it is certainly possible that they are simply a consequence of a relatively high level of DNA sequence conservation for the istA, istB operon.
Immediately downstream of orf243 are two ORFs that encode proteins with significant homology to the TnpA and TnpR proteins that characterize class II transposons such as Tn3. The organization of the transposase genes within the Tn3 family falls into two classes, one in which tnpR and tnpA are transcribed in the same direction and one in which they are divergently transcribed. The opd-linked tnpA and tnpR genes are divergently transcribed, as found in Tn3, Tn1000, Tn2501, and Tn4430.
The transposase (TnpA) proteins of Tn3-related transposons belong to a conserved family of proteins (pfam01562) that are typically around 970 amino acids in length, although one related tnpA gene in Yersinia pestis encodes a protein of only 771 amino acids (accession number AAC62594). The tnpA gene of pPDL2 is unusual because it encodes a protein of only 583 residues. The first 165 residues of the protein show a high level of homology to the N-terminal region of the TnpA family, and the C-terminal 390 residues are highly homologous to the C-terminal region of the TnpA family. Hence, the reduced length is largely accounted for by the absence of a single region of around 400 residues in the center of the protein sequence. An invariant triad D-D-E motif in all transposases has been proposed to be involved directly in catalysis (39), and this motif is completely conserved in the shortened TnpA protein, being represented by Asp237, Asp310, and Glu440. Hence, while the TnpA protein from pPDL2 contains many of the critical residues known to be required for activity, it is certainly possible that the protein is not functional. The stop codon of the tnpA gene overlaps with the predicted start codon of orf243, suggesting that under some conditions at least, the two genes might be translationally coupled.
Transcribed divergently from tnpA is the tnpR gene, which encodes a protein of 148 amino acids. The predicted protein shows homology throughout its length to other TnpR invertase or resolvase proteins belonging to pfam00239, such as that encoded by Tn5501 from Pseudomonas pseudoalcaligenes. However, by far the greatest similarity (82% identical residues) is found to a resolvase (accession number BAA87651) encoded on the Ti plasmid, pTi-Sakura, found in a strain of A. tumefaciens (32).
A search for repeat sequences within the opd region revealed an IR sequence comprising a perfectly matched sequence of 87 bp, one arm of which extends from the 3′ end of opd across the downstream untranslated region and into the 3′ end of orf243 (Fig. 3d). The complementary arm of this sequence is located within the tnpA coding sequence, and this repeat is in turn flanked by two perfect direct repeats of 32 bp (Fig. 3d). The origin of these repeats is unclear, but they suggest that the DNA in this region has undergone major rearrangements. Analysis of the available DNA sequence downstream of P. diminuta opd and F. balustinum opd indicates that the IR that includes the untranslated region between opd and orf243 is conserved in both these sequences, with just one mismatch in 87 bp (26, 30a), suggesting that this complex repeat is not a recent event. There is also a 39-bp perfect match to part of this repeat located just downstream of the A. radiobacter opdA gene.
Given the presence of an insertion sequence and a tnpA-like gene, we examined whether the complete opd gene cluster would transpose in E. coli. A selectable marker (tetracycline resistance) was introduced by cloning the tet gene (amplified from pBR322) into the opd gene on plasmid pSM2 to give pSM3. We then looked for transposition of the cluster from pSM3 either onto pSU18 or into the sacB gene on plasmid pSG335. After repeat subculture of both strains and screening in excess of 1010 cells, no transposition events were detected.
Although we were unable to detect any movement of the element in E. coli, we do not know whether specific conditions, nutritional or environmental, might induce transposition or whether other genetic factors present only in the natural host might be required. Nevertheless, our observations on the linkage of opd to IS elements and transposase genes support the idea that the widespread distribution of the opd gene could be due to its lateral transfer by a combination of transposition and plasmid transfer. Comparative analysis of the opd regions of Flavobacterium plasmid pPDL2 and the P. diminuta plasmid pCMS1 originally suggested that they had a common region of approximately 5.1 kb (20). While the resolution achievable by Southern hybridization is limited, the region delineated by Mulbry and colleagues suggests that only istA, istB, opd, and orf243 are definitely conserved. Demonstration of the conservation of tnpA and tnpR requires further sequencing.
The occurrence of catabolic transposons is now well documented, and they include elements encoding genes for degradation of toluene, naphthalene, chlorobenzene, benzene, and many other xenobiotic chemicals (33, 36). These transposons include both class I composite elements, in which catabolic genes are flanked by IS elements, and class II transposons that are all ancestrally related to the Tn3 family of transposons and are characterized by short (fewer than 50 bp) IRs and the involvement of a transposase (TnpA) and a resolvase (TnpR). However, the detailed structures of these catabolic transposons are extremely varied, and composite elements with features of both class I and class II are also known (22). The evolutionary origins of the opd gene and of the gene cluster that we describe in this paper are presently unknown. However, it is notable that the closest homologues of the istA, istB, and tnpR genes are all found in strains of A. tumefaciens and that an opd gene homologue has now also been reported in an A. radiobacter strain (10). These similarities could indicate an evolutionary origin for these genes in Agrobacterium.
Acknowledgments
We thank W. Mulbry for the provision of plasmids, without which this work would not have been possible.
This work was supported by UK-India Science and Technology Research Fund Project ref. 97AF02 and by a Wellcome Trust International Research Development award to D.S. and M.M. B.M. and S.K. acknowledge support from CSIR Junior Research Fellowships. Work in the laboratory of D.S. was also supported by the Department of Science and Technology, New Delhi, and work in the laboratory of M.M. was supported by the BBSRC through a grant-in-aid to the John Innes Centre.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of sigma54-dependent promoter sequences. Nucleic Acids Res. 27:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 5.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhry, G. R., A. N. Ali, and W. B. Wheeler. 1988. Isolation of a methyl parathion-degrading Pseudomonas sp. that possesses DNA homologous to the opd gene from a Flavobacterium sp. Appl. Environ. Microbiol. 54:288-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geissler, S., and M. Drummond. 1993. A counterselectable pACYC184-based lacZα-complementing plasmid with novel multiple cloning sites; construction of chromosomal deletions in Klebsiella pneumoniae. Gene 136:253-255. [DOI] [PubMed] [Google Scholar]
- 9.Harper, L. L., C. S. Mcdaniel, C. E. Miller, and J. R. Wild. 1988. Dissimilar plasmids isolated from Pseudomonas diminuta MG and a Flavobacterium sp. (ATCC 27551) contain identical opd genes. Appl. Environ. Microbiol. 54:2586-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horne, I., T. D. Sutherland, R. L. Harcourt, R. J. Russell, and J. G. Oakeshott. 2002. Identification of an opd (organophosphate degradation) gene in an Agrobacterium isolate. Appl. Environ. Microbiol. 68:3371-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 12.Karns, J. S., M. T. Muldoon, W. W. Mulbry, M. K. Derbyshire, and P. C. Kearney. 1987. Use of microorganisms and microbial systems in the degradation of pesticides, p. 156-170. In H. M. LeBaron, R. O. Mumma, R. C. Honeycutt, and J. H. Duesing (ed.), Biotechnology in agricultural chemistry. American Chemical Society, Washington, D.C.
- 13.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 15.Mulbry, W., and P. C. Kearney. 1991. Degradation of pesticides by microorganisms and the potential for genetic manipulation. Crop Prot. 10:334-346. [Google Scholar]
- 16.Mulbry, W. W. 1992. The aryldialkylphosphatase-encoding gene adpB from Nocardia sp strain B-1: cloning, sequencing and expression in Escherichia coli. Gene 121:149-153. [DOI] [PubMed] [Google Scholar]
- 17.Mulbry, W. W., and J. S. Karns. 1989. Parathion hydrolase specified by the Flavobacterium opd gene—relationship between the gene and protein. J. Bacteriol. 171:6740-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulbry, W. W., and J. S. Karns. 1989. Purification and characterization of three parathion hydrolases from gram-negative bacterial strains. Appl. Environ. Microbiol. 55:289-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulbry, W. W., J. S. Karns, P. C. Kearney, J. O. Nelson, C. S. Mcdaniel, and J. R. Wild. 1986. Identification of a plasmid-borne parathion hydrolase gene from Flavobacterium sp. by southern hybridization with opd from Pseudomonas diminuta. Appl. Environ. Microbiol. 51:926-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulbry, W. W., P. C. Kearney, J. O. Nelson, and J. S. Karns. 1987. Physical comparison of parathion hydrolase plasmids from Pseudomonas diminuta and Flavobacterium sp. Plasmid 18:173-177. [DOI] [PubMed] [Google Scholar]
- 21.Munnecke, D. M., and D. P. Hsieh. 1976. Pathways of microbial metabolism of parathion. Appl. Environ. Microbiol. 31:63-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakatsu, C., J. Ng, R. Singh, N. Straus, and C. Wyndham. 1991. Chlorobenzoate catabolic transposon Tn5271 is a composite class I element with flanking class II insertion sequences. Proc. Natl. Acad. Sci. USA 88:8312-8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reimmann, C., R. Moore, S. Little, A. Savioz, N. S. Willetts, and D. Haas. 1989. Genetic structure, function and regulation of the transposable element IS21. Mol. Gen. Genet. 215:416-424. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Serdar, C. M., D. T. Gibson, D. M. Munnecke, and J. H. Lancaster. 1982. Plasmid involvement in parathion hydrolysis by Pseudomonas diminuta. Appl. Environ. Microbiol. 44:246-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serdar, C. M., D. C. Murdock, and M. F. Rohde. 1989. Parathion hydrolase gene from Pseudomonas diminuta MG: subcloning, complete nucleotide-sequence, and expression of the mature portion of the enzyme in Escherichia coli. Bio/Technology 7:1151-1155. [Google Scholar]
- 27.Sethunathan, N., and T. Yoshida. 1973. A Flavobacterium sp. that degrades diazinon and parathion. Can. J. Microbiol. 19:873-875. [DOI] [PubMed] [Google Scholar]
- 28.Siddaramappa, R., K. P. Rajaram, and N. Sethunathan. 1973. Degradation of parathion by bacteria isolated from flooded soil. Appl. Microbiol. 26:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solinas, F., A. M. Marconi, M. Ruzzi, and E. Zennaro. 1995. Characterization and sequence of a novel insertion sequence, IS1162, from Pseudomonas fluorescens. Gene 155:77-82. [DOI] [PubMed] [Google Scholar]
- 30.Somara, S., and D. Siddavattam. 1995. Plasmid mediated organophosphate pesticide degradation by Flavobacterium balustinum. Biochem. Mol. Biol. Int. 36:627-631. [PubMed] [Google Scholar]
- 30a.Somara, S., B. Manavathi, C. Tebbe, and D. Siddavattam. 2002. Localisation of identical organophosphorus pesticide degrading (opd) genes on genetically dissimilar indigenous plasmids of soil bacteria: PCR amplification, cloning and sequencing of the god gene from Flavobacterium balustinum. Indian J. Exp. Biol. 40:774-779. [PubMed] [Google Scholar]
- 31.Spain, J. C., O. Wyss, and D. T. Gibson. 1979. Enzymatic oxidation of p-nitrophenol. Biochem. Biophys. Res. Commun. 88:634-641. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, K., Y. Hattori, M. Uraji, N. Ohta, K. Iwata, K. Murata, A. Kato, and K. Yoshida. 2000. Complete nucleotide sequence of a plant tumor-inducing Ti plasmid. Gene 242:331-336. [DOI] [PubMed] [Google Scholar]
- 33.Tan, H. M. 1999. Bacterial catabolic transposons. Appl. Microbiol Biotechnol. 51:1-12. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyndham, R. C., A. E. Cashore, C. H. Nakatsu, and M. C. Peel. 1994. Catabolic transposons. Biodegradation 5:323-342. [DOI] [PubMed] [Google Scholar]
- 37.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 38.Yeo, C. C., and C. L. Poh. 1997. Characterization of IS1474, an insertion sequence of the IS21 family isolated from Pseudomonas alcaligenes NCIB 9867. FEMS Microbiol. Lett. 149:257-263. [DOI] [PubMed] [Google Scholar]
- 39.Yurieva, O., and V. Nikiforov. 1996. Catalytic center quest: comparison of transposases belonging to the Tn3 family reveals an invariant triad of acidic amino acid residues. Biochem. Mol. Biol. Int. 38:15-20. [PubMed] [Google Scholar]