Abstract
Hydroxylamino aromatic compounds are converted to either the corresponding aminophenols or protocatechuate during the bacterial degradation of nitroaromatic compounds. The origin of the hydroxyl group of the products could be the substrate itself (intramolecular transfer mechanism) or the solvent water (intermolecular transfer mechanism). The conversion of hydroxylaminobenzene to 2-aminophenol catalyzed by a mutase from Pseudomonas pseudoalcaligenes JS45 proceeds by an intramolecular hydroxyl transfer. The conversions of hydroxylaminobenzene to 2- and 4-aminophenol by a mutase from Ralstonia eutropha JMP134 and to 4-hydroxylaminobenzoate to protocatechuate by a lyase from Comamonas acidovorans NBA-10 and Pseudomonas sp. strain 4NT were proposed, but not experimentally proved, to proceed by the intermolecular transfer mechanism. GC-MS analysis of the reaction products formed in H218O did not indicate any 18O-label incorporation during the conversion of hydroxylaminobenzene to 2- and 4-aminophenols catalyzed by the mutase from R. eutropha JMP134. During the conversion of 4-hydroxylaminobenzoate catalyzed by the hydroxylaminolyase from Pseudomonas sp. strain 4NT, only one of the two hydroxyl groups in the product, protocatechuate, was 18O labeled. The other hydroxyl group in the product must have come from the substrate. The mutase in strain JS45 converted 4-hydroxylaminobenzoate to 4-amino-3-hydroxybenzoate, and the lyase in Pseudomonas strain 4NT converted hydroxylaminobenzene to aniline and 2-aminophenol but not to catechol. The results indicate that all three types of enzyme-catalyzed rearrangements of hydroxylamino aromatic compounds proceed via intramolecular transfer of hydroxyl groups.
Nitroaromatic compounds can be degraded either oxidatively or reductively by microorganisms (25). Hydroxylamino intermediates are produced from nitroaromatic compounds in reactions catalyzed by nitroreductases in a number of bacteria (25). The hydroxylamino intermediates are then degraded via one of three different routes. In compounds with more than one nitro group, the intermediate is often reduced to the amine, as in the transformation of 2,4,6-trinitrotoluene (16, 28). In a second strategy, used during degradation of 4-nitrobenzoate (4-NBA) by a variety of isolates (6, 9, 27, 29, 35), the hydroxylamino compound is converted to protocatechuate in a reaction catalyzed by a lyase enzyme. A third strategy used by bacteria is the rearrangement of the hydroxylamino intermediates to the corresponding aminophenols (APs) as in the degradation of nitrobenzene (24, 26), 3-nitrophenol (30), chloronitrobenzene (17), 4-nitrotoluene (33), 2,4,6-trinitrotoluene (5, 14), and 2-NBA (10). Most of the APs can be further metabolized by an extradiol-like ring cleavage pathway, as observed in the degradation of nitrobenzene by Pseudomonas pseudoalcaligenes JS45 (19) and Pseudomonas putida HS12 (26), the degradation of 4-nitrotoluene by Mycobacterium sp. strain HL 4-NT-1 (13, 33), and by 3-hydroxyanthranilate 3,4-dioxygenase in Pseudomonas fluorescens strain KU-7 (T. Muraki, T. M., Y. Hasegawa, H. Iwaki, and P. C. K. Lau, Abstr. Am. Soc. Microbiol. Conf. Biodegradation Biotransformation Biocatalysis, abstr. 62, p. 42, 2001).
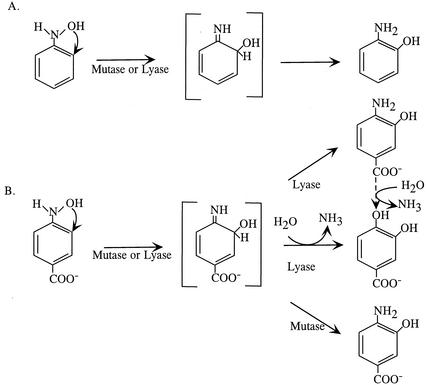
There are various possibilities for the transfer of the hydroxyl groups during conversion of hydroxylamino aromatic intermediates (Table 1). The origin of the hydroxyl group of the products could be the substrate (intramolecular transfer mechanism) or the solvent water (intermolecular transfer mechanism). The acid-catalyzed Bamberger rearrangement of hydroxylaminobenzene (HAB) to 4-AP proceeds via the intermolecular transfer mechanism (18) (Table 1). Recombinant Escherichia coli strains carrying the HAB mutase gene (habB) cloned from P. pseudoalcaligenes JS45 catalyze an intramolecular transfer of the hydroxyl group of HAB to form 2-AP (11). HAB mutase is potentially useful as a biocatalyst for the synthesis of o-APs (23). A second mutase gene, habA, was cloned from strain JS45, and subsequent genetic and biochemical experiments revealed that only habA is expressed in the wild type, but it is not known whether the enzyme catalyzes a similar intramolecular reaction (4). The mutase from Ralstonia eutropha JMP134 catalyzes the transformation of HAB to a mixture of 2- and 4-AP (31). It has been proposed that the reaction mechanism from the mutase from strain JMP134 is analogous to the acid-catalyzed Bamberger rearrangement because the enzyme requires no oxygen or added cofactors for its reaction (31). Considering the fact that the products in the reactions catalyzed by the JMP134 mutase are not limited to the one produced by either the Bamberger rearrangement or by the JS45 mutase (Table 1), the hypothesis must be tested experimentally.
TABLE 1.
Mechanisms of the conversion of hydroxylamino aromatic compounds
| Catalyst | Substrate | Product(s) [%] | Proposed mechanism | Evidence | Reference |
|---|---|---|---|---|---|
| Acid | HAB | 2-AP (Trace), 4-AP (100) | Intermolecular | Labeled product when the reaction is in H218O | 11, 18 |
| Mutase from strain JMP134 | HAB | 2-AP (37), 4-AP (63) | Intermolecular? | Hypothetical | 30, 31 |
| HabB mutase from strain JS45 | HAB | 2-AP (100) | Intramolecular | Nonlabeled product when the reaction is in H218O | 11 |
| Lyase from strains NBA-10 and 4NT | 4-HABA | Protocatechuate (100) | Intermolecular? | Hypothetical | 9, 22 |
4-Hydroxylaminobenzoate (4-HABA) lyase catalyzes the transformation of 4-HABA to protocatechuate in Pseudomonas strain 4NT and several other isolates (6, 9, 29, 35). In the protocatechuate product, one of the hydroxyl groups must originate from water, but the other could be either from water or from the substrate, 4-HABA. Meulenberg and de Bont (22) proposed that the hydroxyl group of 4-HABA is first released and then two hydroxyl groups from water are added to the benzene ring with the release of ammonia to form the product, protocatechuate (Table 1).
We sought to determine here whether the intramolecular mechanism observed in the conversion of HAB by HabB mutase in strain JS45 enzyme also applies to the two types of enzyme-catalyzed reactions that were previously thought to involve intermolecular transfer of hydroxyl groups. We have determined unequivocally the origin of the hydroxyl groups in the reaction products to provide insight into the reaction mechanism.
MATERIALS AND METHODS
Organisms and culture conditions.
P. pseudoalcaligenes JS45 was grown in nitrogen-free mineral medium containing nitrobenzene (2.5 mM) as described previously (24). Pseudomonas sp. strain 4NT was grown on either 4-NBA (1 g liter−1) or on glucose (2 g liter−1) (9). R. eutropha JMP134, kindly supplied by Alan R. Harker (Brigham Young University, Provo, Utah), was grown on succinate (10 mM) with 3-nitrophenol (0.5 mM) as the nitrogen source (30). Mycobacterium strain HL 4-NT-1 was grown on 4-nitrotoluene (0.5 mM) and succinate (10 mM) as reported previously (33).
Preparation of cell extract.
Cells were harvested by centrifugation, suspended in phosphate buffer (20 mM), and disrupted by two to four passages through a French pressure cell at 20,000 lb in−2. The cell extracts for the mutase assays were clarified by centrifugation at 16,000 × g for 20 min at 4°C. The pellet was discarded, and the supernatant fluids were stored at −80°C. The extracts from strain Pseudomonas sp. strain 4NT were prepared as described above, centrifuged 36,000 × g for 45 min at 4°C, and the supernatant fluids were used as the source of the hydroxylaminolyase (9). In some experiments cell extract containing hydroxylaminolyase was treated to remove NADPH by following the desalting-solvent exchange method according to the manufacturer's instructions (three times) in a Centriprep-3 filter (Amicon; Millipore, Bedford, Mass.) at 4°C prior to storage at −80°C.
Transformations.
The H218O enzymatic transformation reactions were performed in an anaerobic chamber (Coy Laboratory Products, Inc., Grass Lake, Mich.) because hydroxylaminoarenes are readily oxidized (3). The mutase reactions were carried out in 0.5 ml of phosphate buffer (20 mM, pH 7.0) containing 33% H218O (nominal) and 20 μl of extract from cells of strains JS45 (0.656 mg of protein), JMP134 (0.36 mg of protein), and HL 4-NT-1 (0.23 mg of protein). The reaction was initiated by adding HAB (4.6 μmol) to the reaction mixture. The formation of AP was monitored by high-pressure liquid chromatography (HPLC). After the reaction was complete the products were extracted with ethyl acetate, concentrated under nitrogen, and analyzed by gas chromatography-mass spectrometry (GC-MS).
The reaction mixture (0.5 ml) for transformations catalyzed by HABA lyase in Pseudomonas strain 4NT was prepared with either normal water or with 33% H218O (nominal) and contained phosphate buffer (20 mM, pH 7.0), NADPH (0.5 mM), crude extract (4.2 mg of protein per ml), and 4-HABA (1.1 μmol). After 2.5 h the reaction was stopped by the addition of 2 M HCl (final concentration, 0.6 mM). The product, protocatechuate (0.6 mM), was extracted into ethyl acetate (four times with equal volumes). Extracts were dried over sodium sulfate and concentrated under nitrogen. The ratio of 16O to 18O in the product was measured by GC-MS analysis as described above. The experiment described above was also carried out with reaction mixtures containing a higher percentage of labeled water to determine rigorously whether an intramolecular reaction mechanism results in the production of protocatechuate. The test and control reactions were carried out in duplicate as described above except that 92% 18O-labeled water (nominal) in 1 mM 4-HABA (final concentration) was used in the reaction mixtures. The reaction mixtures were analyzed by HPLC-MS as described below.
The lyase activity in reactions containing extracts (Centriprep treated) from induced or uninduced cells was determined in 500 μl of phosphate buffer (pH 7.0, 20 mM) containing 0.5 mM concentrations of 4-HABA, HAB, 4-amino-3-hydroxybenzoate, and 2-AP without the addition of NADPH.
Analytical methods.
Protein concentrations were measured by the Pierce BCA protein assay kit (Rockford, Ill.) by using bovine serum albumin as the standard. Substrate transformations and metabolites of HAB were monitored by HPLC with a model 1050 HPLC apparatus (Hewlett-Packard [HP], Palo Alto, Calif.) equipped with an HP model 1040 diode array detector. HAB and related products were separated by using an ABZ column (Supelco, Bellefonte, Pa.) and a mobile phase consisting of acetonitrile-water (30:70 to 65:35 in 8 min, which was then held for 2 min at the final ratio) at a flow rate of 1.2 ml min−1. 4-HABA and protocatechuate were measured by using a previously described method (35). 4-HABA, protocatechuate, and 4-amino-3-hydroxybenzoate were separated on a phenyl column (Alltima C18; Alltech, Deerfield, Ill.) by using an isocratic mobile phase consisting of 20% methanol and 80% water (2% glacial acetic acid; pH 2.5); alternatively, 4-HABA and 4-amino-3-hydroxybenzoate were separated with an isocratic mobile phase consisting of 70% aqueous trifluoroacetic acid (0.1%) and 30% acetonitrile.
GC-MS was performed with an HP model 5890 GC coupled to an HP model 5970 mass selective detector. The injection mode was splitless and the injection inlet temperature was 280°C. The column was a DB-5MS (30 m by 0.25 mm by 0.25 mm; Agilent, Palo Alto, Calif.). The carrier gas was helium at a flow rate of 0.8 ml min−1. The GC oven temperature increased from 55 to 130°C at a rate of 30°C min−1. 4-HABA products were separated by using the same method except that the injection inlet temperature was 250°C, and the column temperature was initially 55°C for 1 min and then increased to 280°C at a rate of 20°C min−1. Metabolites were identified by full-scan electron impact MS with an ionizing voltage of 70 eV. GC-MS analyses of the H218O reagent and reaction mixtures were performed with a splitless injection and a constant GC oven temperature of 100°C.
HPLC-MS separation and analysis was done with a reversed-phase column (Polar; Phenomenex, Torrence, Calif.) and a mobile phase of 95:5 (H2O with 0.1% acetic acid-acetonitrile with 0.1% acetic acid) at a flow rate of 0.5 ml min−1. MS was performed with a ThermoFinnigan Advantage (Schaumburg, Ill.) equipped with electrospray ionization in the negative ionization mode.
Materials.
18O-labeled water (95%) was purchased from Fluka (Milwaukee, Wis.). HAB was provided by S. Nishino (24), and 4-HABA was synthesized by reducing 4-NBA with zinc as previously reported (1). 4-NBA, 2-AP, 4-AP, 4-amino-3-hydroxybenzoate, 3-amino-4-hydroxybenzoate, catechol, aniline, nitrobenzene, 4-nitrotoluene, and 3-nitrophenol were of the highest purity available from Sigma-Aldrich Chemical Company (Milwaukee, Wis.). Protocatechuate was from Pfaltz and Bauer (Waterbury Conn.), and NADPH was from Roche (Indianapolis, Ind.).
RESULTS AND DISCUSSION
Conversion of HAB to APs catalyzed by mutase enzymes.
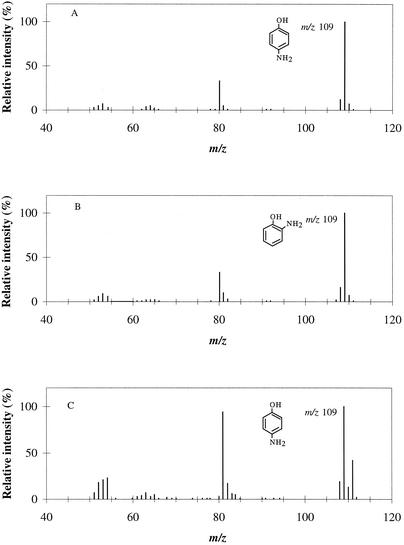
Previous work indicated that ca. 40% 2-AP and 60% 4-AP were formed from HAB in the reaction catalyzed by the mutase from R. eutropha JMP134 (30). When the experiment was conducted in H218O, GC-MS analysis of 2-AP and 4-AP from the enzymatic reaction revealed the molecular ion peaks at m/z 109 (Fig. 1), which indicated that the products contained only 16O, so neither AP isomer contained a hydroxyl group from the solvent water. In the acid-catalyzed reaction, the apparent molecular ion peaks of 4-AP were at m/z 109 and 111 in a ratio (68:32) matching the ratio of H216O to H218O (67:33) in the reaction mixture (11) (Fig. 1). The results indicate that the mutase in R. eutropha JMP134 catalyzes an intramolecular transfer of the hydroxyl group of HAB even when the hydroxyl group is transferred to the 4 position.
FIG. 1.
GC-MS identification of the products of conversion of HAB catalyzed by strain JMP134 mutase in 33% H218O. (A) Mass spectrum of the transformation product (4-AP) with an Rt of 13.8 min; (B) mass spectrum of the product (2-AP) with an Rt of 11.7 min; (C) control, i.e., the mass spectrum of 4-AP obtained in an acid-catalyzed reaction in 33% H218O (these data are from the previous study [11]).
We also examined the conversion of HAB catalyzed by the mutases from Mycobacterium strain HL 4-NT-1 (33) and from P. pseudoalcaligenes JS45 (data not shown). The predominant product in both cases was unlabeled 2-AP with a trace of unlabeled 4-AP. The results presented here clearly indicate that HabA, the mutase expressed in strain JS45 (4), also catalyzes an intramolecular reaction. The observations indicate that the intramolecular transfer of the hydroxyl group is a general mechanism for the mutase-catalyzed conversion of HAB regardless of whether the product is 2- or 4-AP.
Conversion of 4-HABA to protocatechuate catalyzed by the lyase.
Meulenberg and de Bont (22) suggested a hypothetical mechanism involving two hydrolysis reactions for the conversion of 4-HABA to protocatechuate by Comamonas acidovorans NBA-10 (22). We tested the hypothesis by incubating the extracts from cells of Pseudomonas strain 4NT with 4-HABA in reaction mixtures containing 18O-labeled water (33% H218O). The protocatechuate formed in the reaction was identified by HPLC. When the extract of the reaction mixture made with unlabeled water was analyzed by GC-MS, the product eluted at the same time (8.9 min) and showed the same mass spectrum as catechol. Protocatechuate decomposes at ca. 200°C (34), and when authentic protocatechuate was injected in the GC analysis, a compound with a molecular ion at m/z 110 eluted at 8.9 min. The result indicated that protocatechuate underwent a decarboxylation, probably in the injector. Authentic catechol gave the same retention time and mass spectrum. The catechol produced by decarboxylation of protocatechuate formed during the reaction in the 18O-labeled water gave molecular ions at m/z 110 and 112 (68:32), and a molecular ion at m/z 114 was not detected. The relative intensity matched the ratio of H218O to H216O in the reaction mixture. If both hydroxyl groups had been from water, the molecular ions would have been at m/z 110, 112, and 114 (45:44:11). The results indicated that only one hydroxyl group was labeled. Therefore, one hydroxyl group in the product resulted from rearrangement of the hydroxylamine, and the second was introduced during a hydrolysis reaction.
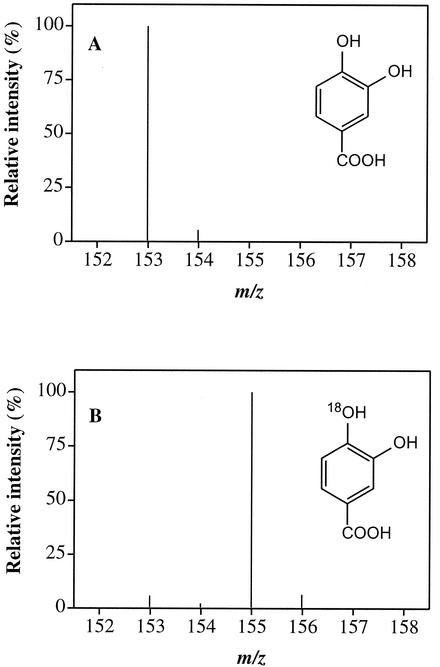
To avoid decarboxylation during the GC-MS analysis and to eliminate the introduction of unlabeled water associated with crude extracts, we used HPLC-MS to analyze for protocatechuate and used lyophilized extracts of Pseudomonas strain 4NT (0.52 mg of protein) to catalyze the reactions (500 μl of phosphate buffer [pH 7.0]). The lyophilized enzyme was less active than the crude extract, so the reactions were carried out for 180 min. Protocatechuate produced in duplicate reactions gave ions at m/z 153, 155, and 157 with mean intensity ratios of 5.5, 94.1, and 0.4, respectively, in the reactions with 18O-labeled water (92%) and with mean ratios of 98.9, 1.1, and 0, respectively, in the control reactions (Fig. 2). If both hydroxyl groups in the product were derived from water, the expected ratios in the reactions containing the 18O-labeled water would be 0.64:14.72:84.64. If sequential intra- and intermolecular reactions are involved the ratio between the intensities of the ions at m/z 153 and 155 should be 8:92, which agrees well with the observed ratio. The absence of an M+4 ion indicates clearly that there was no insertion of two hydroxyl groups from water. The observation that only one labeled hydroxyl group was incorporated in the molecule provides clear evidence that the lyase catalyzes an intramolecular transfer of the hydroxyl group from the hydroxylamino moiety.
FIG. 2.
HPLC-MS analysis of protocatechuate (M-1) enzymatically produced from 4-HABA. (A) Reaction mixture containing normal water; (B) reaction mixture containing 92% H218O.
When cell extracts of Pseudomonas sp. strain 4NT were incubated with 4-HABA (0.54 mM), the major product was protocatechuate (0.4 mM) and a minor amount of 4-amino-3-hydroxybenzoate (0.016 mM) also accumulated. Peres et al. (27) also detected trace amounts of 4-amino-3-hydroxybenzoate in the culture medium of Ralstonia paucula strain SB4 grown on 4-NBA. They concluded that a mutase was responsible for the minor dead-end product. P. putida TW3 growing in the presence of 4-NBA also appears to produce minor amounts of the 4-amino-3-hydroxybenzoate (15). Based on the 18O-labeled H2O experiments that we performed with 4-HABA as a substrate, it appears that the lyase can catalyze a reaction analogous to the one catalyzed by the mutase and could be involved in the production of 4-amino-3-hydroxybenzoate. Experiments by Hughes et al. with a cloned lyase expressed in a heterologous host have shown that the lyase is responsible for the conversion of 4-HABA to 4-amino-3-hydroxybenzoate (15).
Conversion of HAB by lyase.
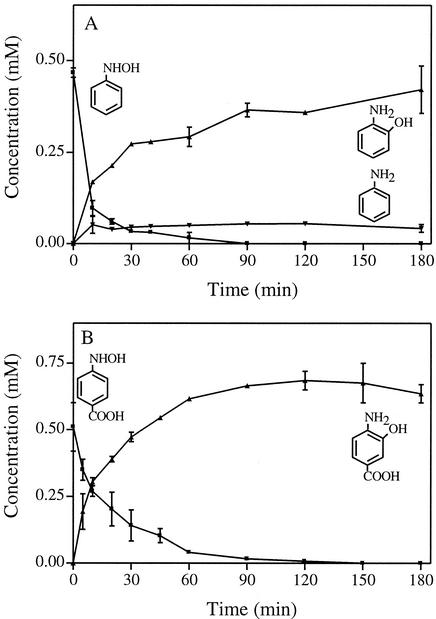
In oxygen uptake experiments, Meulenberg and de Bont (22) demonstrated that the lyase from Pseudomonas sp. strain 4NT has high activity for HABA and low activity for HAB. These authors hypothesized that the low oxygen uptake rate was due to the production of either an AP or a catechol that could not be further oxidized, and the product of HAB transformation remained to be identified. We hypothesized that identification of the product formed from HAB by the lyase could provide evidence for the mechanism of the reaction with HABA. To reduce the possibility of nonspecific reduction of the substrate by flavoproteins (20), we subjected the cell extract to ultrafiltration. We then compared the enzyme activity with or without NADPH and found activity levels of 1.3 and 0.24 μmol min−1 mg of protein−1), respectively. This result suggests that NADPH is not a coenzyme but serves to lower the redox potential of the reaction as proposed by Groenewegen and de Bont (7). The lyase was active under the conditions provided, therefore, the ultrafiltered cell extract of Pseudomonas strain 4NT (0.19 mg of protein) was incubated anaerobically in 0.5 ml of phosphate buffer (pH 7.0, 20 mM) with HAB (0.5 mM). HPLC analysis of the reaction mixture revealed that 2-AP and aniline accumulated, whereas catechol and 4-AP were not detected (Fig. 3A).
FIG. 3.
HPLC analysis of the conversion of HAB by cell lysate of Pseudomonas strain 4-NT (A) and of 4-HABA by P. pseudoalcaligenes JS45 (B). Symbols: ▪, HAB; ▴, 2-AP; ▾, aniline; □, 4-HABA; ○, 4-amino-3-hydroxybenzoate. Each datum point is the mean of duplicate reactions, and the bars indicate the mean standard errors.
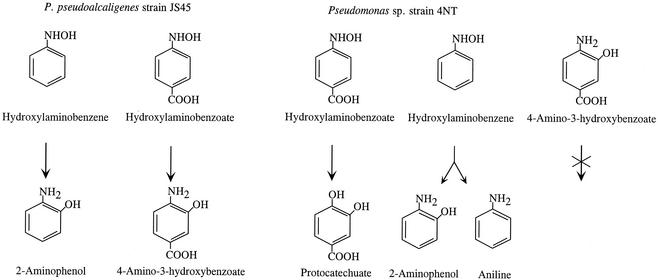
Similar results were obtained with untreated cell extracts and the ultrafiltered extracts with NADPH present. The ability of the lyase to catalyze a mutase reaction provides strong circumstantial evidence that the initial step in the reaction with 4-HABA is the transfer of the hydroxyl group to the adjacent carbon (Fig. 4). Peres et al. (27) have previously observed that a minor amount of p-aminobenzoate was produced by Burkholderia cepacia strain PB4 during the conversion of 4-HABA to protocatechuate. Production of a substituted aniline has also been observed during the enzymatic conversion of hydroxylaminofluorene by alcohol dehydrogenase and during the conversion of 4-hydroxylaminochlorobenzene to the corresponding AP by α-ketoglutarate dehydrogenase (3, 21). Corbett (3) suggest that the enzyme-bound Zn2+ or enzyme-bound Mg2+, respectively, in the active sites served as Lewis acids to reduce the hydroxylamine substituent to an amine.
FIG. 4.
Transformation of hydroxylaminonoarenes. P. pseudoalcaligenes JS45, expressing HabA mutase activity, converts HAB and 4-HABA to the corresponding ortho-AP. Pseudomonas strain 4NT, expressing lyase activity, converts 4-HABA to protocatechuate and HAB to 2-AP and to aniline and does not readily transform 4-amino-3-hydroxybenzoate.
Experiments were performed to determine whether the mutase from P. pseudoalcaligenes JS45 catalyzed the transformation of 4-HABA. HPLC analysis of the reaction mixtures revealed that 4-amino-3-hydroxybenzoate accumulated and that protocatechuate was not produced (Fig. 3B). The observation expands the known substrate range for the mutase from strain JS45. The mutase in R. eutropha did not transform 4-HABA (31). A different isomer, 2-HABA, is converted to 3-hydroxyanthranilate by P. fluorescens strain KU-7 (10). Our results taken with other published results indicate that all mutases studied to date convert the hydroxylaminoarenes to APs. The lyase can convert hydroxylaminoarenes to either catechols or APs depending on the starting compound (Fig. 4). The factors that determine whether the catechols or APs are formed are unknown. Recently, 4-HABA lyase genes have been sequenced from three bacteria (accession numbers AF187879, AF187880, and AF292094). The deduced amino acid sequences of the lyases show no similarity to the deduced amino acid sequences of two JS45 mutases (4). More mechanistic and structural information is needed to reveal the relationships between the two types of enzymes.
Deamination reaction by the lyase.
The catalytic mechanisms of the reactions catalyzed by the hydroxylaminolyases and -mutases have not been investigated in detail. Intermolecular mechanisms were previously proposed for enzymatic hydroxyl transfer reactions (22, 31, 32). However, all of the conversions we examined involve an intramolecular transfer of hydroxyl groups. The results from reactions containing H218O clearly support the proposed intramolecular mechanism for N-hydroxy-2-fluorenylacetamide conversion to o-amidophenols (8). We have previously proposed a mechanism to account for the intramolecular transfer of the hydroxyl group of HAB to the ortho position of the product, 2-AP, by the HabB mutase from strain JS45, and the present results confirm our proposed mechanism (11).
The fact that both mutase and lyase reactions involve transfer of the hydroxyl moiety from the hydroxylamino group to, mainly, the adjacent position of the aromatic ring suggested that the initial steps in the two reactions are similar. We have proposed that the hydrolytic deamination of 2-aminomuconate, an intermediate in the biodegradation of nitrobenzene by strain JS45, proceeds via an imine intermediate (12). The hydrolytic deamination of the intermediate during conversion of 4-HABA could involve a similar mechanism (Fig. 5). The rate-limiting step would be the enzymatic conversion of the substrate to the imine intermediate, which would spontaneously hydrolyze to protocatechuate. Hydrolysis of the imine as the mechanism of ammonia release was proposed as a step during the degradation of 3-nitrophenol by P. putida 2NP8, but no experimental evidence was provided (36).
FIG. 5.
Proposed intramolecular rearrangement mechanisms. (A) Mutases from P. pseudoalcaligenes JS45, R. eutrophus JMP134, and Mycobacterium sp. strain 4-NT, as well as the lyase from Pseudomonas strain 4-NT, convert HAB to the AP by an intramolecular transfer of the hydroxyl group. (B) The lyase from Pseudomonas strain 4-NT catalyzes an intramolecular rearrangement of 4-HABA and a second hydrolase reaction, producing protocatechuate. The mutase from P. pseudoalcaligenes converts HABA to the AP. The imine, in brackets, is proposed as an intermediate in both reactions. Solid arrows represent reactions that have been demonstrated. Dashed arrows represent the hypothetical reaction proposed in references 15 and 23.
Alternatively, the hydroxylamino compound might first be converted to the corresponding AP and the AP group could then be hydrolyzed to produce the dihydroxy compound. Under anaerobic conditions, the lyase converts 4-HABA to protocatechuate that accumulates in the medium. To determine whether the lyase converts HABA to protocatechuate via the AP, we conducted experiments with APs in mixtures containing extracts from cells of Pseudomonas strain 4NT. Negligible amounts of 4-amino-3-hydroxybenzoate were transformed, and no protocatechuate was detected (Table 2). Likewise, in parallel experiments monitoring 2-AP transformation, no catechol was detected by HPLC. The results provide strong circumstantial evidence that HABA is not converted to protocatechuate via 4-amino-3-hydroxybenzoate.
TABLE 2.
Lyase activity from lysates of glucose- and 4-NBA-grown cells of Pseudomonas sp. strain 4NTa
| Assay substrate | Mean sp act (nmol min−1 mg of protein−1) ± SD
|
|
|---|---|---|
| Glucose | 4-NBA | |
| 4-HABA | 3.7 ± 4.7* | 211 ± 19.3† |
| HAB | 3.0 ± 1.3** | 337 ± 9.9† |
| 4-A-3-HOBAb | ≤0.1* | ≤0.1* |
| 2-AP | 0.4 ± 0.5* | ≤0.1* |
Reaction mixtures contained 0.5 mM substrate and lysate (0.55 [∗], 0.055 [†], or 0.285 [∗∗] mg of protein as indicated) in 500 μl of phosphate buffer (20 mM, pH 7.0), and assays were performed in an anaerobic chamber. The reactions were terminated by the addition of acetonitrile (4°C) and analyzed by HPLC. The values reported are averages of duplicate measurements ± the standard deviations.
4-A-3-HOBA, 4-amino-3-hydroxybenzoate.
Our results support the conclusions of Peres et al. (27), who reported previously that B. cepacia strain PB4 and R. paucula strain SB4 expressing lyase activity were unable to transform 4-amino-3-hydroxybenzoate. Although it is possible that the AP participates as an enzyme-bound intermediate (5, 15, 22), the conversion of the substituted AP to the corresponding dihydroxy compound has never been demonstrated. Cain and Cartwright (2) first reported that trace amounts of 4-amino-3-hydroxybenzoate accumulated in p-NBA-grown cultures of Nocardia and concluded that 4-amino-3-hydroxybenzoate was not an intermediate in the metabolic pathway of p-NBA to protocatechuate. Peres et al. (27) detected traces of 3-hydroxy-4-acetamidobenzoate, and both Peres et al. (27) and Hughes et al. (15) reported that the oxidative dimerization product, 2-aminophenoxazin-3-one-7-carboxylate, accumulates in the medium during the conversion of 4-NBA to protocatechuate. These observations indicate clearly that at least small amounts of 4-amino-3-hydroxybenzoate are produced during the reaction. Peres et al. (27) indicate that it is a dead-end metabolite, whereas Hughes et al. (15) suggest that it is an intermediate in the pathway. Our results seem to support the conclusion that 4-amino-3-hydroxybenzoate is a dead-end metabolite in strain 4NT (Fig. 4). The aminohydroxybenzoate could form from the putative imine intermediate (Fig. 5). The observation that Pseudomonas sp. strain 4NT lacks appropriate enzymes for subsequent degradation of aminohydroxybenzoate seems to be consistent with these conclusions.
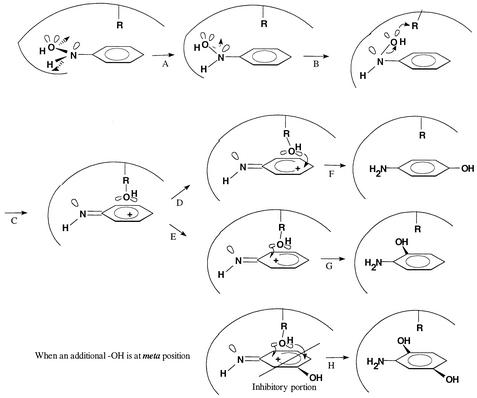
According to the stereochemistry, the O-N-H bonds of HAB should be coplanar with the benzene ring to keep the molecule at the lowest energy level (Fig. 6). With the coplanar structure, the hydroxyl group could be readily transferred to the adjacent carbon of the benzene ring. In the acid-catalyzed conversion of HAB to 4-AP, the hydroxyl group of the substrate is released first; a hydroxyl group from the solvent water then attacks the C-4 position of the aromatic ring to form a 4-AP (the intermolecular transfer mechanism). The previously proposed catalytic mechanism for strain JS45 mutase (11) could not explain how a hydroxyl group is transferred across the distance of several bonds in the reaction catalyzed by the mutase in strain JMP134. We suggest here a working hypothesis for a three-dimensional interaction between the substrate and the active site of the mutase from strain JMP134 (Fig. 6). In the scheme, the transfer of the hydroxyl group is mediated by an amino acid residue above the plane of the aromatic ring. Such a configuration would allow the hydroxyl group to attach either at the para or ortho position of the aromatic ring. A bimolecular mechanism where the hydroxyl group of one molecule of substrate is transferred to a second substrate molecule would also explain the results, but we believe the mechanism proposed above is more likely.
FIG. 6.
Hypothetical model for hydroxyl transfer mechanism during conversion of HAB catalyzed by the mutase from strain JMP134. (A) Mutase turns the O-N-C group away from the plane of the benzene ring to a position perpendicular to the ring; (B) the −OH group approaches an amino acid residue (R) of the enzyme; (C) an R-OH intermediate forms above the benzene ring; (D and F) the −OH moves to the para position to form 4-AP; (E and G) the −OH moves to the ortho position to form 2-AP; (H) the stereochemical effect of a hydroxyl group at the meta position inhibits the R-OH group accessible to the para position, resulting in the formation of the ortho isomer only.
The mutase in strain JMP134 also catalyzes the conversion of the physiological substrate 3-hydroxylaminophenol only to 2,5-dihydroxyaniline (30, 31). The observation can be explained by the stereochemical effect of the hydroxyl group at the meta position, which would restrict the formation of 2,4-dihydroxyaniline. The highly schematic pathway in Fig. 6 reflects our lack of understanding about how the nitrogen atom and the benzene ring change or interact during the catalytic process.
Acknowledgments
The work was supported in part by the U.S. Air Force Office of Scientific Research and the Strategic Environmental Defense Research Program. Part of Z. He's work was done under an appointment to the Research Participation Program at the U.S. Air Force Research Laboratory, Tyndall Air Force Base, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and Tyndall Air Force Base.
We thank Jed Pitera, Don Hilvert, Glenn Johnson, Chris Antworth, and Venkateswarlu Kadiyala for helpful discussions. We also thank Peter Williams for providing a preprint of reference 15.
REFERENCES
- 1.Bauer, H., and S. M. Rosenthal. 1944. 4-Hydroxylaminobenzenesulfonamide, its acetyl derivatives and diazotization reaction. J. Am. Chem. Soc. 66:611-614. [Google Scholar]
- 2.Cain, R. B., and N. J. Cartwright. 1960. Intermediary metabolism of nitrobenzoic acids by bacteria. Nature 185:868-869. [Google Scholar]
- 3.Corbett, M. D. 1995. Bioorganic chemistry of the arylhydroxylamine and nitrosoarene functional groups, p. 151-182. In J. C. Spain (ed.), Biodegradation of nitroaromatic compounds. Plenum Press, Inc., New York, N.Y.
- 4.Davis, J. K., G. C. Paoli, Z. He, L. J. Nadeau, C. C. Somerville, and J. C. Spain. 2000. Sequence analysis and initial characterization of two isozymes of hydroxylaminobenzene mutase from Pseudomonas pseudoalcaligenes JS45. Appl. Environ. Microbiol. 66:2965-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiorella, P. D., and J. C. Spain. 1997. Tranformation of 2,4,6-trinitrotoluene by Pseudomonas pseudoalcaligenes JS52. Appl. Environ. Microbiol. 63:2007-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groenewegen, P. E. J., P. Breeuwer, J. M. L. M. van Helvoort, A. A. M. Langenhoff, F. P. de Vries, and J. A. M. de Bont. 1992. Novel degradative pathway of 4-nitrobenzoate in Comamonas acidivorans NBA-10. J. Gen. Microbiol. 138:1599-1605. [DOI] [PubMed] [Google Scholar]
- 7.Groenewegen, P. E. J., and J. A. M. de Bont. 1992. Degradation of 4-nitrobenzoate via 4-hydroxylaminobenzoate and 2,3-dihyroxybenzoate in Comamonas acidivorans NBA-10. Arch. Microbiol. 158:381-386. [Google Scholar]
- 8.Gutmann, H. R., and R. R. Erickson. 1972. The conversion of the carcinogen N-hydroxy-2-fluoroenylacetamide to o-amidophenols by rat liver in vitro. J. Biol. Chem. 247:660-666. [PubMed] [Google Scholar]
- 9.Haigler, B. E., and J. C. Spain. 1993. Biodegradation of 4-nitrotoluene by Pseudomonas sp. strain 4NT. Appl. Environ. Microbiol. 59:2239-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasegawa, Y., T. Muraki, T. Tokuyama, H. Iwaki, M. Tatsuno, and P. C. K. Lau. 2000. A novel degradative pathway of 2-nitrobenzoate via 3-hydroxyanthranilate in Pseudomonas fluorescens strain KU-7. FEMS Microbiol. Lett. 190:185-190. [DOI] [PubMed] [Google Scholar]
- 11.He, Z., L. J. Nadeau, and J. C. Spain. 2000. Characterization of hydroxylaminobenzene mutase from pNBZ139 cloned from Pseudomonas pseudoalcaligenes JS45 A highly associated SDS-stable enzyme catalyzes an intramolecular transfer of hydroxyl groups. Eur. J. Biochem. 267:1110-1116. [DOI] [PubMed] [Google Scholar]
- 12.He, Z., and J. C. Spain. 1998. A novel 2-aminomuconate deaminase in the nitrobenzene degradation pathway of Pseudomonas pseudoalcaligenes JS45. J. Bacteriol. 180:2502-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He, Z., and J. C. Spain. 2000. Reactions involved in the lower pathway for degradation of 4-nitrotoluene by Mycobacterium strain HL-4NT-1. Appl. Environ. Microbiol. 66:3010-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, J. B., C. Wang, K. Yesland, A. Richardson, R. Bhadra, G. Bennett, and F. Ruldoph. 1998. Bamberger rearrangement during TNT metabolism by Clostridium acetobutylicum. Environ. Sci. Technol. 32:494-500. [Google Scholar]
- 15.Hughes, M. A., M. J. Baggs, J. Al-Dulayymi, M. S. Baird, and P. A. Williams. 2002. Accumulation of 2-aminophenoxazin-3-one-7-carboxylate during growth of Pseudomonas putida TW3 on 4-nitro-substituted substrates requires 4-hydroxylaminobenzoate lyase (PnbB). Appl. Environ. Microbiol. 68:4965-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, G. R., B. F. Smets, and J. C. Spain. 2001. Oxidative transformation of aminodinitrotoluene isomers by multicomponent dioxygenases. Appl. Environ. Microbiol. 67:5460-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katsivela, E., V. Wray, D. H. Pieper, and R. M. Wittich. 1999. Initial reactions in the biodegradation of 1-chloro-4-nitrobenzene by a newly isolated bacterium, strain LW1. Appl. Environ. Microbiol. 65:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kukhtenko, I. I. 1971. Study of the mechanism of the rearrangement of n-phenylhydroxylamine into p-aminophenol. Zh. Organicheskoi Khimii 7:330-333. [Google Scholar]
- 19.Lendenmann, U., and J. C. Spain. 1996. 2-Aminophenol 1,6-dioxygenase: a novel aromatic ring cleavage enzyme purified from Pseudomonas pseudoalcaligenes JS45. J. Bacteriol. 178:6227-6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leskovac, V., J. Svircevic, S. Trivic, M. Popovic, and M. Radulovic. 1989. Reduction of aryl-nitroso compounds by pyridine and flavin coenzymes. Int. J. Biochem. 21:825-834. [DOI] [PubMed] [Google Scholar]
- 21.Maskos, Z., and G. W. Winston. 1993. Alcohol dehydrogenase-dependent reduction of 2-nitrofluorene and rearrangement of N-hydroxy-2-aminofluorene. Biochemistry 32:12768-12773. [DOI] [PubMed] [Google Scholar]
- 22.Meulenberg, R., and J. A. M. de Bont. 1995. Microbial production of catechols from nitroaromatic compounds, p. 37-52. In J. C. Spain (ed.), Biodegradation of nitroaromatic compounds. Plenum Press, Inc., New York, N.Y.
- 23.Nadeau, L. J., Z. He, and J. C. Spain. 2000. Production of 2-amino-5-phenoxyphenol from 4-nitrobiphenyl ether using nitrobenzene nitroreductase and hydroxylaminobenzene mutase from Pseudomonas pseudoalcaligenes strain JS45. J. Ind. Microbiol. Biotechnol. 24:301-305. [Google Scholar]
- 24.Nishino, S. F., and J. C. Spain. 1993. Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl. Environ. Microbiol. 59:2520-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishino, S. F., J. C. Spain, and Z. He. 2000. Strategies for aerobic degradation of nitroaromatic compounds: process discovery to field application, p. 7-61. In J. C. Spain, J. B. Hughes, and H.-J. Knackmuss (ed.), Biodegradation of nitroaromatic compounds and explosives. Lewis Publishers, Boca Raton, Fla.
- 26.Park, H.-S., and H.-S. Kim. 2000. Identification and characterization of the nitrobenzene catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12. J. Bacteriol. 182:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peres, C., R. Russ, H. Lenke, and S. Agathos, N. 2001. Biodegradation of 4-nitrobenzoate, 4-aminobenzoate and their mixtures: new strains, unusual metabolites and insights into pathway regulation. FEMS Microbiol. Ecol. 37:151-159. [Google Scholar]
- 28.Preuss, A., J. Fimpel, and G. Diekert. 1993. Anaerobic transformation of 2,4,6-trinitrotoluene. Arch. Microbiol. 159:345-353. [DOI] [PubMed] [Google Scholar]
- 29.Rhys-Williams, W., S. C. Taylor, and P. A. Williams. 1993. A novel pathway for the catabolism of 4-nitrotoluene by Pseudomonas. J. Gen. Microbiol. 139:1967-1972. [DOI] [PubMed] [Google Scholar]
- 30.Schenzle, A., H. Lenke, P. Fischer, P. A. Williams, and H.-J. Knackmuss. 1997. Catabolism of 3-nitrophenol by Ralstonia eutropha JMP134. Appl. Environ. Microbiol. 63:1421-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schenzle, A., H. Lenke, J. C. Spain, and H.-J. Knackmuss. 1999. 3-Hydroxylaminophenol mutase from Ralstonia eutropha JMP134 catalyzes a Bamberger rearrangement. J. Bacteriol. 181:1444-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spain, J. C. 1995. Bacterial degradation of nitroaromatic compounds under aerobic conditions, p. 19-35. In J. C. Spain (ed.), Biodegradation of nitroaromatic compounds. Plenum Press, Inc., New York, N.Y.
- 33.Spiess, T., F. Desiere, P. Fischer, J. C. Spain, H.-J. Knackmuss, and H. Lenke. 1998. A new 4-nitrotoluene degradation pathway in a Mycobacterium strain. Appl. Environ. Microbiol. 64:446-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stecher, E. P. G. 1968. The Merck index, 8th ed. Merck & Co., Rathway, N.J.
- 35.Yabannavar, A. V., and G. J. Zylstra. 1995. Cloning and characterization of the genes for p-nitrobenzoate degradation from Pseudomonas pickettii YH105. Appl. Environ. Microbiol. 61:4284-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao, J.-S., A. Singh, X.-D. Huang, and O. P. Ward. 2000. Biotransformation of hydroxylaminobenzene and aminophenol by Pseudomonas putida 2NP8 cells grown in the presence of 3-nitrophenol. Appl. Environ. Microbiol. 66:2336-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]