Abstract
We tested a previously described protocol for fluorescence in situ hybridization of marine bacterioplankton with horseradish peroxidase-labeled rRNA-targeted oligonucleotide probes and catalyzed reporter deposition (CARD-FISH) in plankton samples from different lakes. The fraction of Bacteria detected by CARD-FISH was significantly lower than after FISH with fluorescently monolabeled probes. In particular, the abundances of aquatic Actinobacteria were significantly underestimated. We thus developed a combined fixation and permeabilization protocol for CARD-FISH of freshwater samples. Enzymatic pretreatment of fixed cells was optimized for the controlled digestion of gram-positive cell walls without causing overall cell loss. Incubations with high concentrations of lysozyme (10 mg ml−1) followed by achromopeptidase (60 U ml−1) successfully permeabilized cell walls of Actinobacteria for subsequent CARD-FISH both in enrichment cultures and environmental samples. Between 72 and >99% (mean, 86%) of all Bacteria could be visualized with the improved assay in surface waters of four lakes. For freshwater samples, our method is thus superior to the CARD-FISH protocol for marine Bacteria (mean, 55%) and to FISH with directly fluorochrome labeled probes (mean, 67%). Actinobacterial abundances in the studied systems, as detected by the optimized protocol, ranged from 32 to >55% (mean, 45%). Our findings confirm that members of this lineage are among the numerically most important Bacteria of freshwater picoplankton.
The taxonomic composition of picoplankton communities profoundly differs in freshwater and marine systems. Cultivation-independent molecular approaches have demonstrated that bacteria from large phylogenetic lineages of typical freshwater microbes, e.g., the β-subdivision of the proteobacteria, are virtually absent in the marine picoplankton (15, 23). For other bacterial lineages the evidence is less conclusive. Comparative analysis of 16S rDNA genes indicate that uncultured members of the class Actinobacteria are ubiquitous in lakes of various trophic state, size or geographic location (16, 36), but actinobacterial sequence types are also known from marine systems (31).
A direct microscopic visualization of freshwater Bacteria by fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes is still difficult, and frequently only a low fraction of all picoplankton cells (<50%) can be visualized by FISH (5, 10, 15, 19). We hypothesize that this may be due to high abundances of Actinobacteria rather than of groups that are not targeted by the commonly used probe for the domain Bacteria, EUB338 (9). Cells from the actinobacterial lineage are found in the smallest microbial size fractions (16) even at highly eutrophic conditions (29). They are, therefore, probably also low in total ribosome content, which limits their detectability by FISH with directly fluorochrome labeled oligonucleotides. So far, there is only one report about the abundances of Actinobacteria in lake picoplankton (16). In that study, FISH with specifically designed helper oligonucleotides (14) visualized large populations of this lineage in a mountain lake.
Recently, a novel FISH protocol was developed for the reliable visualization of small bacterial cells in the marine picoplankton (26). This approach is based on hybridization with horseradish peroxidase (HRP)-labeled oligonucleotide probes and subsequent tyramide signal amplification (4). It has been termed catalyzed reporter deposition (CARD)-FISH and yields significantly higher signal intensities than FISH with fluorescently monolabeled oligonucleotide probes. Since the fractions of hybridized bacterial cells by CARD-FISH in marine samples were on average twice as high as by FISH with directly fluorochrome labeled probes (26), this technique might also improve the analysis of freshwater microbial communities.
However, one crucial step of the above CARD-FISH protocol is a directed permeabilization of microbial cell walls after the embedding of cells in an agarose matrix. The published procedure for CARD-FISH of marine Bacteria might not sufficiently permeabilize the presumably gram-positive cell walls of aquatic Actinobacteria for the penetration of enzyme-labeled probes. Therefore, we attempted to develop a modified CARD-FISH protocol for the specific visualization of limnetic Actinobacteria, and we investigated the potential of this technique for FISH of freshwater picoplankton in different lakes.
MATERIALS AND METHODS
Sampling and study sites.
Sample for initial methods development originated from experimental enrichments of Actinobacteria from a typical freshwater lineage in continuous culture (29, 30). Subsequently, the protocol was refined and tested on surface water samples (0.5 m) from four different lakes. Gossenköllesee is a shallow oligotrophic, high-mountain lake situated in the Tyrolean Alps (Austria) at an altitude of 2,417 m above mean sea level (AMSL) (28). Piburgersee is an oligo-mesotrophic Tyrolean alpine lake (913 m AMSL) (25). In these lakes, samples were collected during June 2002. Lake Fuchskuhle is a small meso- to acidotrophic humic forest lake located in the Brandenburg-Mecklenburg lake district, Germany, at an altitude of 59 m AMSL. The lake was artificially divided into four basins with different catchment areas during 1990 (32). Samples were collected from the southwest compartment of the lake during May 2002. Lake Cadagno is a mesotrophic meromictic mountain lake situated at 1,923 m AMSL in the Piora Valley in the south of Switzerland (33). Samples from Lake Cadagno were collected during July 2001.
Sample fixation and preparation.
Subsamples from the actinobacterial enrichments were prefixed with alkaline Lugol's iodine solution for 30 min followed by 2% (vol/vol) final concentration of formaldehyde (FA) for 30 min. Portions of 10 ml were then filtered onto polycarbonate membrane filters (type GTTP; pore size, 0.2 μm; diameter, 47 mm; Millipore, Eschborn, Germany), rinsed with double-distilled water (ddH2O), and stored at −20°C until further processing (29). Samples from Lake Cadagno and Fuchskuhle were prefiltered through cellulose nitrate membrane filters (pore size, 3 μm; Sartorius, Göttingen, Germany). One set of samples from all lakes was fixed with particle-free, molecular biology grade FA (2%, final concentration) (Sigma-Aldrich, Steinheim, Germany) and a second set by addition of 96% ethanol (EtOH) to a final concentration of 50%. Samples were fixed for 24 h at 4°C. Portions of 10 to 100 ml of FA-fixed and equivalent (double) volumes of EtOH-fixed samples from the various lakes were filtered onto membrane filters (type GTTP, Millipore). The preparations were washed with 10 ml of ddH2O and air dried. The samples were then stored at −20°C until further processing.
Permeabilization.
In order to avoid cell loss during permeabilization (26), the air dried filters were embedded in low gelling point agarose (concentration, 0.2%, MetaPhor, FMC Bioproducts, Rockland, Maine) and subsequently dried at 35°C for 10 min. Samples from the actinobacterial enrichment culture were subjected to various permeabilization methods. The tested treatments included (i) no enzyme pretreatment, (ii) digestion by lysozyme (Sigma-Aldrich), 10 mg ml−1, dissolved in 0.05 M EDTA (pH 8.0) and 0.1 M Tris-HCl (pH 7.4) for 60 min at 37°C; (iii) by higher concentrations of lysozyme (15 and 20 mg ml−1) for 30 and 60 min each; (iv) treatment with of 0.2, 0.5 and 1 M HCl for 5, 10, and 30 min at room temperature or at 37°C followed by incubations with lysozyme (10 mg ml−1) for 60 min; and (v) digestion by lysozyme (10 mg ml−1) followed by incubation with different concentrations of achromopeptidase (E.C. 3.4.21.50, Sigma-Aldrich) (30, 60, 307, 768, 1,535, and 3,070 U ml−1, i.e., dilutions from 1 to 100). Achromopeptidase incubations were performed at 37°C in a buffer containing 0.01 M NaCl, 0.01 M Tris-HCl (pH 8.0) for 15 min. Different incubation times were tested at several enzyme concentrations. Finally, lysozyme digestion (60 min, 10 mg ml−1) followed by achromopeptidase digestion (30 min, 60 U ml−1) was selected for the evaluation of samples from enrichments and the various environments. This was compared with pretreatments by lysozyme only, as proposed for CARD-FISH of marine picoplankton (26). After the enzyme treatments, the filters were thoroughly washed in ddH2O and incubated in 0.01 M HCl for 10 min to inactivate endogenous peroxidase. The filters were again thoroughly washed with ddH2O, dehydrated with 96% ethanol, and air dried. The pretreated preparations were stored at −20°C until further processing.
Hybridizations with oligonucleotide probes.
Whole-cell in situ hybridizations of sections from the above described polycarbonate filters were performed using the oligonucleotide probes EUB338 (most Bacteria), EUB I to III (Bacteria including Verrucomicrobia and Planctomycetes) (9), NON338 (antisense EUB338, negative control), HGC69a (Actinobacteria, 23S rRNA-targeted) (1), and HGC236 (Actinobacteria, 16S rRNA-targeted) (16). All probes were purchased from ThermoHybaid, Interactiva Division (Ulm, Germany). Untreated filter sections (chord length, approximately 5 mm) were hybridized with oligonucleotide probes that were 5′ monolabeled with the indocarbocyanine dye Cy3. Hybridization with directly fluorochrome labeled probes were performed as described previously (27). For probes EUB338, NON338, and HGC69a a formamide concentration of 20 to 35% (vol/vol) was used in the hybridization buffers, and 10% was used for the directly fluorochrome-labeled probe HGC236.
Pretreated filter sections (i.e., preparations subjected to permeabilization) were hybridized with 5′-HRP-labeled oligonucleotide probes (ThermoHybaid, Interactiva Division) according to the protocol of Pernthaler et al. (26). The hybridization buffer used for EUB338, NONEUB, and EUB I to III contained 0.9 M NaCl, 20 mM Tris-HCl (pH 7.4), 10% dextran sulfate (wt/vol), 55% (vol/vol) formamide, 1% blocking reagent (Boehringer, Mannheim, Germany), and 0.05% Triton X-100 (vol/vol). The blocking reagent was prepared in maleic acid buffer (100 mM maleic acid, 150 mM NaCl, pH 7.5). A 30% (vol/vol) formamide concentration was used for probes HGC69a and HGC236. Four hundred microliters of hybridization buffer and 4 μl of probe working solution (50 ng μl−1) was put in a 0.5-ml reaction vial and 10 to 15 filter sections were placed in the buffer. The reaction vial was incubated in a hybridization oven at 35°C on a rotation shaker for a minimum of 2 h. Next the filter sections were transferred to 50 ml of prewarmed washing buffer and incubated at 37°C for 10 min. The washing buffer for EUB338, NON338, and EUB I to III was 20 mM NaCl, 5 mM EDTA (pH 8.0), 20 mM Tris-HCl (pH 7.4), and 0.02% (wt/vol) sodium dodecyl sulfate. For probes HGC69a and HGC236, NaCl concentrations of 112 mM were used.
Tyramide signal amplification.
After the washing step, filters hybridized with HRP-labeled probes were transferred to 15 ml of 1× phosphate-buffered saline (PBS) (pH 7.3) amended with 0.05% Triton X-100 for 15 min at room temperature to equilibrate the probe-delivered HRP. Then, the filters were dabbed onto blotting paper to remove excess buffer and transferred to substrate mix and incubated at 37°C for at least 10 min in dark. The substrate mix consisted of 1 part Cy3-labeled tyramide (NEN Life Science Products, Boston, Mass.) and 100 parts freshly prepared amplification diluent (1× PBS [pH 7.3], 0.0015% H2O2, 0.1% blocking reagent). The filters were then washed with 1× PBS amended with 0.05% Triton X-100 for 15 min at room temperature followed by thorough washing with ddH2O and then with 96% ethanol for 1 min. Then, the filters were air dried and mounted on glass slides using a previously described mountant mix amended with 4′,6′-diamidino-2-phenylindol (DAPI) (final concentration, 1 μg ml−1) (26). Prior to evaluation the slides could be stored at −20°C up to several days without loss of fluorescence intensity. All preparations were done in triplicate.
Microscopic evaluation.
The stained filter sections were inspected on a Zeiss Axioplan II imaging microscope equipped with a 100x Plan Apochromat oil objective lens (Carl Zeiss, Jena, Germany) and a HBO 100-W Hg vapor lamp. The filter sets were Chroma HQ 41007 (Chroma Tech. Corp. Brattleboro, Vt.) for Cy3 and Zeiss01 for DAPI. First, Cy3-stained cells were counted in one microscopic field, and this was followed by determination of DAPI-stained cells. At least 1,000 DAPI-stained cells were counted in >10 microscopic fields randomly selected across the filter sections. Images of Cy3 and DAPI fluorescence were captured using a SPOT slow-scan cooled charge coupled device camera (resolution, 1,033 by 1,315 pixels; Diagnostic Instruments, Sterling Heights, Mich.) mounted on the Axioplan II microscope (Carl Zeiss), and a PC-based image acquisition software (Diagnostics Instruments).
Data analysis.
Statistical analysis of the observed differences was performed using Friedman's nonparametric analysis of variance for dependent samples on the pooled data set from all lakes followed by Dunnett's multiple comparisons against a control group. We tested if the hybridized fractions of Bacteria and Actinobacteria significantly differed in the various assays from the percentages of cells visualized by FISH with directly fluorochrome-labeled probes (of FA-fixed samples). We also tested for differences between pairs of treatments using the Student-Newman-Keul test for post hoc multiple comparisons of pooled data (e.g., CARD-FISH pretreatment and fixation). We did not test for differences between lakes, since this was not the scope of the study and the individual data sets were too small. The statistical analysis software SigmaStat (SPSS, Inc., Chicago, Ill.) was used for test calculations.
RESULTS
Pretreatment optimization.
Only few cells could be visualized in the enrichment or lake samples after CARD-FISH without any permeabilization pretreatment (data not shown). In samples from the actinobacterial enrichments the fraction of hybridized cells after CARD-FISH with probe EUB338 ranged between 57.4 and 62.1% after pretreatment with lysozyme (10 mg ml−1 for 60 min). Higher lysozyme concentrations (15 and 20 mg ml−1) did not increase the fraction of hybridized cells, but rather caused a visible disintegration of cells. Incubation with 1 M HCl for 5 min at room temperature followed by lysozyme digestion (10 mg ml−1) increased the fraction of hybridized cells by less than 5% (data not shown). Several other treatments did not have significant effects on the fraction of hybridized cells, and the probe signal was completely lost in samples treated with 1 M HCl at 37°C for 10 min or more.
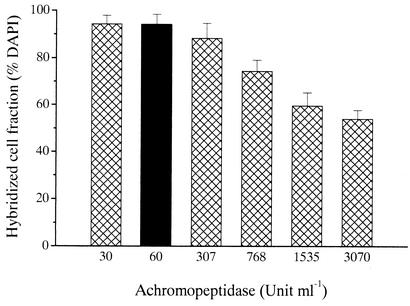
Pretreatment with lysozyme (10 mg ml−1 for 60 min) followed by incubation with achromopeptidase at concentrations between 30 and 3,070 U ml−1 (15 min) showed pronounced effects on visualization by CARD-FISH (Fig. 1). The fraction of hybridized cells decreased from 95% ± 4% (mean ± standard error) at 30 U ml−1 of achromopeptidase to 54% ± 4% at 3,070 U ml−1. In addition, cell loss was observed at concentrations ≥307 U ml−1. Subsequently, different incubation times of achromopeptidase digestion were tested (15, 30, and 60 min). Lysozyme pretreatment (10 mg ml−1, 60 min) followed by digestion for 30 min with achromopeptidase at a concentration of 60 U ml−1 was found optimal in terms of hybridized cell fraction and reproducibility (Fig. 2), and no cell loss was observed under these conditions. This pretreatment was then used for the further evaluation of the enrichment and environmental samples with specific FISH probes for Actinobacteria. Interestingly, achromopeptidase treatment followed by incubation with lysozyme did not increase the fraction of hybridized cells, compared to lysozyme treatment only (data not shown).
FIG. 1.
CARD-FISH detection rate of bacteria with the general bacterial probe EUB338 in samples from an actinobacterial enrichment culture after incubations (15 min, 37°C) at different dilutions of achromopeptidase. The black bar represents the enzyme concentration selected for subsequent sample evaluation. Error bars, standard deviations.
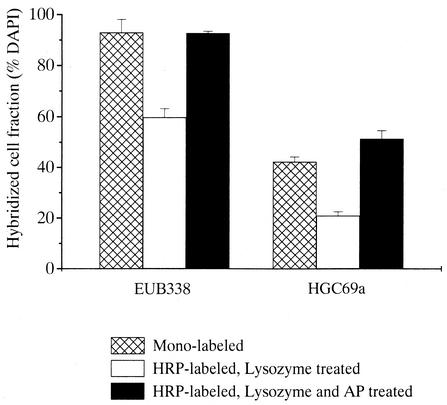
FIG. 2.
Detection rate of bacteria (probe EUB338) and actinobacteria (probe HGC69a) by FISH with monolabeled probes or CARD-FISH with HRP-labeled probes and different pretreatments in samples from an actinobacterial enrichment culture. Error bars, standard deviations.
Modifications of the CARD-FISH protocol.
Table 1 depicts the optimized protocol for the visualization of freshwater bacterioplankton by CARD-FISH. Several important modifications have been introduced compared to a recently published protocol for the visualization of marine Bacteria (26). During the permeabilization step, a treatment with achromopeptidase as described above was included, followed by incubation in 0.01 M HCl to inactivate endogenous peroxidase. The tyramide signal amplification step was performed in the presence of blocking reagent in the substrate mix, which reduces unspecific binding of labeled tyramides and thus of background fluorescence. The commercial amplification diluent was replaced by a custom mix. Finally, tyramide signal amplification was carried out at 37°C rather than at 20°C, which greatly enhanced fluorescence signal intensity. Except for the additional enzyme digestion, these modifications also improve the performance of CARD-FISH in marine systems (A. Pernthaler, unpublished data).
TABLE 1.
Improved CARD-FISH protocol for freshwater bacterioplankton
| Stage | Step no. | Description |
|---|---|---|
| Embedding | 1 | Prepare samples on membrane filters. |
| 2 | Dip filters in 0.2% low-gelling-point agarose and place filters face up into glass slides and air dry at 35°C (10 min). | |
| 3 | Dehydrate in 96% ethanol (room temperature [RT], 1 min). | |
| 4 | Air dry filters.a | |
| Permeabilization | 5 | Incubate in lysozyme (37°C, >60 min). |
| 6 | Wash with double distilled water (ddH2O) (1 min, RT). | |
| 7 | Incubate in achromopeptidase (37°C, >30 min). | |
| 8 | Wash with ddH2O. | |
| 9 | If necessary, incubate in 0.01 M HCl for 10 min at RT to inactivate endogenous peroxidase. | |
| 10 | Wash thoroughly with ddH2O. | |
| 11 | Wash with 96% ethanol (1 min, RT). | |
| 12 | Air dry filters and cut in sections.a | |
| Hybridization | 13 | Place filter sections in reaction vial (0.5 ml, 10 to 15 sections per vial). |
| 14 | Mix 400 μl of hybridization buffer and 4 μl of probe working solution and add to filter sections. Incubate at 35°C for at least 2 h. | |
| 15 | Wash filter sections in prewarmed washing buffer (10 min, 37°C); do not air dry filter sections after washing. | |
| Tyramide signal | 16 | Remove excess liquid with blotting paper, but don't let filter sections run dry. |
| amplification | 17 | Incubate in 1× PBS amended with 0.05% of Triton X-100 (15 ml, RT, 15 min). |
| 18 | Dab filter sections on blotting paper, but don't let run dry. | |
| 19 | Incubate in substrate mix (1 parts of CY3-labeled tyramide and 100 parts of amplification buffer [1× PBS, 0.0015% H2O2, 0.1% blocking reagent]) at 37°C for 10 min in dark. | |
| 20 | Dab filter sections on blotting paper. | |
| 21 | Wash in 15 ml of 1× PBS amended with 0.05% Triton X-100 (15 min, RT). | |
| 22 | Wash in ddH2O (RT, 1 min). | |
| 23 | Wash in 96% ethanol (RT, 1 min). | |
| 24 | Air dry filter sections.a | |
| 25 | Counter stain with DAPI.a |
Preparations may be stored at −20°C for several days to weeks without loss in signal.
Quantification of Bacteria and Actinobacteria in enrichments.
In samples from the actinobacterial enrichments, the fraction of hybridized cells after FISH with a directly fluorochrome labeled probe EUB338 was 93% ± 5% of total cells (Fig. 2). The percentage of cells that hybridized with a HRP-labeled EUB338 was only 60% ± 3% after lysozyme treatment (10 mg ml−1 for 60 min), whereas 93% ± 1% of all DAPI-stained cells were visualized in samples pretreated with lysozyme and achromopeptidase. A total of 42% ± 2% of cells hybridized with a directly fluorochrome labeled probe specific for Actinobacteria (HGC69a), but only 21% ± 2% with a HRP-labeled probe HGC69a after lysozyme treatment. Treatment with lysozyme and achromopeptidase increased the fraction of Actinobacteria hybridized by CARD-FISH to 51% ± 3%.
Quantification of Bacteria in environmental samples.
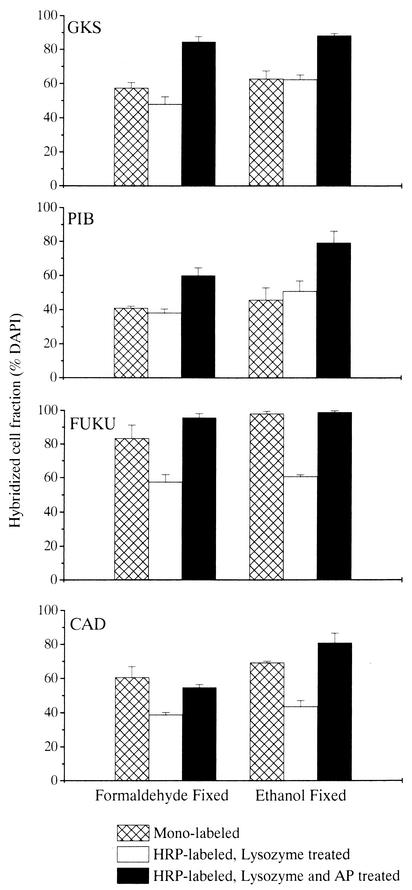
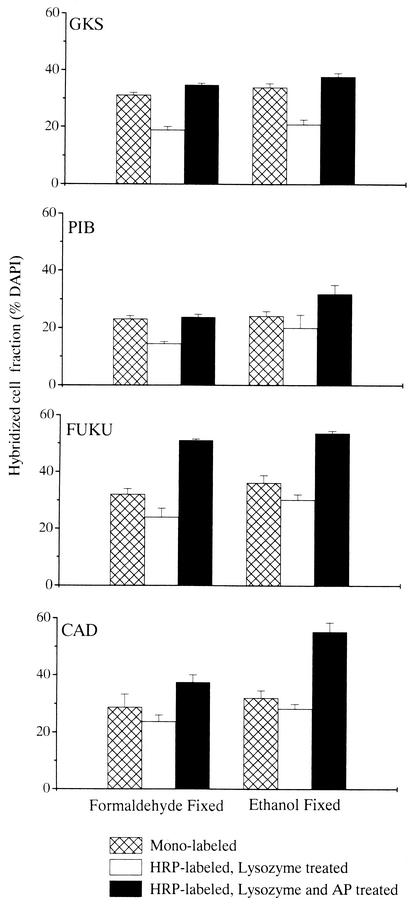
The original CARD-FISH protocol resulted in significantly lower fractions of hybridized bacterial cells in the environmental samples than the pretreatment with lysozyme and achromopeptidase (Fig. 3) (Student-Newman-Keul test, q = 7.38; P < 0.05). In FA-fixed samples, CARD-FISH after pretreatment with lysozyme only yielded significantly lower fractions of hybridized cells than FISH with directly fluorochrome labeled probes (Table 2). In contrast, significantly more bacterial cells were visualized in the lake samples after CARD-FISH and pretreatment with lysozyme and achromopeptidase than after FISH with fluorescently monolabeled probes, irrespective of fixation (Table 2). In three of the four FA-fixed environmental samples (Gossenköllesee, Piburgersee and Lake Fuchskuhle), and in EtOH-fixed samples of Gossenköllesee and Piburgersee, pretreatment with lysozyme and achromopeptidase and CARD-FISH resulted in higher fractions of cells hybridizing with probe EUB338 than FISH with mono-labeled EUB338 (P < 0.05) (Fig. 3). Pretreatment with lysozyme and achromopeptidase did not only increase the relative abundances but also the fluorescence intensities of hybridized cells (Fig. 4a and b).
FIG. 3.
Detection rate of bacteria (probe EUB338) by FISH with monolabeled probes, CARD-FISH and pretreatment with lysozyme, or CARD-FISH and pretreatment with lysozyme and achromopeptidase (AP) in samples from four lakes. Abbreviations: GKS, Gossenköllesee; PIB, Piburgersee; FUKU, Lake Fuchskuhle; CAD, Lake Cadagno. Error bars, standard deviations.
TABLE 2.
Comparisons of FISH detection rates in various assays with the control treatment (i.e., detection rates by FISH with monolabeled probes in formaldehyde-fixed samples)a
| Treatment | Probe
|
|||
|---|---|---|---|---|
| EUB338
|
HGC69a
|
|||
| q′ | P | q′ | P | |
| CARD-FISH, Lys + AP, EtOH | 3.825 | <0.05 | 3.21 | <0.05 |
| CARD-FISH, Lys + AP, FA | 2.687 | <0.05 | 2.32 | NS |
| CARD-FISH, Lys, EtOH | 0.949 | NS | 1.77 | NS |
| CARD-FISH, Lys, FA | 2.598 | <0.05 | 2.53 | <0.05 |
| FISH, monolabeled probe, EtOH | 2.012 | NS | 2.25 | NS |
Abbreviations: FA, formaldehyde fixation; EtOH, ethanol fixation; Lys, lysozyme pretreatment; Lys + AP, lysozyme and achromopeptitase pretreatment; NS, not significant.
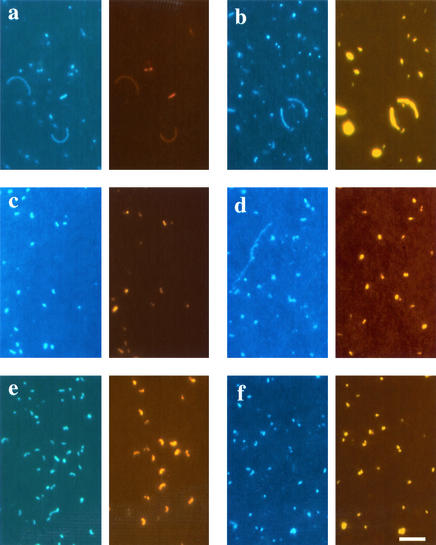
FIG. 4.
Photomicrographs of DAPI-stained and hybridized bacteria from the different lakes. Within each lettered panel, the left image depicts DAPI staining, and the right depicts FISH staining. (a and b) Piburgersee, probe EUB338; (a) FISH with monolabeled probe; (b) CARD-FISH and pretreatment with lysozyme and achromopeptidase (AP). (c to f) CARD-FISH with probe HGC69a; (c) Piburgersee, lysozyme pretreatment; (d) Piburgersee, pretreatment with lysozyme and AP; (e) Fuchskuhle, pretreatment with lysozyme and AP; (f) Lake Cadagno, pretreatment with lysozyme and AP. Scale bar, 5 μm.
Relative abundances of Bacteria in EtOH-fixed samples pretreated with lysozyme and achromopeptidase of Piburgersee and Lake Cadagno after CARD-FISH with probe mix EUB I to III were 81% ± 2% and 82% ± 2%, respectively, which is comparable to the fraction of cells hybridized by CARD-FISH with probe EUB338 (79% ± 4% and 81% ± 3%, respectively) (means ± standard deviations) (Fig. 3). Neither fixation nor pretreatment with lysozyme and achromopeptidase was found to affect bacterial total abundances (data not shown). In all lake samples, the fraction of cells that hybridized with the negative control probe NON338 was <1% irrespective of probe labeling, fixation, or pretreatment.
Quantification of Actinobacteria in environmental samples.
The probes HGC69a and HGC236 (mono-labeled and HRP-labeled) were used to quantify Actinobacteria in lake samples. Samples from Gossenköllesee and Piburgersee contained comparable fractions of hybridized cells with either probe after pretreatment with lysozyme and achromopeptidase and EtOH fixation (Gossenköllesee: HGC69a, 38% ± 1%; HGC236, 36% ± 1%; Piburgersee: HGC69a, 32% ± 2%; HGC236, 30% ± 4%). Probe HGC69a was used for further quantification of Actinobacteria in different lakes and treatments.
Relative abundances of Actinobacteria after CARD-FISH and lysozyme pretreatment were significantly underestimated in formalin-fixed environmental samples, compared to visualization by either FISH with mono-labeled probes or CARD-FISH after pretreatment with lysozyme and achromopeptidase (Fig. 5; Table 2). The relative percentage of Actinobacteria visualized with CARD-FISH after pretreatment with lysozyme and achromopeptidase was higher than after FISH with directly fluorochrome labeled probes in two of the lakes (Lake Fuchskuhle and Lake Cadagno) and equally high in the other two systems. At least in two cases (Piburgersee and Lake Cadagno), higher fractions of hybridized actinobacterial cells were observed after fixation with EtOH. In the pooled data set from all four lakes, significantly more Actinobacteria in EtOH-fixed samples were hybridized by the improved CARD-FISH protocol than by FISH with directly fluorochrome labeled probes (Table 2). Figure 4c and d depict Bacteria after CARD-FISH staining with HGC69a and pretreatment with either lysozyme only or with both lysozyme and achromopeptidase. Lysozyme-treated samples contained fewer bright and many weakly stained Actinobacteria, whereas cells were uniformly stained after pretreatment with lysozyme and achromopeptidase. In Lake Fuchskuhle and Lake Cadagno, remarkable abundances of Actinobacteria were found (54% ± 1% and 55% ± 3%, respectively [EtOH-fixed samples]). Interestingly, different actinobacterial morphotypes were observed in samples from the various lakes (Fig. 4).
FIG. 5.
Detection rate of actinobacteria (probe HGC69a) by FISH with monolabeled probes, CARD-FISH and pretreatment with lysozyme, or CARD-FISH and pretreatment with lysozyme and achromopeptidase (AP) in samples from four lakes. Abbreviations: GKS, Gossenköllesee; PIB, Piburgersee; FUKU, Lake Fuchskuhle; CAD, Lake Cadagno. Error bars, standard deviations.
DISCUSSION
CARD-FISH in freshwater samples.
Our results suggest that the recently published protocol for whole cell hybridization and fluorescence signal amplification of marine picoplankton (26) is not adequate for a general use in freshwater samples. Although the fluorescence intensities of hybridized lake Bacteria were higher after CARD-FISH than after FISH with monolabeled probes, the fraction of cells visualized by an HRP-labeled general bacterial probe (EUB338) was significantly lower than by FISH staining with a monolabeled fluorescent probe (Table 2). Evidence from phylogenetic analysis (16, 36) and previous FISH data (16) both suggested that this might have been due to high numbers of Actinobacteria in the picoplankton of lakes. A typical feature of Actinobacteria isolated from soils is the gram-positive cell wall, which is more robust than cell walls of gram-negative Bacteria and contains a substantially higher fraction of peptidoglycan. We thus hypothesized that the permeabilization pretreatment developed for CARD-FISH of marine bacterioplankton cells was not adequate for freshwater communities.
Actinobacteria from typical aquatic lineages had not been isolated at the time of investigation, and therefore we could not use pure cultures for the testing of permeabilization strategies. Instead, we developed the protocol on samples from an earlier experiment (30). In that study, a bacterial phylotype closely affiliated with uncultured freshwater actinobacteria (>98% sequence similarity) was enriched to high relative abundances (Fig. 2) (29). In addition, the total fraction of bacteria hybridized with monolabeled FISH probes was also very high in these enrichments (Fig. 2), probably because the microbial assemblage was grown under very eutrophic conditions. Samples from the cultivation system therefore represented an ideal model for the initial stage of method development (Fig. 1 and 2), since we could determine the accurate abundances of actinobacteria with monolabeled probes and optimize the CARD-FISH protocol accordingly.
Permeabilization with achromopeptidase.
Although lysozyme hydrolizes the glycosidic bonds of peptidogycans in either Gram-staining cell wall types, it is probably only partly disintegrating the murein multilayers of fixed gram-positive bacterioplankton cells. This would explain why only a fraction of Actinobacteria could be detected by CARD-FISH after lysozyme pretreatment (mean, 53% of cells detected after pretreatment with lysozyme and achromopeptidase, Fig. 5). Several cultured gram-positive genera, e.g., Actinomyces are known to be rather resistant to lysozyme digestion (3). Thus, we introduced a second digestion step using achromopeptidase. This commercially available bacteriolytic preparation is specific for lysine and hydrolyzes lysyl bonds in peptidoglycan (35). Enzymes purified from commercial achromopeptidase like the α-lytic protease have been found to cleave the bonds between N-acetylmuramic acid molecules and the peptides that link neighboring peptidoglycan strands of gram-positive Staphylococcus aureus (22). Another enzyme purified from achromopeptidase, a β-lytic protease, was reported to cleave the linkage between the peptide subunit and the interpeptide bond in peptidoglycan of gram-positive (and to a lesser extend also of gram-negative) cell walls (21).
Permeabilization with achromopeptidase for CARD-FISH was only successful if achromopeptidase was applied after the treatment with lysozyme, but not before (data not shown). Cell wall predigestion is thus improving the action of the achromopeptidase, probably because it enhances enzyme accessibility to the peptide bonds. It should be noted that in contrast to the lysozyme digestion, the achromopeptidase pretreatment was a very sensitive step, and either higher enzyme concentrations (Fig. 1) or shorter incubation times resulted in reduced quality of the CARD-FISH staining and lower fractions of hybridized cells. At higher concentrations the enzyme thus probably also digests the intracellular target proteins that are required for tyramide deposition.
Abundant freshwater Bacteria with gram-positive cell walls?
The significantly higher fraction of hybridized Actinobacteria after CARD-FISH and pretreatment with lysozyme and achromopeptidase indicates that aquatic representatives of this lineage might indeed possess a gram-positive cell wall. This is also supported by our finding that ethanol fixation could in some cases further increase the hybridized cell fraction (Fig. 5). Commonly, gram-positive Bacteria are conserved with ethanol rather than aldehydes for subsequent FISH analysis, because it is denaturing rather than cross-linking cell wall components. This raises the interesting question why bacteria with this type of cell wall might be successful in the water column of lakes. One reason could be that this feature would protect Actinobacteria from digestion by protistan picoplankton feeders, e.g., heterotrophic nanoflagellates (18). Grazing mortality is the most important bacterial elimination process in many freshwater systems, and in laboratory studies, bacterial genotypes affiliated with a typical freshwater actinobacterial lineage were rapidly enriched in the presence of flagellates (29).
Bacterial groups potentially undetected by our approach.
After improvement of the permeabilization pretreatment, the vast majority of lake picoplankton (80 to 99%) could be readily hybridized by EUB338 (Fig. 3). This indicates that the abundances of Archaea in the studied systems were probably low. We did not find any differences between FISH with probe EUB338 or with a probe mix (EUB I to III) that detects several groups not targeted by the former probe (9). It either suggests that neither members of the Verrucomicrobia nor of the Planctomycetes formed substantial populations in the surface waters of the different lakes at the time point of sampling. Alternatively, cell wall permeabilization may have been inadequate for these groups. 16S rRNA sequences affiliated with the class Verrucomicrobia have been reported from a number of lakes (36). However, PCR-based methods produce qualitative diversity evidence and often do not reflect the quantitative importance of genotypes in microbial communities (7, 11). Nevertheless, since our evidence is only indirect, it would be premature to draw general conclusions about the importance of Archaea, Verrucomicrobia, and Planctomycetes in the bacterioplankton of lakes.
Outlook.
Our results show that various morphotypes of Actinobacteria formed a substantial fraction of the bacterioplankton in lakes of different trophic state, geographic location, dissolved organic carbon composition and catchment characteristics (Fig. 4 and 5). In all studied systems the fraction of cells hybridizing with a general actinobacterial probe was >30%, and in surface waters of Lakes Fuchskuhle and Cadagno they even constituted the majority of the total microbial community. In contrast, this group is rare in the water column of some marine environments, e.g., in the coastal North Sea (12). At present we can only speculate about the ecological role of Actinobacteria in different types of freshwaters. Members of this lineage might be able to utilize refractory humic substances, as is known for other gram-positive Bacteria, including actinobacterial genera (13, 20). Humic substances are a prominent component of the dissolved organic carbon pool in many systems (6, 24) (e.g., in Lake Fuchskuhle) and an important carbon source for aquatic Bacteria (2, 34). In combination with methods for tracking per-cell substrate uptake (8, 17), CARD-FISH after pretreatment with lysozyme and achromopeptidase may provide a future means to follow such hypotheses about the role of Actinobacteria in different freshwater systems.
Acknowledgments
This study was supported by the German Ministry of Education and Research (BMBF 01 LC0021/TP4) and the Max Planck Society. R.S. was supported by a grant from the German Academic Exchange Service.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bano, N., M. A. Moran, and R. E. Hodson. 1997. Bacterial utilization of dissolved humic substances from a freshwater swamp. Aquat. Microb. Ecol. 12:233-238. [Google Scholar]
- 3.Barsotti, O., J. J. Morrier, J. Freney, F. Renaud, G. Benay, D. Decoret, and J. Dumnont. 1988. Achromopeptidase for rapid lysis of oral anaerobic Gram-positive rods. Oral Microbiol. Immunol. 3:86-88. [DOI] [PubMed] [Google Scholar]
- 4.Bobrow, M. N., T. D. Harris, K. J. Shaughnessy, and G. J. Litt. 1989. Catalyzed reporter deposition, a novel method of signal amplification—application to immunoassays. J. Immunol. Methods 125:279-285. [DOI] [PubMed] [Google Scholar]
- 5.Bouvier, T. C., and P. A. del Giorgio. 2002. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol. Oceanogr. 47:453-470. [Google Scholar]
- 6.Cole, J. J. 1999. Aquatic microbiology for ecosystem scientists: new and recycled paradigms in ecological microbiology. Ecosystems 2:215-225. [Google Scholar]
- 7.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 10.del Giorgio, P. A., and T. C. Bouvier. 2002. Linking the physiologic and phylogenetic successions in free-living bacterial communities along an estuarine salinity gradient. Limnol. Oceanogr. 47:471-486. [Google Scholar]
- 11.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eilers, H., J. Pernthaler, J. Peplies, F. O. Glöckner, G. Gerdts, and R. Amann. 2001. Isolation of novel pelagic bacteria from the German Bight and their seasonal contribution to surface picoplankton. Appl. Environ. Microbiol. 67:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esham, E. C., W. Y. Ye, and M. A. Moran. 2000. Identification and characterization of humic substances-degrading bacterial isolates from an estuarine environment. FEMS Microbiol. Ecol. 34:103-111. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs, B. M., F. O. Glockner, J. Wulf, and R. Amann. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray, N. D., R. Howarth, R. W. Pickup, J. G. Jones, and I. M. Head. 2000. Use of combined microautoradiography and fluorescence in situ hybridization to determine carbon metabolism in mixed natural communities of uncultured bacteria from the genus Achromatium. Appl. Environ. Microbiol. 66:4518-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iriberri, J., I. Azua, A. Labirua-Iturburu, I. Artolozaga, and I. Barcina. 1994. Differential elimination of enteric bacteria by protists in a freshwater system. J. Appl. Bacteriol. 77:476-483. [DOI] [PubMed] [Google Scholar]
- 19.Klammer, S., T. Posch, B. Sonntag, C. Griebler, B. Mindl, and R. Psenner. Dynamics of bacterial abundance, biomass, activity, and community composition in the oligotrophic Traunsee and the Traun river (Austria). Water Air Soil Pollut., in press.
- 20.Kontchou, C. Y., and R. Blondeau. 1992. Biodegradation of soil humic acids by Streptomyces viridosporus. Can. J. Microbiol. 38:203-208. [DOI] [PubMed] [Google Scholar]
- 21.Li, S. L., S. Norioka, and F. Sakiyama. 1998. Bacteriolytic activity and specificity of Achromobacter beta-lytic protease. J. Biochem. 124:332-339. [DOI] [PubMed] [Google Scholar]
- 22.Li, S. L., S. Norioka, and F. Sakiyama. 1997. Purification, staphylolytic activity, and cleavage sites of alpha-lytic protease from Achromobacter lyticus. J. Biochem. 122:772-778. [DOI] [PubMed] [Google Scholar]
- 23.Methé, B. A., W. D. Hiorns, and J. P. Zehr. 1998. Contrasts between marine and freshwater bacterial community composition: analyses of communities in Lake George and six other Adirondack lakes. Limnol. Oceanogr. 43:368-374. [Google Scholar]
- 24.Nürnberg, G. K., and M. Shaw. 1998. Productivity of clear and humic lakes: nutrients, phytoplankton, bacteria. Hydrobiologia 382:97-112. [Google Scholar]
- 25.Pechlaner, R. 1979. Response of the eutrophied Piburger See to reduced external loading and removal of monimolimnetic water. Arch. Hydrobiol. Beih. Erg. Limnol. 37:293-305. [Google Scholar]
- 26.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 28.Pernthaler, J., F. O. Glöckner, S. Unterholzner, A. Alfreider, R. Psenner, and R. Amann. 1998. Seasonal community and population dynamics of pelagic Bacteria and Archaea in a high mountain lake. Appl. Environ. Microbiol. 64:4299-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pernthaler, J., T. Posch, K. Simek, J. Vrba, A. Pernthaler, F. O. Glöckner, U. Nübel, R. Psenner, and R. Amann. 2001. Predator-specific enrichment of actinobacteria from a cosmopolitan freshwater clade in mixed continuous culture. Appl. Environ. Microbiol. 67:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posch, T., K. Simek, J. Vrba, J. Pernthaler, J. Nedoma, B. Sattler, B. Sonntag, and R. Psenner. 1999. Predator-induced changes of bacterial size-structure and productivity studied on an experimental microbial community. Aquat. Microb. Ecol. 18:235-246. [Google Scholar]
- 31.Rappe, M. S., D. A. Gordon, K. L. Vergin, and S. J. Giovannoni. 1999. Phylogeny of actinobacteria small subunit (SSU) rRNA gene clones recovered from marine bacterioplankton. Syst. Appl. Microbiol. 22:106-112. [Google Scholar]
- 32.Simek, K., D. Babenzien, T. Bittl, R. Koschel, M. Macek, J. Nedoma, and J. Vrba. 1998. Microbial food webs in an artificially divided acidic bog lake. Int. Rev. Hydrobiol. 83:3-18. [Google Scholar]
- 33.Tonolla, M., A. Demarta, R. Peduzzi, and D. Hahn. 1999. In situ analysis of phototrophic sulfur bacteria in the chemocline of meromictic Lake Cadagno (Switzerland). Appl. Environ. Microbiol. 65:1325-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tranvik, L. J. 1988. Availability of dissolved organic-carbon for planktonic bacteria in oligotrophic lakes of differing humic content. Microb. Ecol. 16:311-322. [DOI] [PubMed] [Google Scholar]
- 35.Tsunasawa, S., T. Masaki, M. Hirose, M. Soejima, and F. Sakiyama. 1989. The primary structure and structural characteristics of Achromobacter lyticus protease-I, a lysine-specific serine protease. J. Biol. Chem. 264:3832-3839. [PubMed] [Google Scholar]
- 36.Zwart, G., B. C. Crump, M. Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]