Abstract
Enzymatic reductive dehalogenation of tri-, tetra-, penta-, and hexachlorobenzenes was demonstrated in cell extracts with low protein concentration (0.5 to 1 μg of protein/ml) derived from the chlorobenzene-respiring anaerobe Dehalococcoides sp. strain CBDB1. 1,2,3-trichlorobenzene dehalogenase activity was associated with the membrane fraction. Light-reversible inhibition by alkyl iodides indicated the presence of a corrinoid cofactor.
Chlorinated benzenes are highly persistent pollutants which are ubiquitously distributed in the environment and impose a significant risk for human health (24, 27). Reductive dechlorination of multiple chlorinated benzenes was detected in various anaerobic mixed cultures (3). The isolation of the first anaerobic bacterium capable of reductive dechlorination of chlorinated benzenes, Dehalococcoides sp. strain CBDB1, was previously reported (2). Strain CBDB1 is able to grow with trichlorobenzene (TCB), hydrogen, and acetate, indicating that it conserves energy by using TCB as the terminal electron acceptor in a respiratory process (2). Recently, it was shown that strain CBDB1 also dechlorinates chlorinated dioxins (5). Several anaerobic bacteria couple the reductive dehalogenation of chlorinated aliphatic or aromatic compounds to ATP synthesis via an electron transport chain, a process which is referred to as dehalorespiration (8). Reductive dehalogenases of species dehalorespiring with chlorinated ethenes, phenols, or benzoates have been isolated and characterized (6, 10, 12, 16, 18, 20, 21, 25). In the present study, reductive dehalogenation of TCB and highly chlorinated benzenes was demonstrated in cell extracts of strain CBDB1 and an initial characterization of 1,2,3-TCB dehalogenase activity was carried out.
Dehalococcoides strain CBDB1 was cultivated under strictly anaerobic conditions in flasks sealed with Teflon-lined rubber septa in completely defined medium containing vitamins including 50 ng of vitamin B12/liter, 5 mM acetate, 15 μM 1,2,3-TCB, and 15 μM 1,2,4-TCB (1). The headspace was flushed with N2-CO2 (80%-20%), and hydrogen was added to 8% of the gas phase. Two weeks after inoculation, cultures were supplied with additional 10 mM 1,2,3-TCB (nominal concentration), added as a hexadecane solution (2, 7). Three weeks after inoculation, cultures reached their maximum dechlorination activities. Total protein concentrations of cultures were very low and amounted to 0.5 to 1 μg/ml, corresponding to cell numbers of 107/ml. Cells were harvested at this stage by centrifugation at 10,000 × g for 20 min at 4°C. The barely visible sediment was resuspended in 50 mM Tris-HCl, pH 7.5, containing 1.5 mM titanium(III) citrate (28). Five to 10 ml of the resulting cell suspension (0.5 to 1 μg of protein/ml) was subjected to three cycles of French press treatment at 28 MPa, and the lysate was centrifuged at 10,000 × g for 20 min at 4°C. The supernatant (crude extract) contained 0.3 to 0.5 μg of protein/ml.
Dehalogenase activity was routinely measured with reduced methyl viologen (MV) as the artificial electron donor (17). The final assay contained 100 mM Tris-HCl (pH 7.5), 1 mM MV, 2 mM titanium(III) citrate, and chlorobenzene congeners at concentrations of 50 μM (TCBs; 1,2,3,4-tetrachlorobenzene [1,2,3,4-TeCB]; 1,2,3,5-TeCB) or 15 μM (chlorinated benzenes with very low solubility in water: 1,2,4,5-TeCB; pentachlorobenzene [PeCB]; hexachlorobenzene [HCB]). 1,2,3-TCB was used as a model substrate. To test different electron donors, MV in the assay solution was replaced by 1 mM ethyl viologen (EV), 1 mM benzyl viologen (BV), 1 mM anthraquinone 2,6-disulphonic acid (AQDS), or 1 mM NADH. Reactions were started by addition of 200 μl of crude extract corresponding to about 100 ng of protein. After 5 to 120 min of incubation at 25°C (when necessary, incubation was extended to 24 h), reactions were stopped by extracting the reaction mixture with hexane. Extracts were analyzed by gas chromatography and flame ionization detection (2). 1,2,3-TCB dechlorination in whole-cell suspensions was determined with the same assay.
1,2,3-TCB, 1,2,4-TCB, and 1,2,3,4-TeCB were obtained from Merck-Schuchard (Hohenbrunn, Germany). 1,3,5-TCB, 1,2,4,5-TeCB, and PeCB were from Aldrich (Steinheim, Germany). 1,2,3,5-TeCB was purchased from Riedel-de Haën (Seelze, Germany). HCB was from Fluka (Neu-Ulm, Germany). Viologens and AQDS were from Aldrich. Solvents were from Roth (Karlsruhe, Germany) and Merck-Schuchard. Alkyl iodides were obtained from Acros (Nidderau, Germany) and Aldrich. Buffers and other chemicals were from AppliChem (Darmstadt, Germany), Fluka, and Merck-Schuchard.
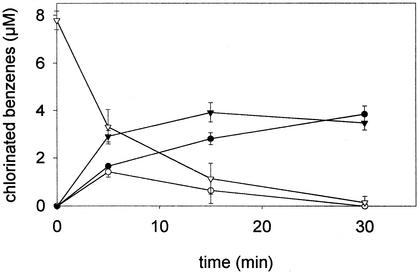
Although the amount of protein obtained from cultures of strain CBDB1 was extremely low, dechlorination of all tested chlorobenzene congeners except 1,3,5-TCB could be demonstrated in crude extracts (Table 1). Final dechlorination products were 1,3-dichlorobenzene (1,3-DCB), 1,4-DCB, and 1,3,5-TCB. PeCB dechlorination proceeded via the formation of 1,2,3,5-TeCB and 1,2,4,5-TeCB (Fig. 1). 1,2,3,5-TeCB was further dechlorinated to 1,3,5-TCB, whereas 1,2,4,5-TeCB temporarily accumulated due to its slow dechlorination rate (Table 1). HCB was dechlorinated to 1,3,5-TCB, 1,3-DCB, and 1,4-DCB via PeCB. With heat-inactivated crude extracts, no dechlorination could be detected. To our knowledge, this is the first report of reductive dechlorination of PeCB and HCB by cell extracts of a pure culture. Previously, it was demonstrated that strain CBDB1 in culture reductively dechlorinates 1,2,3-TCB, 1,2,4-TCB, and all TeCB isomers with hydrogen as the electron donor (2). In contrast, PeCB and HCB were not dechlorinated by the pure culture (2). Experiments are in progress to find the reasons for the difference between results obtained with PeCB and HCB in vitro and in culture.
TABLE 1.
Dehalogenase activity in crude extracts with highly chlorinated benzenes as electron acceptorsa
| Added chlorobenzene congener | First dechlorination product(s) | Specific activity ± SD (nkat/mg of protein)b |
|---|---|---|
| 1,2,3-TCB | 1,3-DCB | 11 ± 0.7 |
| 1,2,4-TCB | 1,3-DCB, 1,4-DCB | 0.3 ± 0.0 |
| 1,3,5-TCB | <0.05 | |
| 1,2,3,4-TeCB | 1,2,4-TCB | 355 ± 21 |
| 1,2,3,5-TeCB | 1,3,5-TCB | 76 ± 1.7 |
| 1,2,4,5-TeCB | 1,2,4-TCB | 3 ± 0.1 |
| PeCB | 1,2,3,5-TeCB, 1,2,4,5-TeCB | 171 ± 12 |
| HCB | PeCB | 0.4 ± 0.0 |
Means of results of triplicate assays ± standard deviation.
One nanokatal is defined as 1 nmol of dechlorination products formed per s at 25 °C.
FIG. 1.
Product formation from PeCB by cell extracts of strain CBDB1. PeCB, open triangles; 1,2,4,5-TeCB, solid triangles; 1,2,3,5-TeCB, open circles; 1,3,5-TCB, solid circles. Values are means of results of triplicate assays ± standard deviation.
Reductive dehalogenases from other dehalorespiring bacteria dechlorinating chlorobenzoate, chlorophenol, or chlorinated ethenes have specific activities between 2.5 and 25 nkat/mg in crude extracts with MV as the electron donor (8). 1,2,3-TCB dehalogenase activity in crude extracts of strain CBDB1 showed a similar specific activity of 11 nkat/mg, but much higher specific activities were obtained for dechlorination of 1,2,3,4-TeCB (355 nkat/mg) and PeCB (171 nkat/mg). Whether all detected dechlorination reactions are mediated by one enzyme or several enzymes is not yet known.
In crude extracts of strain CBDB1, 1,2,3-TCB dehalogenase activity was highest with MV (Eo′ = −446 mV [14]) as the electron donor. With EV (Eo′ = −480 mV [14]) and BV (Eo′ = −360 mV [14]), 89 and 12% of the MV-supported activity were measured, respectively. With AQDS (Eo′ = −184 mV [4]) and NADH (Eo′ = −320 mV) or in the absence of any added electron donor, no dehalogenation was observed within 24 h. A higher activity with MV as the electron donor than with EV was also described for the tetrachloroethene (PCE) dehalogenases of Dehalospirillum multivorans and Desulfitobacterium sp. strain PCE-S (14, 15). For D. multivorans, a hampered access of the larger molecule EV to the electron-accepting site of the dehalogenase was assumed (14). Whereas no dechlorination with BV was detected in crude extracts of D. multivorans and Desulfitobacterium chlororespirans Co23 (11, 14), BV supported dehalogenase activity in strain PCE-S (15), Dehalococcoides ethenogenes (I. Nijenhuis and S. H. Zinder, Abstr. 100th Gen. Meet. Am. Soc. Microbiol. 2000, abstr. Q-126, p. 569, 2000), and strain CBDB1. The results suggest that the cofactors of the dehalogenases from D. multivorans and D. chlororespirans Co23 possess more negative redox potentials than the cofactors of the dehalogenases from strain PCE-S, D. ethenogenes, and strain CBDB1, although steric effects cannot be excluded. For D. ethenogenes, it was suggested that no quinone is involved in the respiratory electron transport (Nijenhuis and Zinder, Abstr. 100th Gen. Meet. Am. Soc. Microbiol. 2000). In crude extracts of strain CBDB1, the quinone analog AQDS did not serve as electron donor for dechlorination, also suggesting that quinones are not involved in the transport of electrons to TCB.
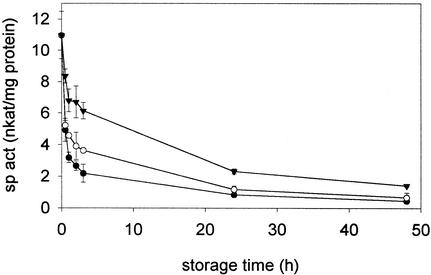
To assess oxygen sensitivity of 1,2,3-TCB dehalogenase, crude extracts of strain CBDB1 were stored at 4°C under (i) reduced anaerobic conditions [Eh < −200 mV; anaerobic gas phase; extracts reduced with titanium(III) citrate (28)], (ii) anoxic conditions [Eh ≥ 0 V; anaerobic gas phase; extracts without titanium(III) citrate], or (iii) aerobic conditions [aerobic gas phase; extracts without titanium(III) citrate]. Crude extracts lost most of the initial TCB dehalogenase activity during 48 h of storage. Anoxic and even more anaerobic storage conditions increased the stability of the enzyme significantly (Fig. 2). The pH optimum of 1,2,3-TCB dehalogenase activity in crude extracts was 6.1. Crude extracts lost dehalogenase activity after 1 h of incubation at 60°C or above.
FIG. 2.
Aerotolerance and stability of TCB dehalogenase activity. Crude extracts were stored at 4°C under reduced anaerobic conditions (Eh < −200 mV) (triangles), anoxic conditions (Eh ≥ 0 V) (open circles), or aerobic conditions (solid circles). Values are means of results of triplicate assays ± standard deviation. sp act, specific activity.
Reductive dehalogenases of most dehalorespiring species are light-reversibly inhibited by alkyl iodides, indicating the involvement of corrinoids as cofactors (8, 10, 12, 15, 17). In fact, corrinoids have been detected in purified enzymes (6, 10, 16, 19, 21). In whole cells and crude extracts of strain CBDB1, the presence of 10 μM ethyl iodide or propyl iodide in the dark reduced 1,2,3-TCB dehalogenase activity to 50 to 75% of that of control samples without inhibitor. Activity could be restored to 78 to 96% of control activity by 10 min of exposure to the light of a 250-watt lamp. The results suggest the involvement of a corrinoid cofactor also in dehalogenation of chlorobenzenes by strain CBDB1.
1,2,3-TCB dehalogenase activity of strain CBDB1 was found to be associated with the membrane fraction which was obtained after ultracentrifugation of the crude extract at 120,000 × g for 1 h. Treatment of whole cells with 0.01% Triton X-100 or 10 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) resulted in solubilization of TCB dehalogenase activity. Solubilization by such low concentrations of the nonionic detergent Triton X-100 indicates a peripheral attachment to the membrane (22). This was also suggested for trichloroethene dehalogenase from D. ethenogenes, which could be extracted from the membrane with 0.1% Triton X-100 or 1 M NaCl (13).
The orientation of reductive dehalogenases of dehalorespiring bacteria in the cell membrane is still under investigation. For PCE dehalogenase of strain PCE-S, experiments with the hydrophilic electron donor MV indicate a cytoplasmic orientation of the enzyme (15). In contrast to PCE dechlorination in strain PCE-S, dechlorination of 1,2,3-TCB in strain CBDB1 with MV as the electron donor did not increase after cell disruption. Also, permeabilization of whole cells with 0.4% toluene did not enhance dechlorination. MV has been shown to permeate cell membranes only to a very limited extent in Escherichia coli (9). Therefore, our findings match with the hypothesis that TCB dehalogenase of strain CBDB1 is oriented to the outside of the membrane, as was also suggested for the reductive dehalogenases of D. ethenogenes (13) and Desulfitobacterium dehalogenans (23, 26).
Acknowledgments
We thank G. Wagner for technical assistance and G. Jayachandran for helpful discussions and establishing the GC conditions for quantitation of TeCB, PeCB, and HCB.
This work was supported by the Deutsche Forschungsgemeinschaft project number AD 178/1.
REFERENCES
- 1.Adrian, L., W. Manz, U. Szewzyk, and H. Görisch. 1998. Physiological characterization of a bacterial consortium reductively dechlorinating 1,2,3- and 1,2,4-trichlorobenzene. Appl. Environ. Microbiol. 64:496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 3.Adrian, L., and H. Görisch. 2002. Microbial transformation of chlorinated benzenes under anaerobic conditions. Res. Microbiol. 153:131-137. [DOI] [PubMed] [Google Scholar]
- 4.Benz, M., B. Schink, and A. Brune. 1998. Humic acid reduction by Propionibacterium freudenreichii and other fermenting bacteria. Appl. Environ. Microbiol. 64:4507-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Görisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 6.Christiansen, N., B. K. Ahring, G. Wohlfarth, and G. Diekert. 1998. Purification and characterization of the 3-chloro-4-hydroxy-phenylacetate reductive dehalogenase of Desulfitobacterium hafniense. FEBS Lett. 436:159-162. [DOI] [PubMed] [Google Scholar]
- 7.Holliger, C., G. Schraa, A. J. M. Stams, and A. J. B. Zehnder. 1992. Enrichment and properties of an anaerobic mixed culture reductively dechlorinating 1,2,3-trichlorobenzene to 1,3-dichlorobenzene. Appl. Environ. Microbiol. 58:1636-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holliger, C., G. Wohlfarth, and G. Diekert. 1999. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383-398. [Google Scholar]
- 9.Jones, R. W., and P. B. Garland. 1977. Sites and specificity of the reaction of bipyridylium compounds with anaerobic respiratory enzymes of Escherichia coli. Biochem. J. 164:199-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krasotkina, J., T. Walters, K. A. Maruya, and S. W. Ragsdale. 2001. Characterization of the B12- and iron-sulfur-containing reductive dehalogenase from Desulfitobacterium chlororespirans. J. Biol. Chem. 276:40991-40997. [DOI] [PubMed] [Google Scholar]
- 11.Löffler, F. E., R. A. Sanford, and J. M. Tiedje. 1996. Initial characterization of a reductive dehalogenase from Desulfitobacterium chlororespirans Co23. Appl. Environ. Microbiol. 62:3809-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magnuson, J. K., R. V. Stern, J. M. Gossett, S. H. Zinder, and D. R. Burris. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, E., G. Wohlfarth, and G. Diekert. 1997. Studies on tetrachloroethene respiration in Dehalospirillum multivorans. Arch. Microbiol. 166:379-387. [DOI] [PubMed] [Google Scholar]
- 15.Miller, E., G. Wohlfarth, and G. Diekert. 1997. Comparative studies on tetrachloroethene reductive dechlorination mediated by Desulfitobacterium sp. strain PCE-S. Arch. Microbiol. 168:513-519. [DOI] [PubMed] [Google Scholar]
- 16.Miller, E., G. Wohlfarth, and G. Diekert. 1998. Purification and characterization of tetrachloroethene reductive dehalogenase of strain PCE-S. Arch. Microbiol. 169:497-502. [DOI] [PubMed] [Google Scholar]
- 17.Neumann, A., G. Wohlfarth, and G. Diekert. 1995. Properties of tetrachloroethene and trichloroethene dehalogenase of Dehalospirillum multivorans. Arch. Microbiol. 163:276-281. [Google Scholar]
- 18.Neumann, A., G. Wohlfarth, and G. Diekert. 1996. Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J. Biol. Chem. 271:16515-16519. [DOI] [PubMed] [Google Scholar]
- 19.Neumann, A., A. Siebert, T. Trescher, S. Reinhardt, G. Wohlfarth, and G. Diekert. 2002. Tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans: substrate specificity of the native enzyme and its corrinoid cofactor. Arch. Microbiol. 177:420-426. [DOI] [PubMed] [Google Scholar]
- 20.Ni, S., J. K. Fredrickson, and L. Xun. 1996. Purification and characterization of a novel 3-chlorobenzoate-reductive dehalogenase from the cytoplasmic membrane of Desulfomonile tiedjei DCB-1. J. Bacteriol. 177:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schumacher, W., C. Holliger, A. J. B. Zehnder, and W. R. Hagen. 1997. Redox chemistry of cobalamin and iron-sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett. 409:421-425. [DOI] [PubMed] [Google Scholar]
- 22.Scopes, R. K. 1994. Protein purification: principles and practice, 3rd ed., p. 38-42. Springer Verlag, New York, N.Y.
- 23.Smidt, H. 2001. Molecular characterization of anaerobic dehalogenation by Desulfitobacterium dehalogenans. Ph.D. thesis. Wageningen University, Wageningen, The Netherlands.
- 24.U. S. Environmental Protection Agency. 2002. Persistent organic pollutants (POPs). Publication no. EPA160-F-02-001. U.S. Environmental Protection Agency, Washington, D.C.
- 25.Van de Pas, B. A., H. Smidt, W. R. Hagen, J. van der Oost, G. Schraa, A. J. M. Stams, and W. M. de Vos. 1999. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J. Biol. Chem. 274:20287-20292. [DOI] [PubMed] [Google Scholar]
- 26.Van de Pas, B. A., S. Jansen, C. Dijkema, G. Schraa, W. M. de Vos, and A. J. M. Stams. 2001. Energy yield of respiration on chloroaromatic compounds in Desulfitobacterium dehalogenans. Appl. Environ. Microbiol. 67:3958-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization. 1991. Chlorobenzenes other than hexachlorobenzene. Environmental Health Criteria 128, World Health Organization, Geneva, Switzerland.
- 28.Zehnder, A. J. B., and K. Wuhrmann. 1976. Titanium(III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science 194:1165-1166. [DOI] [PubMed] [Google Scholar]