Abstract
Some Bacillus subtilis strains, including natto (fermented soybeans) starter strains, produce a capsular polypeptide of glutamate with a γ-linkage, called poly-γ-glutamate (γ-PGA). We identified and purified a monomeric 25-kDa degradation enzyme for γ-PGA (designated γ-PGA hydrolase, PghP) from bacteriophage ΦNIT1 in B. subtilis host cells. The monomeric PghP internally hydrolyzed γ-PGA to oligopeptides, which were then specifically converted to tri-, tetra-, and penta-γ-glutamates. Monoiodoacetate and EDTA both inhibited the PghP activity, but Zn2+ or Mn2+ ions fully restored the enzyme activity inhibited by the chelator, suggesting that a cysteine residue(s) and these metal ions participate in the catalytic mechanism of the enzyme. The corresponding pghP gene was cloned and sequenced from the phage genome. The deduced PghP sequence (208 amino acids) with a calculated Mr of 22,939 was not significantly similar to any known enzyme. Thus, PghP is a novel γ-glutamyl hydrolase. Whereas phage ΦNIT1 proliferated in B. subtilis cells encapsulated with γ-PGA, phage BS5 lacking PghP did not survive well on such cells. Moreover, all nine phages that contaminated natto during fermentation produced PghP, supporting the notion that PghP is important in the infection of natto starters that produce γ-PGA. Analogous to polysaccharide capsules, γ-PGA appears to serve as a physical barrier to phage absorption. Phages break down the γ-PGA barrier via PghP so that phage progenies can easily establish infection in encapsulated cells.
Bacterial capsules that are exposed to the outermost cell surface play important roles in cell attachment for colonization, biofilm formation, and protecting cells from serum and phagocytosis (11, 12, 20, 22). Since capsules coat phage receptors on the cell surface, they also function as physical barriers against bacteriophages. The protective function of capsules against phage infection has been demonstrated with the capsular polysaccharides (CPSs) of several types of bacteria (5, 6, 9, 23, 37). However, CPS barriers are not always effective for all types of phages. Some Escherichia coli phages possess an enzyme that can degrade CPSs (4, 16, 18, 20, 24). For instance, E. coli phage 29 has an endo-N-acetylneuraminidase in its spike that hydrolyzes repeating poly-α-2,8-linked sialosyl units of K1 capsules (7, 16). Some phages of Klebsiella and Streptococcus strains also have CPS degradation activities associated with their particles (10, 25, 27).
Some strains of B. subtilis, B. licheniformis, and B. anthracis produce a unique capsule polymer of γ-linked glutamate, poly-γ-glutamate (γ-PGA) (30). The γ-PGA of B. subtilis contains almost equimolar amounts of l- and d-glutamate and the ratio of the d-isomer increases to about 80% when cells are cultured in the presence of MnCl2 (21). The capABC genes encoding the γ-PGA synthetic system were originally identified in B. anthracis (19). Thereafter, their homologues were found in many Bacillus strains including B. subtilis (2, 3, 34). B. subtilis CapABC proteins are structurally and functionally equivalent to the B. anthracis counterparts (2, 3, 34), but the B. subtilis genes are referred to as pgsABC or ywsC-ywtAB (2, 3, 34). The ywsC, ywtA, and ywtB genes have been renamed capB, capC, and capA, respectively (35). To avoid confusion with pgsA that has been assigned to a gene for phosphatidylglycerophosphate synthase (15), we also refer to the capsule synthetic genes of B. subtilis NAFM5 as capABC (DDJB accession number AB039950). Unlike B. anthracis, which forms γ-PGA in response to carbon dioxide (30), B. subtilis exclusively produces the capsule during the stationary growth phase through regulation by the ComQXPA quorum-sensing machinery (32). Because γ-PGA is very viscous, this capsule polypeptide, like CPSs, could perform a barrier function against phagocytosis and phage infection. Indeed, Makino et al. (19) have demonstrated a protective function of B. anthracis γ-PGA against phagocytosis by leukocytes. No evidence yet supports the notion of a hypothetical barrier function against phages. Rather, incidents contradictory to a barrier function are often experienced in natto factories. Although B. subtilis starters produce voluminous amounts of γ-PGA during fermentation, natto products are often contaminated with phages and then tend to rapidly lose γ-PGA viscosity when mixed for serving. An early study (14) identified and partially purified a γ-PGA depolymerase in a B. natto (= B. subtilis) culture infected with phage NP-1 cl, but did not investigate the role of the enzyme in phage infection and the corresponding gene. These findings indicated that phages produce a γ-PGA-degradation enzyme and that this enzyme may contribute to the infection of encapsulated host cells by eliminating the capsule.
To further characterize the γ-PGA degradation enzyme, we isolated phage ΦNIT1 from a natto product, purified its γ-PGA hydrolase (PghP) to homogeneity, and identified the enzyme gene in the phage genome. We found that the enzyme is a novel γ-glutamyl hydrolase that randomly hydrolyzes γ-PGA into oligo-γ-glutamates and then specifically into tri-, tetra-,and penta-γ-glutamate. We showed that ΦNIT1 has an apparent advantage over a phage that lacks PghP in terms of proliferation on encapsulated host cells. These findings support the notion that γ-PGA protects cells from phage absorption and that PghP degrades the capsular barrier to allow phage progenies to infect encapsulated host cells.
MATERIALS AND METHODS
Strains, plasmids, phages and media. B. subtilis NAFM5 (Rifr) is a spontaneous rifampin-resistant mutant of the natto starter strain Miyagino (Miura Natto starter) cured of cryptic plasmids pLS20 and pTA1015 as described (21). Bacteriophages ΦNIT1, FSG, KKP, MOP, ONPC, ONPB, P-1, SUP, SS2P, and THP were isolated from independent natto products with suspected phage contamination. B. subtilis typing phages (BS5, CS1, F, Phi105, Phi29, Phi3T, PS10, PS50, S-a, and SPP1) and their host strains were obtained from Ackermann et al. (1). E. coli and B. subtilis strains were cultured in Luria-Bertani (LB) medium (26). B. subtilis NAFM5 was cultured on GSP for the production of γ-PGA as described (21). E. coli was transformed and selected on LB agar containing an appropriate antibiotic as described previously (32).
Assay for PghP.
γ-PGA (5 × 106 Da) was extracted and purified from B. subtilis NAFM5 culture incubated on GSP with or without 0.1 mM MnCl2 (21). The d-glutamate contents of the polypeptides from MnCl2-free and 0.1 mM MnCl2 media were 56 and 80%, respectively, as determined according to Nagai et al. (21). A standard mixture for the PghP assay (1 ml; 1 mg of γ-PGA with 56% of d-glutamate, 10 mM sodium phosphate [pH 7.5], 150 mM NaCl, enzyme) was incubated at 37°C for the indicated periods. The viscosity of reaction mixtures was measured using a falling ball viscometer (Haake) equipped with a syringe (0.5 ml) containing a stainless steel ball (3 mm in diameter) and is expressed as falling velocity (centimeters per second) of the ball. Degradation products of γ-PGA were detected by agarose gel electrophoresis. Portions (10 μl) of reaction mixtures sampled after the indicated incubation periods were resolved by electrophoresis at 6 V/cm for 30 min on 1.0% agarose gels using TAE (40 mM Tris-hydroxylaminomethane, 1 mM EDTA, 0.14% [vol/vol] acetic acid) running buffer. Resolved degradation products on the gels were visualized by staining with methylene blue (0.23% [wt/vol] methylene blue, 23% [vol/vol] ethanol, 0.008% [wt/vol] KOH) for 10 min, followed by destaining with water.
Purification of PghP.
B. subtilis NAFM5 cultured overnight in 20 ml of LB medium was inoculated into 1 liter of fresh LB medium along with phage ΦNIT1 at a multiplicity of infection of 5 × 10−4. The culture was vigorously shaken at 37°C for 5 h. Cells and cell debris were removed by centrifugation, and then the enzyme was precipitated from the supernatant by adding ammonium sulfate (40 to 80% saturation), dissolved in 35 ml of buffer A (25 mM sodium phosphate buffer, pH 6.8, and 10 mM NaCl) and dialyzed against buffer A. After centrifugation at 100,000 × g for 1 h, dialyzed proteins were loaded on a DEAE-Sepharose column equilibrated with buffer A. Active fractions eluted from the column using a gradient of NaCl (0 to 0.4 M) in buffer A were pooled and dialyzed against 10 mM sodium phosphate buffer, pH 6.8 (buffer B). Proteins were precipitated with ammonium sulfate (70% saturation), dialyzed against the same buffer, and fractionated by DEAE-Sepharose column chromatography as described above. Active fractions were combined, dialyzed against buffer B containing 1 M ammonium sulfate, and then loaded onto a Butyl-Toyopearl column (Tosoh). Active proteins were eluted using a reverse gradient (1 to 0 M) of ammonium sulfate in buffer B. Fractions containing the enzyme were pooled, dialyzed in buffer B, and fractionated by chromatography through a MonoQ column (Amersham Biosciences). The enzyme was eluted from the column using a gradient of 0 to 0.4 M NaCl in buffer B, concentrated with a Centriprep-10 (Millipore), and finally purified by Superose 12 (Amersham Biosciences) gel filtration column chromatography using buffer C (10 mM sodium phosphate, pH 6.9, 150 mM NaCl).
Analysis of reaction products.
γ-PGA was incubated with purified PghP (25 ng) in the standard reaction mixture (1.5 ml) at 37°C. A portion (0.27 ml) was withdrawn from the reaction mixture after various incubation periods, boiled for 15 min to terminate the reaction, and fractionated through Superdex peptide HR10/30 (Amersham Biosciences) using buffer C. Degradation products were detected by absorbance at 210 nm. The molecular mass of reaction products was estimated from their elution volumes relative to those of glutamate (Mr = 147), di-γ-glutamate (Mr = 276; Bachem), tetra-γ-glutamate (Mr = 535; Hokkaido System Science),and gastrin (Mr = 2,126; Bachem). A mass spectrometer (Apex II 70e; Bruker Daltonics) also determined the molecular masses of reaction products. Salts in reaction samples were removed by filtration through TSK-GEL ALPHA-2500 (Tosoh) before mass spectrometry. Amounts of l-glutamate were measured using NAD-l-glutamate dehydrogenase as described previously (36).
Cloning of pghP.
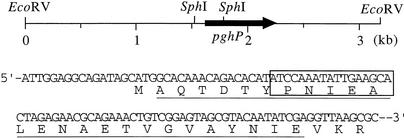
We initially identified restriction fragments of ΦNIT1 DNA carrying pghP appropriate for cloning, by Southern blotting. ΦNIT1 DNA was digested with various restriction enzymes and blotted onto a Hybond N+ membrane (Amersham Biosciences) after resolution by agarose gel electrophoresis (26). Southern blotting using 32P-end-labeled oligonucleotides, with 5′-TA(C/T)CCGAA(C/T)ATTGA(A/G)GC-3′ as the probe, identified a 250-bp SphI fragment and an 3.2-kb EcoRV fragment that carry the relevant region of pghP: the nucleotide probes correspond to the 6th to 11th residues (YPNIEA) of the determined amino terminal sequence for PghP. The SphI fragment was extracted from an agarose gel and cloned into plasmid pKF19 (Takara Shuzo) at the SphI site. E. coli DH5α (Bethesda Research Laboratories) transformants harboring the 250-bp pghP fragment on the plasmid were screened by colony hybridization using 32P-end-labeled oligonucleotides as described previously (26). A 3.2-kb EcoRV fragment carrying pghP was also extracted from an agarose gel and cloned into plasmid pKF19 at the HincII site and then transformed into E. coli DH5α. Transformants harboring pghP on the plasmid were selected by colony hybridization using the 32P-labeled 250-bp SphI fragment as the probe. The pghP sequence was determined using plasmid pNAG201 harboring pghP in a positive clone.
Phage titration.
Either phage ΦNIT1 (pghP+) or BS5 (pghP−) (104 PFU/ml) was inoculated into log-phase (optical density at 600 nm = 0.3) or stationary-phase cultures (24 h incubation) of B. subtilis NAFM5 shaken in 20 ml of GSP medium at 37°C. The incubation was continued for the indicated periods at 37°C with shaking. Aliquots (0.5 ml each) were withdrawn from the cultures and mixed with a few drops of chloroform. After removing cells and cell debris by centrifugation and subsequent filtration through a membrane filter (pore size, 0.45 μm), phage samples were diluted as required with LB medium. Portions (0. 1 ml) of diluted phage were added to 3 ml of soft LB agar containing 0.8% (wt/vol) agarose and 2 × 107 cells of strain NAFM5. Soft agar was quickly mixed and overlaid on bottom LB layers (1.5% [wt/vol] agarose) in plates. After incubation at 37°C overnight, the number of plaques that developed was counted. Strain NAFM5 does not produce γ-PGA in LB medium.
Nucleotide and amino acid sequencing.
Nucleotides were sequenced using a Dye terminator cycle sequencing kit (Applied Biosystems) and an ABI 310A DNA sequencer (Applied Biosystems). The pghP sequence was deposited in DDJB/EMBL/GenBank databases under accession number AB091475. Purified PghP (10 pmol) was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (17) and blotted onto a polyvinylidene difluoride membrane (Millipore). The amino terminus was then determined using a HP G10000A protein sequencer (Hewlett-Packard).
RESULTS
Identification of a γ-PGA-degradation enzyme in a culture infected with ΦNIT1.
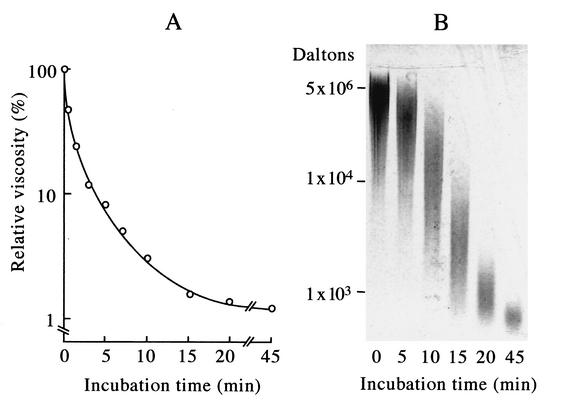
Bacteriophage ΦNIT1 was isolated from a natto product that contained small amounts of γ-PGA, an indication of phage contamination. To examine whether this phage produces a γ-PGA degradation enzyme, we cultured B. subtilis NAFM5, a derivative of a natto starter, along with phage ΦNIT1 at a multiplicity of infection of 5 × 10−4 in LB medium at 37°C for 5 h. Incubation with the culture supernatant caused γ-PGA to rapidly lose viscosity (Fig. 1A) and undergo fragmentation as confirmed by agarose gel electrophoresis (Fig. 1B). B. subtilis strains produce an unidentified γ-PGA degradation enzyme during the late stationary growth phase (31). However, neither viscometry nor agarose gel electrophoresis detected γ-PGA degradation activity in the culture supernatant or in extracts of host cells. In addition, γ-PGA degradation activity was undetectable by the phage particles. These results indicated that the γ-PGA degradation enzyme is specifically synthesized in host cells harboring the phage and that the enzyme is not associated with the phage particle. We designated the enzyme PghP, for γ-PGA hydrolase of phage.
FIG. 1.
γ-PGA degradation activity in a B. subtilis NAFM5 culture infected with ΦNIT1 phage measured by viscometry (A) and by agarose gel electrophoresis (B). Reactions proceeded as described in Materials and Methods, and portions of mixtures were withdrawn after the indicated incubation periods. (A) Viscosity was measured using a falling-ball viscometer and is expressed as relative viscosity to the mixture at zero time. (B) Samples (10 μl) were resolved by 1.0% agarose gel electrophoresis, and degradation products were visualized by staining with methylene blue. Positions of intact γ-PGA (5 × 106 Da) and of 104 Da and 103 Da γ-PGA (fractionated from partial hydrolysates of γ-PGA by PghP through gel filtration and high-performance liquid chromatography) (21) are indicated at left.
Purification and characterization of PghP.
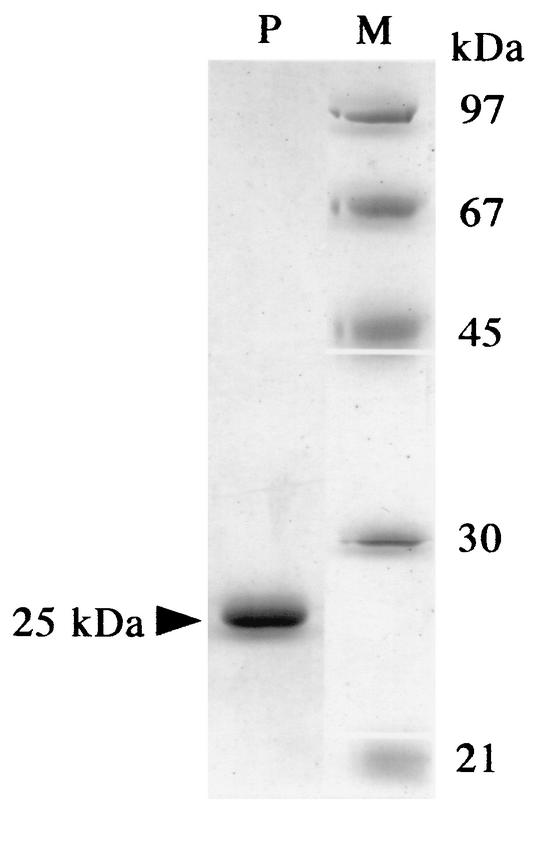
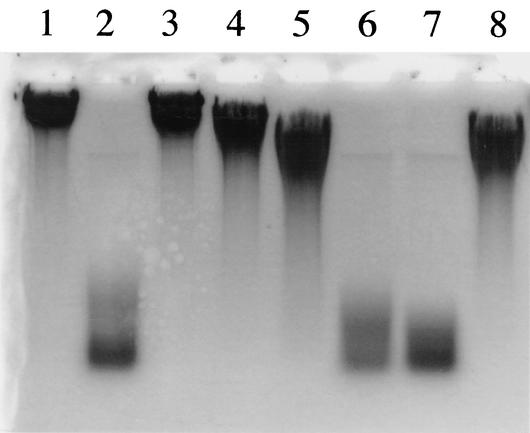
We purified PghP from a ΦNIT1-infected NAFM5 culture through five chromatographic steps as described in Materials and Methods. The purified enzyme migrated as a single band of 25-kDa protein on SDS-PAGE (Fig. 2). This value was the same as the Mr (25,000) determined for native PghP by Superose 12 gel filtration chromatography, indicating that PghP is a monomer of 25 kDa. The first 25 amino acids of the amino-terminal sequence of PghP was AQTDTYPNIEALENAETVGVAYNIE. As described below, the deduced nucleotides corresponding to residues 6 to 11 (YPNIEA) of the amino terminus were used as probes in Southern blotting and in phgP cloning. EDTA (1 mM) powerfully inhibited the enzyme, and an excess of MnCl2 and ZnCl2, but not MgCl2 and CaCl2, fully restored the activity inhibited by the chelator (Fig. 3). Monoiodoacetate also inhibited the enzyme (Fig. 3).
FIG. 2.
SDS-PAGE of purified PghP. PghP (1 μg, lane P) from Superose 12 column chromatography was analyzed together with molecular mass markers (1 μg each, lane M) by SDS-10% PAGE (17). Molecular markers: phosphorylase b (97 kDa), bovine serum albumin (67 kDa), ovalbumin (45 kDa), carbonic anhydrase (30 kDa), and catalase (21 kDa).
FIG. 3.
Effects of monoiodoacetate, EDTA, and divalent cations on PghP activity. Reactions proceeded at 37°C for 60 min in PghP assay mixture (see Materials and Methods) containing 50 mM Tris-HCl (pH 7.5) instead of 50 mM sodium phosphate (pH 7.5), and reaction products were analyzed using agarose gel electrophoresis. Lane 1, no enzyme (control). Lanes 2 through 8 contained 45 ng of purified PghP/ml plus the following: lane 2, no further addition; lane 3, 1 mM EDTA; lane 4, 1 mM EDTA + 5 mM CaCl2; lane 5, 1 mM EDTA + 5 mM MgCl2; lane 6, 1 mM EDTA + 5 mM MnCl2, lane 7, 1 mM EDTA + 5 mM ZnCl2; lane 8, 1 mM monoiodoacetate. Reaction mixtures for lanes 4 to 7 were incubated for 5 min before adding chlorides.
Analysis of degradation products.
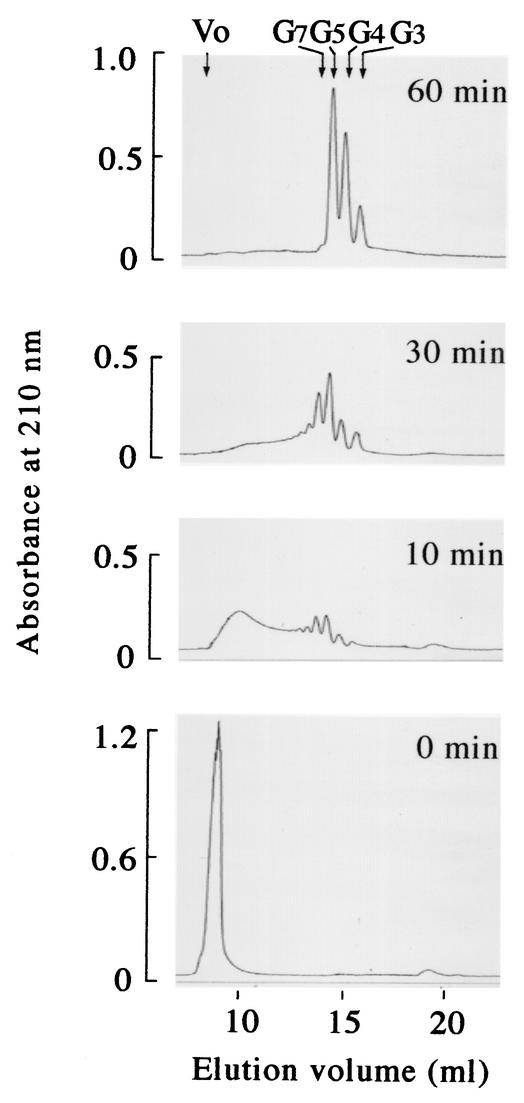
Glutamate was undetectable even after prolonged incubation or after incubation with more of the purified enzyme, implying that this enzyme does not degrade γ-PGA into the monomer. We accordingly determined the sizes of the smallest degradation products by gel filtration chromatography. Like crude enzyme (Fig. 1A and B), purified PghP rapidly degraded γ-PGA of 5 × 106 Da into smaller fragments. The substrate was degraded into fragments of diverse sizes within 10 min (Fig. 4 [10 min]). The average molecular mass of the products in the first peak was approximately 105 Da as determined by comparison with the elution volume relative to those of pullulans in gel filtration chromatography (21). The smallest peptide was a trimer, and relatively large amounts of pentamers and heptamers were generated (Fig. 4 [10 min]). Degradation intermediates larger than octamers were further degraded to accumulate trimers, tetramers, pentamers, and heptamers within a further 20 min of incubation (Fig. 4 [30 min]). By 60 min, only trimers, tetramers, and pentamers were detected (Fig. 4 [60 min]). These oligo-γ-glutamates were not further cleaved by prolonged incubation or by more of the enzyme. Hydrolysis of heptamers did not yield monomers or dimers, and the amounts of trimers and tetramers significantly increased relative to that of pentamers (Fig. 4 [30 and 60 min]), suggesting that heptamers are specifically converted into trimers and tetramers. Mass spectroscopic analyses confirmed the presence of these oligomers in the 60-min reaction mixture. We propose a hydrolytic mechanism for PghP on oligo-γ-glutamates in the Discussion.
FIG. 4.
Analysis of γ-PGA degradation products generated by PghP over reaction time. Reactions proceeded under standard assay conditions (see Materials and Methods). Samples (0.2 ml) after various incubation periods were applied onto Superdex peptide columns, and reaction products were monitored by absorbance at 210 nm. Molecular sizes of oligomers were estimated from elution volumes relative to those of glutamate (Mr = 147), di-γ-glutamate (Mr = 276), tetra-γ-glutamate (Mr = 535), and gastrin (Mr = 2,126). Abbreviations: Vo, void volume; G3, tri-γ-glutamate; G4, tetra-γ-glutamate; G5, penta-γ-glutamate; G7, hepta-γ-glutamate.
Cloning and structure of pghP.
Southern blotting using oligonucleotides deduced from the determined amino terminal sequence of PghP showed that the 5′-region of pghP is located on a 250-bp SphI segment of a 3.2-kb EcoRV fragment of ΦNIT1 DNA (Fig. 5). We initially cloned the SphI segment and sequenced the nucleotides to confirm that the segment contains the amino terminal region of pghP that is identical to the determined sequence, except for the first methionine, which appears to be eliminated posttranslationally (Fig. 5). We subsequently cloned the 3.2-kb EcoRV fragment and determined the entire sequence of pghP (DDJB/EMBL/GenBank accession number AB091475) located between 1.6 and 2.3 kb (Fig. 5). The predicted Mr of PghP (203 amino acids) was 22,939, which was in agreement with the Mr (25,000) of purified PghP. A BLAST search using the deduced PghP sequence did not reveal any similar proteins.
FIG. 5.
Location of pghP on an EcoRV fragment and the amino-terminal region of PghP. Deduced and determined amino-terminal sequences match perfectly (underline). Possible ribosome-binding site (GGAGG) precedes initiation ATG codon at 9 bp upstream. Nucleotides 5′-TA(C/T)CCGAA(C/T)ATTGA(A/G)GC-3′ used in Southern blotting and in cloning pghP correspond to region between residues 7 and 12 (boxed) of deduced amino-terminal sequence.
Infection of encapsulated cells with phage ΦNIT1.
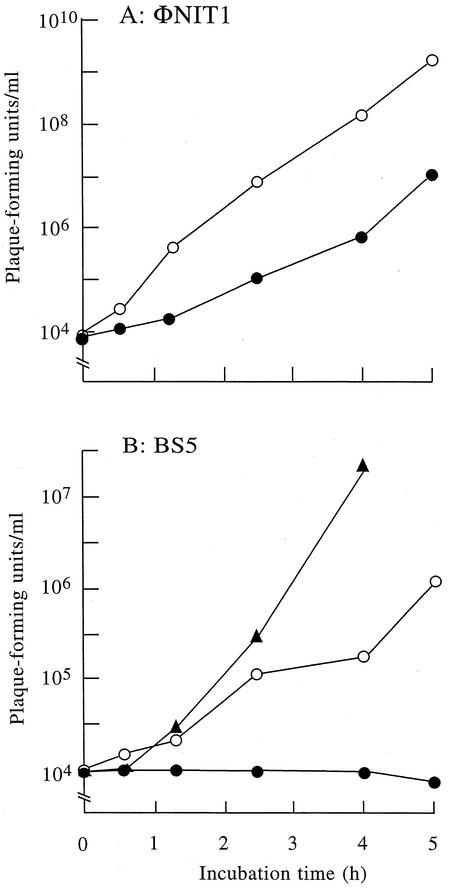
For the above experiments we incubated phage ΦNIT1 in log-phase cultures in which strain NAFM5 does not produce capsule (32). Thus, whether phage ΦNIT1 could infect encapsulated cells remained unclear. To investigate the role of PghP in the infection of encapsulated host cells, we compared the growth of phage ΦNIT1 with that of a phage lacking PghP. To achieve this, we required a phage that can infect strain NAFM5 but which does not produce PghP. We tested the ability of 10 B. subtilis typing phages (1) to infect strain NAFM5 and to produce PghP. We found that typing phage BS5 proliferates in NAFM5 cells but does not produce PghP. Phage BS5 produced smaller plaques than phage ΦNIT1, implying a smaller burst size or longer lytic cycle of this phage. We titrated the progenies of phages ΦNIT1 and BS5 during incubation in log (noncapsulated)- and stationary (encapsulated)-phase cultures of strain NAFM5. Phage ΦNIT1 exponentially produced progenies in the log-phase culture after a short lag time (Fig. 6A). Although a longer lag time was required for PghP synthesis, this phage also produced significant numbers of progenies (107 PFU/ml after a 5-h incubation) in the stationary-phase culture (Fig. 6A). Phage BS5 also developed progenies in the log-phase culture at lower rates than ΦNIT1 but did not generate any progeny in stationary phase cultures even after 5 h (Fig. 6B). When PghP was added to the stationary-phase culture along with phage BS5 at a concentration of 1 μg/ml, which roughly corresponds to the enzyme concentration in phage-infected cultures, the phage became infective to the stationary-phase cells of strain NAFM5 and multiplied as in log-phase culture (Fig. 6B). These results indicated that the stationary-phase cells are still susceptible to BS5 but γ-PGA prevents the phage from gaining access to cell surface receptors.
FIG. 6.
Effects of γ-PGA capsule on growth of phages with or without PghP. B. subtilis NAFM5 was shaken in GSP medium (22) at 37°C. When the optical density at 600 nm of the cultures reached 0.3 (noncapsulated [open circles]) or after 24 h (encapsulated stationary phase [closed circles]), phages ΦNIT1 (PghP+) (A) and BS5 (PghP−) (B) were added at concentrations of 104 PFU/ml and incubation was continued at 37°C. Purified PghP (1 μg/ml) was added to stationary-phase cultures together with BS5 (closed triangle). After indicated periods, phages in cultures were titrated against strain NAFM5 as the indicator.
Distribution of PghP-producing phages.
PghP appears to be necessary for phages to infect encapsulated hosts. Presumably phages that contaminate natto products must form PghP to infect B. subtilis starters that produce large quantities of γ-PGA capsule during natto fermentation. To examine this hypothesis, we propagated nine phages isolated from independent natto products in strain NAFM5 and assayed PghP activities in the culture supernatants. We found as much PghP activity as that of phage ΦNIT1 in all the culture supernatants (data not shown). Thus, PghP may be widely distributed in B. subtilis phages, and when a phage capable of producing PghP appears in fermenting natto, it would easily propagate and spoil the natto product. To investigate whether PghP occurs in other B. subtilis phages isolated from different sources, we assayed PghP activity in cultures incubated with each of ten B. subtilis typing phages that were screened without considering the γ-PGA productivity of the hosts (1). Among the typing phages, four (CS1, S-a, SP10, and SP50) produced PghP.
DISCUSSION
Irrespective of their chemical composition, bacterial capsules that cover the cell surface would perturb bacteriophage access to their cell surface receptors. We showed that like CSPs (5, 6, 9, 23, 27, 37), the γ-PGA capsule of B. subtilis also functions as a physical barrier against phages (Fig. 6B). In addition, analogous to phages that can infect CPS-encapsulated cells by dismantling the capsule with a degradation enzyme (4, 7, 10, 16, 24, 25, 27, 37), phage ΦNIT1 achieves absorption by eliminating γ-PGA on host cells with PghP (Fig. 6A and B). In contrast to CPS degradation enzymes that are associated with phage particles (4, 10, 16, 18, 25, 27), PghP is produced in the absence of such association. The different locations of degradation enzymes may correlate with distinct mechanisms of capsule synthesis by host cells. In contrast to E. coli that forms capsule independently of growth phase (8), B. subtilis strains form γ-PGA under the high-cell-density conditions of stationary phase (32). Phage ΦNIT1, which does not have PghP as its component, may not easily access receptors on encapsulated cells. However, an infective phage produces the enzyme in host cells that will be released outside host cells during lysis, which would eliminate γ-PGA on neighboring cells and thereby allow its progenies to easily access cell surface receptors. PghP would efficiently degrade capsules when the cell density is high, particularly in colonies or layers on solid surfaces such as that of fermenting soybeans, where diffusion of the enzyme is limited.
Phages often cause serious damage in natto factories. Problems associated with phage contamination are not only poor fermentation but also the rapid decrease in viscosity of natto products (an important factor in natto quality). Contamination with a few phages does not cause an apparent failure of fermentation but can cause a significant decrease in slime viscosity when mixed for serving. Phage ΦNIT1 produces about 1 mg of PghP per liter under appropriate culture conditions, and 1 μg of the enzyme completely hydrolyzes 1.5 mg of γ-PGA within 1 min to the final products. Our present results showed that PghP, which is produced at high levels and has powerful degradation activity, is responsible for the rapid decay of γ-PGA slime in natto that is contaminated with phage.
Other reported enzymes that cleave the γ-glutamyl linkage include γ-PGA depolymerase of B. anthracis (33), γ-PGA hydrolase of Myrothecium sp. (29), carboxypeptidase G (EC 3.4.17.11) of Pseudomonas sp. (28), animal γ-glutamyl hydrolase (EC 3.4.19.9) (13, 38), and animal glutamate carboxypeptidase II (EC 3.4.17.21) (13). Among these, carboxypeptidase G, γ-glutamyl hydrolase, and glutamate carboxypeptidase II have been well characterized and their corresponding genes have been cloned. γ-Glutamyl hydrolase is a thiol enzyme that externally and internally hydrolyzes the γ-glutamyl tail of folyl-γ-PGAs (38). Glutamate carboxypeptidase II is an endopeptidase of folyl-γ-PGA that requires Zn2+, but not cysteine, for activity (13). Pseudomonas carboxypeptidase G (EC 3.4.17.11) is also a zinc metalloenzyme that releases carboxyl-terminal glutamate residues from γ-glutamyl peptides and folyl-γ-PGAs (28). Besides the carboxypeptidases, many peptidyl and nonpeptidyl C-N hydrolases use Zn2+ or Mn2+ to activate the hydroxyloxide of water that makes a nucleophilic attack on the carbon atom of a C-N bond. PghP requires both cysteine and Zn2+ or Mn2+ for activity (Fig. 3), implying that they participate in the reaction mechanism. PghP has two cysteine residues at positions 91 and 156. However, these regions do not show any local homology with the region of γ-glutamyl hydrolase containing the essential cysteine residue (Cys110) (38). The cysteine residue(s) of PghP might be involved either in formation of a thiolester enzyme-substrate intermediate or in binding to metal ions. This requires elucidation.
PghP internally and randomly degrades γ-PGA and large oligo-γ-glutamates (Fig. 1B and Fig. 4 [10 min]). Within 30 min of incubation, trimers, tetramers, pentamers, and heptamers become dominant (Fig. 4 [30 min]). PghP therefore appears inert to trimers, tetramers, and pentamers, thus accumulating these peptides as the final product (Fig. 4 [60 min]). High levels of heptamers relative to larger oligopeptides suggest that PghP hydrolyzes heptamers more slowly than it does the other larger oligopeptides. The absence of monomers and dimers in the final product indicates the specific conversion of hexamers to two trimers. Heptamers and octamers may also be specifically cleaved into trimers and tetramers and into trimers and pentamers or into two tetramers. Thus, the cleavage of oligo-γ-glutamates by PghP is not random. The specific hydrolysis of oligo-γ-glutamates suggests that this enzyme binds to oligo-γ-glutamates at six γ-glutamyl residues and therefore may have low binding-affinity for oligopeptides that are smaller than pentamers. Assuming that hydrolysis occurs at the center of the binding site, this substrate-binding mode is quite consistent with the product profiles of PghP (Fig. 4 [60 min]). Alternatively, PghP might recognize the γ-linkages of either d- or l-glutamate in γ-PGA. However, this is less likely because PghP generated tri-, tetra-, and penta-γ-glutamates from γ-PGA consisting of 80% d-glutamate in the same amounts as those from the polypeptide consisting of 56% d-glutamate (Fig. 4 [60 min]).
The γ-PGA degradation enzyme of phage NP-1 cl produces dimers and trimers as the final product (14). Perhaps the substrate-binding site of this enzyme corresponds to a tetrapeptide. It may therefore be able to bind pentamers and tetramers and to cleave them into dimers and trimers and into two dimers, respectively. Our preliminary Southern blot experiments showed that ΦNIT1 pghP hybridizes with the counterparts of phages KKP, MOP, ONPC, ONPB, SUP, SS2P, THP, and SP50, but not with those of phages CS1, FSG, P-1, S-a, and SP10 (data not shown). Phage PghPs appear to be heterogeneous with respect to primary structure as well as mode of oligopeptide cleavage. B. subtilis typing phages are not related to each other or to natto phages in terms of morphology, restriction profiles of genomic DNA, and host range (1). Nevertheless, the pghP gene also occurs in some typing phages. In conclusion, PghP is widely distributed in B. subtilis phages and significantly contributes to the infection of B. subtilis strains that produce γ-PGA.
Acknowledgments
We thank K. Nagashima for providing phage ΦNIT1 and H.-W. Ackerman for the gift of typing phages and their host strains.
This study was supported in part by a grant-in-aid from the Ministry of Agriculture, Forestry and Fisheries.
REFERENCES
- 1.Ackermann, H.-W., R. R. Azizbekyan, R. L. Bernier, H. de Barje, S. Saindouk, J.-R. Valéro, and M.-X. Yu. 1995. Phage typing of Bacillus subtilis and B. thuringensis. Res. Microbiol. 146:643-657. [DOI] [PubMed] [Google Scholar]
- 2.Ashiuchi, M., C. Nawa, T. Kamei, J.-J. Song, S.-P. Hong, M.-H. Sung, K. Soda, T. Yagi, and H. Misono. 2001. Physical and biochemical characterization of poly-γ-glutamate synthase complex of Bacillus subtilis. Eur. J. Biochem. 268:5321-5328. [DOI] [PubMed] [Google Scholar]
- 3.Ashiuchi, M., K. Soda, and H. Misono. 1999. A poly-γ-glutamate synthetic complex of Bacillus subtilis IFO3336: gene cloning and biochemical analysis of poly-γ-glutamate produced by Escherichia coli clone cells. Biochem. Biophys. Res. Commun. 263:6-12. [DOI] [PubMed] [Google Scholar]
- 4.Bayer, M. E., H. Thurow, and M. H. Bayer. 1979. Penetration of the polysaccharide capsule of Escherichia coli (Bi161/42) by bacteriophage K29. Virology 94:95-118. [DOI] [PubMed] [Google Scholar]
- 5.Benedi, V. J., B. Ciurana, and J. M. Tomas. Isolation and characterization of Klebsiella pneumoniae unencapsulated mutants. J. Clin. Microbiol. 27:82-87. [DOI] [PMC free article] [PubMed]
- 6.Bernheimer, H. P., and J. G. Tiraby. 1976. Inhibition of phage infection by pneumococcus capsule. Virology 73:308-309. [DOI] [PubMed] [Google Scholar]
- 7.Bessler, W., F. Fehmel, E. Freund-Molbert, H. Knufermann, and S. Stirm. 1975. Escherichia coli capsule bacteriophages. IV. Free capsule depolymerase 29. J. Virol. 15:976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bliss, J. M., and R. P. Silver. 1996. Coating the surface: a model for expression of capsular polysialic acid in Escherichia coli K1. Mol. Microbiol. 21:221-231. [DOI] [PubMed] [Google Scholar]
- 9.Burt, S., S. Meldrum, D. R. Woods, and D. T. Jones. 1978. Colonial variation, capsule formation, and bacteriophage resistance in Bacteroides thetaiotaomicron. Appl. Environ. Microbiol. 35:439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cescutti, P., and S. Paoletti. 1994. On the specificity of a bacteriophage-borne endoglycanase for the native capsular polysaccharide produced by Klebsiella pneumoniae SK1 and its derived polymers. Biochem. Biophys. Res. Commun. 198:1128-1134. [DOI] [PubMed] [Google Scholar]
- 11.Cross, A. S., K. S. Kim, D. C. Wright, J. C. Sadoff, and P. Gemski. 1986. Role of lipopolysaccharide and capsule in the serum resistance of bacteremic strains of Escherichia coli. J. Infect. Dis. 154:497-503. [DOI] [PubMed] [Google Scholar]
- 12.Czuprynski, C. J., and A. K. Sample. 1990. Interactions of Haemophilus-Actinobacillus-Pasteurella bacteria with phagocytic cells. Can. J. Vet. Res. 54(Suppl.):S34-S40. [PubMed] [Google Scholar]
- 13.Galivan, J., T. J. Ryan, K. Chave, M. Rhee, R. Yao, and D. Yin. 2000. Glutamyl hydrolase: pharmacological role and enzymatic characterization. Pharmacol. Ther. 85:207-215. [DOI] [PubMed] [Google Scholar]
- 14.Hongo, M., and A. Yoshimoto. 1970. Bacteriophage of Bacillus natto. part III. Action of phage-induced γ-polyglutamic acid depolymerase on γ-polyglutamic acid and the enzymatic hydrolyzates. Agric. Biol. Chem. 34:1055-1063. [Google Scholar]
- 15.Kunst, F., et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 16.Kwiatkowski, B., B. Boschek, H. Thiele, and S. Stirm. 1982. Endo-N-acetylneuraminidase associated with bacteriophage particles. J. Virol. 43:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Long, G. S., J. M. Bryant, P. W. Taylor, and J. P. Luzio. 1995. Complete nucleotide sequence of the gene encoding bacteriophage E endosialidase: implications for K1E endosialidase structure and function. Biochem. J. 309:543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makino, S., I. Uchida, N. Terakado, C. Sasakawa, and M. Yoshikawa. 1989. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J. Bacteriol. 171:722-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCallum, K. L., D. H. Laakso, and C. Whitfield. 1989. Use of a bacteriophage-encoded glycanase enzyme in the generation of lipopolysaccharide O side chain deficient mutants of Escherichia coli O9:K30 and Klebsiella O1:K20: role of O and K antigens in resistance to complement-mediated serum killing. Can. J. Microbiol. 35:994-999. [DOI] [PubMed] [Google Scholar]
- 21.Nagai, T., K. Koguchi, and Y. Itoh. 1997. Chemical analysis of poly-γ-glutamic acid produced by plasmid-free Bacillus subtilis (natto): Evidence that plasmids are not involved in poly-γ-glutamic acid production. J. Gen. Appl. Microbiol. 43:139-143. [DOI] [PubMed] [Google Scholar]
- 22.Ohman, D. E., K. Kalai, C. J. McPherson, C. A. DeVries, S. Ma, D. J. Wozniak, and M. J. Franklin. 1996. Regulation of the alginate (algD) operon in Pseudomonas aeruginosa, p. 472-483. In T. Nakazawa, K. Furukawa, D. Haas, and S. Silver (ed.) Molecular biology of pseudomonads. ASM Press, Washington, D.C.
- 23.Ohshima, Y., F. Schumacher-Perdreau, G. Peters, and G. Pulverer. 1988. The role of capsule as a barrier to bacteriophage adsorption in an encapsulated Staphylococcus simulans strain. Med. Microbiol. Immunol. 177:229-233. [DOI] [PubMed] [Google Scholar]
- 24.Pelkonen, S., J. Aalto, and J. Finne. 1992. Differential activities of bacteriophage depolymerase on bacterial polysaccharide: binding is essential but degradation is inhibitory in phage infection of K1-defective Escherichia coli. J. Bacteriol. 174:7757-7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rieger-Hug, D., and S. Stirm. 1981. Comparative study of host capsule depolymerases associated with Klebsiella bacteriophages. Virology 113:363-378. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Saxelin, M. L., E. L. Nurmiaho, M. P. Korhola, and V. Sundman. 1979. Partial characterization of a new C3-type capsule-dissolving phage of Streptococcus cremoris. Can. J. Microbiol. 25:1182-1187. [DOI] [PubMed] [Google Scholar]
- 28.Sherwood, R., R. G. Melton, S. M. Alwan, and P. Hughes. 1985. Purification and properties of carboxypeptidase G2 from Pseudomonas aeruginosa sp. strains RS-16. 148:447-453. [DOI] [PubMed]
- 29.Tanaka, T., O. Hirata, T. Futamura, K. Uotani, A. Satoh, M. Taniguchi, and S. Oi. 1993. Purification and characterization of poly(γ-glutamic acid) hydrolase from a filamentous fungus, Myrothecium sp. TM-422. Biosci. Biotech. Biochem. 57:2148-2153. [Google Scholar]
- 30.Thorne, C. B. 1993. Bacillus anthracis. P. 113-124. In A. L. Sonenshein, J. A. Hock, and R. Losick (ed.), Bacillus subtilis and other Gram-positive bacteria, American Society for Microbiology, Washington, D.C.
- 31.Throne, C. B., C. G. Gómez, H. E. Noyes, and R. D. Housewright. 1954. Production of glutamyl polypeptide by Bacillus subtilis. J. Bacteriol. 68:307-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran, L.-S. P., T. Nagai, and Y. Itoh. 2000. Divergent structure of the comQXPA quorum-sensing components: molecular basis of strain-specific communication mechanism in Bacillus subtilis. Mol. Microbiol. 37:1159-1171. [DOI] [PubMed] [Google Scholar]
- 33.Uchida, I., S. Makino, C. Sasakawa, M. Yoshikawa, C. Sugimoto, and N. Terakado. 1993. Identification of a novel gene, dep, associated with depolymerization of the capsule polymer in Bacillus anthracis. Mol. Microbiol. 9:487-496. [DOI] [PubMed] [Google Scholar]
- 34.Urushibata, Y., S. Tokuyama, and Y. Tahara. 2002. Characterization of the Bacillus subtilis ywsC gene, involved in γ-polyglutamic acid production. J. Bacteriol. 184:337-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urushibata, Y., S. Tokuyama, and Y. Tahara. 2002. Difference in transcription levels of cap genes for γ-polyglutamic acid production between Bacillus subtilis IFO16449 and Marburg 168. J. Biosci. Bioeng. 93:252-254. [DOI] [PubMed] [Google Scholar]
- 36.Valero, E., and F. Garcia-Carmona. 1998. A continuous spectrophotometric method based on enzymatic cycling for determining L-glutamate. Anal. Biochem. 259:265-271. [DOI] [PubMed] [Google Scholar]
- 37.Wilkinson, B. J., and K. M. Holmes. 1979. Staphylococcus aureus cell surface: capsule as a barrier to bacteriophage adsorption. Infect. Immun. 23:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao, R., E. Schneider, T. J. Ryan, and J. Galivan. 1996. Human γ-glutamyl hydrolase: cloning and characterization of the enzyme expressed in vitro. Proc. Natl. Acad. Sci. USA 93:10134-10138. [DOI] [PMC free article] [PubMed] [Google Scholar]