Abstract
Approximately 10% of Streptococcus pyogenes strains inhibit the growth of all nine indicators in a standardized streptococcal bacteriocin typing scheme. The present study has shown that this inhibitory profile, referred to as bacteriocin producer (P)-type 777 activity, is due to the type A1 lantibiotic streptin. Two major forms of streptin were purified to homogeneity from 95% acidified (pH 2) methanol extracts of S. pyogenes M25 cells by using a series of reversed-phase chromatographic separations. The fully processed form of streptin (streptin 1) is a 23-amino-acid peptide with a mass of 2,424 Da. The 2,821-Mr form of the peptide (streptin 2) has three additional amino acids (TPY) at the N terminus. Strain M25 extracts also contained small quantities of the streptin 1 and streptin 2 peptides in various stages of dehydration. Streptin 1 and streptin 2 were each capable of specifically inducing streptin production when added to strain M25 cultures. The streptin gene cluster resembled that of other type A1 lantibiotics but appeared to lack a streptin-specific proteinase gene. Although the streptin structural gene (srtA) was widespread within S. pyogenes, being detected in 40 of 58 strains, each representing a different M serotype, only 10 of these srtA-positive strains produced active streptin. The failure of some strains to express streptin was attributed to an ∼4.5-kb deletion in their streptin loci, encompassing genes putatively encoding proteins involved in streptin processing (srtB and srtC) and transport (srtT). In other strains, srtA transcription appeared to be defective. No direct association could be detected between the production of streptin and the production of the lantibiotic-like hemolysin streptolysin S in strain M25.
Bacteriocin production by Streptococcus pyogenes was first documented in 1971 (30). Streptococcin A-FF22 (SA-FF22), the inhibitory product of S. pyogenes strain FF22, was later found to have characteristics similar to those of nisin, the “prototype” of the lantibiotic class of bacteriocins produced by certain strains of Lactococcus lactis (32).
A bacteriocin “fingerprinting” scheme for the typing of beta-hemolytic streptococci uses a set of nine standard indicator bacteria (I1 to I9) to detect patterns of inhibitory activity produced in a deferred antagonism test on blood agar medium (29). In practice, these patterns are converted to numerical code designations called bacteriocin production (P) types. For example, S. pyogenes strain FF22 is P-type 436 (29). Testing of a set of 54 S. pyogenes, each strain representing a different M protein serotype, indicated that P-type patterns 655, 614, and 777 were given only by the M-type 4, 57, and 60 strains, respectively (29). Subsequent studies have shown that a homologue of the S. salivarius lantibiotic salivaricin A (21) is produced uniquely in S. pyogenes by M-type 4 strains (unpublished data). Similarly, all tested M-type 57 strains of S. pyogenes appear to produce a unique bacteriocin-like inhibitory substance (BLIS), in this case a plasmid-encoded, heat-labile protein (26).
More recent testing of 73 M-prototype S. pyogenes strains showed 10 of these (types 2, 11, 12, 25, 28, 60, 66, 67, 71, and 76) to be P-type 777 (28). There was no apparent correlation detected between the production of P-type 777 activity and the disease association of the S. pyogenes. It was noted, however, that except for M-types 12, 67, and 71, all of the P-type 777-producing strains were opacity factor positive (13). The inhibitory spectrum of the P-type 777 strains was shown to include a wide variety of streptococci and, surprisingly, one strain of the gram-negative species Bacteroides intermedius (13) (now Prevotella intermedia [24]). Hynes and Tagg (14) partially purified the P-type 777 BLIS from the serotype M25 S. pyogenes strain M25 and showed that there was no apparent link between its production and the production of streptolysin S (SLS) (14). Furthermore, in all of nine Tn916-derived BLIS-negative mutants of strain M25 the production of SLS did not appear to have been affected (14). In other experiments, all of six protease-negative variants derived after treatment of strain M25 with nitrosoguanidine, although remaining unchanged in their production of SLS, were found to have become BLIS negative (14). This indicated a potential involvement of streptococcal proteinase in production of P-type 777 activity.
Recently, a novel locus consisting of 10 open reading frames was identified in an S. pyogenes strain of undefined M-type (16). It was noted by the authors that the structural gene (srtA) for the putative lantibiotic (named streptin) had “significant homology with nisA.” Associated with the loss of streptin production after Tn916 mutagenesis, there appeared to be an associated loss of SLS activity, causing the authors of that study to speculate that streptin gene products may have a role in SLS synthesis, modification, maturation, or secretion (16).
In the present study the purification of streptin is documented. We further demonstrate that streptin is responsible for P-type 777 BLIS activity in S. pyogenes and reaffirm our previous observation that its production is not directly linked to that of SLS.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The S. pyogenes isolates used in the present study were strains M25 (wild type, streptin+ [P-type 777]) (14), Tn216 (streptin− derivative of strain M25 generated by Tn916 mutagenesis [P-type 000]) (14), SF370 (M-type 1, streptin− [P-type 000]) (8), Blackmore (SLS+ SLO−) and C203U (SLS− SLO+) (31), M-type 60 strains 76068 (P-type 000) and 73220 (P-type 777), M-type 11 strains 74823 (P-type 000) and 71948 (P-type 777), M-type 4 strain 148 (P-type 655), and M-type 57 prototype strain (P-type 614) were from the laboratory culture collection. The standard indicator strains (I1 to I9) and the set of 73 prototype S. pyogenes (M-types 1 to 81) have been described previously (28). S. equi subsp. zooepidemicus strain 4881 was the source of the muralytic BLIS zoocin A (25). Routine culture was at 37°C in 5% CO2 in air on BaCa (Columbia agar base [CAB; Life Technologies, Ltd., Paisley, United Kingdom], supplemented with 5% [vol/vol] human blood and 0.1% [wt/vol] CaCO3). Incubation to enhance streptin production was at 30°C in air. Other culture media utilized in the present study were Ba (CAB supplemented with 5% [vol/vol] human blood), Ba supplemented with cholesterol (0.005 g/ml), and Todd-Hewitt broth (THB; Difco, Becton Dickinson, Sparks, Md.).
Streptin assays.
A series of doubling dilutions of the test preparation was prepared in distilled water purified with a MilliQ system (MQ water; Millipore, Inc., Molsheim, France). A total of 20 μl of each dilution was placed on the surface of a dry BaCa plate. The drops were allowed to dry, and the agar surface was sterilized with chloroform vapour. The indicator organism, Micrococcus luteus T-18 (indicator I1), was applied by charging a sterile swab with an 18-h THB culture and applying evenly over the surface of the agar. The titer (in arbitrary units [AU]/milliliter) of the inhibitory activity in the original test preparation was defined as the reciprocal of the highest dilution to inhibit the growth of the indicator lawn.
DNA extraction.
Chromosomal DNA for Southern analysis was extracted by the method of Upton et al. (34). Briefly, confluent growth from one-half of an agar plate culture was collected in TE buffer (10 mM Tris-HCl [pH 8] containing 10 mM EDTA), and the cells were harvested by centrifugation (8,000 × g for 10 min) and suspended in 0.3 ml of a lysis mixture (50 mM Tris-HCl [pH 8.0] containing 10 mM EDTA and 2% [vol/vol] Triton X-100) was added. Lysis was achieved by adding 30 U of mutanolysin (Sigma) and then 0.3 mg of pronase (Sigma), followed by incubation of the preparation at 37°C for 2 h. The suspension was extracted once with 0.6 ml of phenol, three times with 0.6 ml of phenol-chloroform (0.3 ml of phenol plus 0.3 ml of chloroform), and once with 0.6 ml of chloroform-isoamyl alcohol (24:1). Nucleic acids were precipitated from the aqueous phase with 2 volumes of 100% ethanol at −70°C and collected by centrifugation (12,000 × g for 20 min), and the pellet was washed with 70% (vol/vol) ethanol, air dried, and dissolved in TE buffer. In a number of experiments the solutions were then incubated with 50 μg of RNase A (Sigma)/ml at 37°C for 30 min and then extracted with phenol-chloroform, and the DNA was precipitated, washed, and dissolved in TE buffer as described above. Streptococcal DNA for use as a template in PCRs was isolated as described elsewhere (2). The streptococci were grown overnight on BaCa. One loopful of growth was suspended in 300 μl of 0.8% NaCl and heated for 30 min at 60°C. Cells were centrifuged and resuspended in 100 μl of TE (10 mM Tris, 1 mM EDTA [pH 8]) containing 300 U of mutanolysin (Sigma) and 30 μg of hyaluronidase (Sigma)/ml for 30 min at 37°C. Samples were then heated at 100°C for 10 min and briefly centrifuged to pellet debris. Then, 1 μl of supernatant was used as a template for each 50-μl PCR mixture.
Inverse PCR and sequencing of the streptin locus.
Identification of the Tn916 insertion site in the streptin operon of strain Tn216 was achieved by using inverse PCR (33). Approximately 5 μg of chromosomal DNA from S. pyogenes strain Tn216 was digested 4 h at 37°C with HindIII (20-μl total volume). Then, 5 μl of this digest was ligated for 18 h at 12°C with T4 ligase (2 μl of T4 buffer, 1 μl of T4 ligase, and 12 μl of MQ water). Next, 1 μl of the ligation mix was used as a template in a PCR of 30 cycles, consisting of an annealing temperature of 60°C and elongation time of 7 min at 68°C with primers based on the Tn916 sequence (GenBank accession no. U09422), i.e., primers 1537 (positions 17954 to 17984; 5′-TACGCGGCCGCACATAGAATAAGGCTTTACG) and 1546 (positions 12238 to 12209; 5′-CCTGCGGCCGCGCTTCCTAATTCTGTAATC). This amplified an ∼7-kb fragment of the streptin operon. The PCR products were sequenced directly with a Perkin-Elmer ABI 377A sequencer. Primary sequence data was collated with SeqEd sequencer software, and sequence alignments, translation, and general analyses were performed by using either DNAMAN (Lynnon BioSoft, Vaudreuil, Canada) or Lasergene 99 expert sequence analysis software (DNASTAR, Inc., Madison, Wis.). Sequences were analyzed for homology by comparison to sequences in DNA and protein databases by using the BLAST facilities on the National Center for Biotechnology Information (NCBI) server (http://www.ncbi.nlm.nih.gov) and the University of Oklahoma server (http://www.genome.ou.edu/strep).
Production of streptin.
A biphasic culture system was used to obtain streptin in a liquid form. This consisted of 50 ml of BaCa base in a 500-ml Schott bottle on which a lawn culture of strain M25 was grown 24 h at 30°C prior to overlaying the culture with 150 ml of THB supplemented with 1.8 × 10−6 mol liter−1 CaCO3 and 0.1% (wt/vol) maltose and reincubation for 24 h. The cells from 2 liters of THB from biphasic cultures of strain M25 were harvested by centrifugation (15,300 × g for 10 min) and treated with 500 ml of 95% (vol/vol) methanol for 18 h at 4°C. The cells were again collected, and the streptin activity was extracted by treatment for 18 h at 4°C with 200 ml of acidified (pH 2) methanol. Removal of the methanol by rotary evaporation gave 10 ml of crude streptin. Five 2-ml aliquots of the crude streptin were passed through a pretreated (5 ml of 80% [vol/vol] methanol, followed by 5 ml of MQ water) Sep-Pak C-18 classic cartridge (Millipore). Serial washes of the cartridge with 4-ml quantities of 50, 60, 70, 80, and 95% (pH 2) methanol were followed by treatment with 4 ml of MQ water. Fractions corresponding to each wash step were pooled. The streptin present in the 80% (vol/vol) methanol-eluted fraction (partially purified streptin) containing inhibitory activity against indicator I1 was further purified by high-pressure liquid chromatography (HPLC; ICI Instruments) by using a Brownlee C8 column (RP-300, Aquapore Octyl, 300 A, 7 U). Streptin activity was eluted by using isocratic (32%) acetonitrile. Peptides corresponding to three absorbance peaks with inhibitory activity against indicator I1 were further purified by either C8 or C18 (Phenomenex Jupiter C18 column, 5 U, 300 A, 250 by 4.6 mm) HPLC fractionation. The active fractions corresponding to two major peaks from six consecutive C8 reversed-phase HPLC runs of partially purified streptin were refractionated by using either a C18 or a C8 column (as specified in Results) to yield purified streptin 1 and streptin 2 preparations.
Peptide analysis.
Amino acid composition analysis, mass spectrometry, and N-terminal amino acid sequencing were done by the Protein Microchemistry Facility, Department of Biochemistry, University of Otago. The amino acid compositions of phenylthiocarbamyl derivatives were determined by using a narrow-bore binary reversed-phase HPLC system (10). Detection of lanthionine was done by comparison with lanthionine peaks in the standard run. Mass spectrometry was with a matrix-assisted laser desorption ionization-time of flight mass analyzer (Finnigan Lasermat 2000; Thermo Bioanalysis), and N-terminal sequencing was done by automated Edman degradation on a 470A pulsed liquid protein sequencer (Applied Biosystems, Inc.) (11).
Crude and purified streptin preparations were digested with an equal volume of trypsin (1 g/liter) in 0.1 M phosphate buffer (pH 7). Each preparation was incubated at 37°C for 3 h, followed by heating at 80°C for 30 min to inactivate the enzyme prior to assaying the preparation against indicator I1.
srtA detection by using colony and dot blots.
The distribution of srtA among M-prototype S. pyogenes and various known BLIS-producing bacteria was determined by dot blotting. The DNA applied to these membranes was derived by a method based on that of Upton et al. (34), but with the use of only a single phenol-chloroform extraction of each DNA sample. A 5-μl portion of each DNA sample was applied to a nylon membrane (Hybond-N+ [Amersham Pharmacia Biotech, Inc.]) via a vacuum manifold, followed by 100 μl of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Denaturation of the DNA was done by exposure to two 2-min washes of 0.4 M NaOH, followed by two 2-min washes with 1 M Tris-HCl. The membrane was then exposed to UV light for ca. 5 min and probed with a digoxigenin (DIG)-dUTP (Roche Diagnostics, Ltd., Lewes, England)-labeled srtA probe, derived with the PCR primers srtAF (positions 2919 to 2940; 5′-AAGACTTTGATCTCGATTTGAA) and srtAR (positions 3020 to 2998; 5′-AAACTAATTTCCAACAAGAACCA) corresponding to srtA of the streptin locus (12,040 nucleotides; GenBank accession no. AB030831). The PCR was carried out by using Taq polymerase (Roche) for 30 cycles, with denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s.
Membranes containing colony blots of S. pyogenes were prepared by inoculating the test strains into 150-μl aliquots of THB (in a 96-well microtiter plate). After 18 h of incubation at 37°C in 5% CO2 in air, samples of these cultures were inoculated by using a plate replicator onto Hybond-N+ nylon membranes laid on top of CAB medium. After incubation for 18 h at 37°C in 5% CO2 in air, each membrane was placed on top of three layers of filter paper saturated with a zoocin A preparation (obtained from an 18-h CAB broth culture [CAB was dissolved in distilled water and then the agar removed by centrifugation at 15,300 × g for 10 min prior to autoclaving] of S. equi subsp. zooepidemicus strain 4881 incubated at 37°C in 5% CO2 in air) with a titer 4 AU/ml against S. pyogenes strain FF22 (indicator I2), followed by incubation for 18 h at room temperature in a sealed container. The membranes were then layered onto filter papers soaked in 10% sodium dodecyl sulfate and left for 45 min to allow cell lysis. The released DNA was fixed by exposure to 0.5 M NaOH in 1.5 M NaCl (fix solution) for 20 min (colony side up) on soaked Whatman paper, followed by flooding with fix solution for 20 min. Each membrane was then boiled twice for 20 min in 500 ml of 0.5% sodium dodecyl sulfate, rinsed in 2× SSC, and probed with an srtA probe as described above. The membranes were subsequently stripped by exposure to 0.4 M NaOH solution for 30 min at room temperature and then reprobed by using a DIG-labeled salA PCR amplicon. This probe was generated with the primers salAF (positions 604 to 629; 5′-GATATTTTGAACAATGCTATCGAAGA) and salAR (positions 897 to 921; 5′-ACTAATAGAAGTATCTAGTATGTCG) based on the S. salivarius 20P3 sal locus (10,610 nucleotides; GenBank accession no. AY005472).
Long template PCR.
PCR amplification of the entire streptin locus was done with the PCR primer pair srtIF (positions 352 to 369; 5′-GAGAACCGCCGTTTGGCC) and srtGR (positions 10740 to 10720; 5′-GTAAACCTCCACGTTGACTCC) based on the S. pyogenes strain BL-T streptin locus (12,040 nucleotides; GenBank accession no. AB030831) with an annealing temperature of 60°C and by using the Expand Long Template PCR kit (Roche) according to the manufacturer's instructions.
RNA extraction and analysis.
RNA was extracted by a modification of the methods of Lunsford (18) and Chomczynski and Sacchi (4). The cells used as the RNA source were in each case harvested from a 10- to 16-h (30°C) lawn culture of the test strain on BaCa in 9-cm-diameter petri dishes. Approximately one-quarter of the cellular growth was collected on a cotton swab and resuspended into 1 ml of 0.85% NaCl. After centrifugation (5,000 × g for 5 min at 4°C), the cell pellet was resuspended in 200 μl of spheroplasting buffer (20 mM Tris-HCl [pH 6.8] plus 10 mM MgCl2 and 26% [wt/vol] raffinose) containing 500 U of mutanolysin and 0.1 mg of spectinomycin/ml and then incubated at 37°C for 30 min. Spheroplasts were collected by centrifugation (5,000 × g for 5 min at 4°C), and the RNA was extracted by using a Qiagen RNeasy kit and a Qiashredder column (Qiagen, Ltd., Crawley, England) as recommended by the manufacturer. RNA was quantitated by measuring the absorbance at 260 nm.
RNA samples (15 μg per lane) were subjected to electrophoresis (80 V for 2 to 3 h) through 1% agarose supplemented with 0.02 M guanidine thiocyanate liter−1 by using TBE buffer (0.09 M Tris base, 0.088 M boric acid, 0.002 M EDTA [pH 8]). The RNA was then capillary blotted onto a Hybond-N+ nylon membrane (using the manufacturer's instructions). A PCR product generated with the primers srtAF and srtAR and labeled with [α-32P]dCTP by using Ready-To-Go DNA labeling beads (Amersham Pharmacia Biotech, Inc.) was used for Northern analysis of the srtA transcripts. Hybridization reactions were carried out at 65°C for 18 h as described elsewhere (5). The RNA marker used to determine transcript sizes was 3 μg of the 0.24- to 9.5-kb RNA ladder (Gibco-BRL/Life Technologies).
Evaluation of the effect of streptin on srtA transcription levels.
THB cultures (18 h at 30°C) of the S. pyogenes strains M25, Tn216, SF370, 148, and M-57 were used to inoculate lawn cultures on BaCa, either supplemented with partially purified streptin (adjusted to a final concentration of titer 2 AU/ml against indicator I1) (induced culture) or not supplemented (control). After incubation for 14 h at 30°C, the cells were resuspended in 0.85% NaCl, and the total RNA was extracted as described above.
Autoinduction of streptin production.
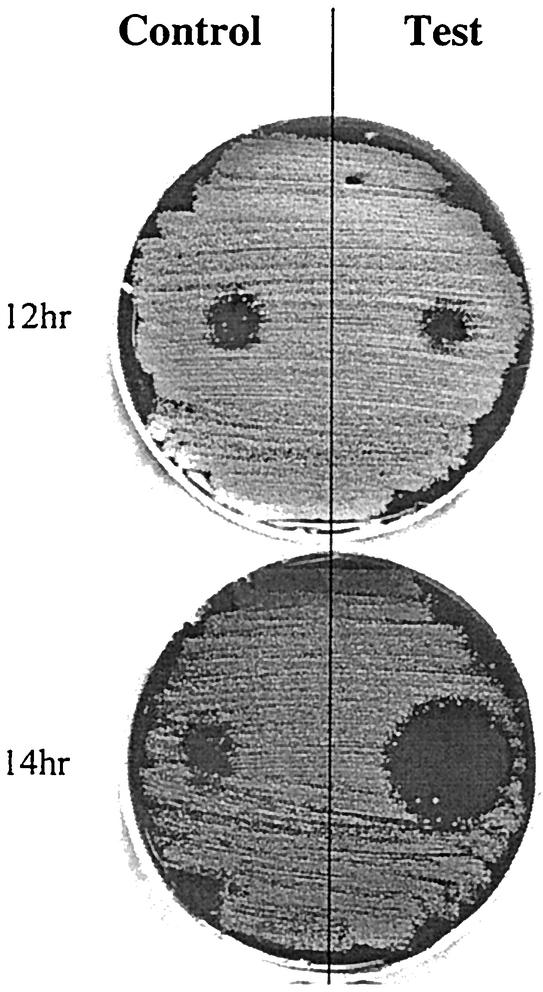
The autoinducibility of streptin production was assessed by using a novel plate assay (Fig. 1). A 20-μl sample of purified streptin 1 or streptin 2 (titer 2 against indicator I1) was spotted centrally onto the test side of a BaCa plate and left to dry. A lawn culture of strain M25 was then seeded evenly over the entire agar surface by using a swab charged by dipping into an 18-h THB culture. A series of replica plates were set up in this manner, and each one was incubated at 30°C for a specific time (12, 14, or 16 h), after which the cellular growth was removed and the agar surface was sterilized by exposure to chloroform vapor for 30 min. Purified streptin 1 or streptin 2 (20 μl of the titer 2 preparation) was then spotted onto the control side of the agar and allowed to dry prior to swabbing an 18 h THB culture of indicator I1 evenly over the surface of the agar. After incubation (18 h at 37°C), a relatively larger zone of inhibition on the test side compared to the control was taken to indicate enhanced (autoinduced) streptin production.
FIG. 1.
Autoinduction of streptin production by the plate assay method. HPLC-purified streptin 1 (20 μl) was spotted onto the test side of the BaCa plates and allowed to dry into the agar. A lawn of strain M25 was inoculated onto the surface and incubated for the times indicated. The bacterial growth was then removed, and the plate surface was sterilized with chloroform vapor. HPLC-purified streptin 1 preparation (20 μl) was spotted onto the control side of the plate. A lawn of indicator I1 was then inoculated onto the plate and grown for 18 h. An increased zone of inhibition on the induced side compared to the control side indicates autoinduction of streptin production.
Elimination of streptolysin O (SLO)-associated hemolysis on cholesterol-supplemented Ba.
Ba was supplemented with 5 mg of cholesterol/ml to eliminate hemolysis due to SLO (9). Control S. pyogenes were strains M25 (SLS+ SLO+), Blackmore (SLS+ SLO−), and C203U (SLS− SLO+). Strains that produce SLO but not SLS are nonhemolytic on this medium. Hemolysis due to the action of SLS was evident after 24 to 36 h of incubation at 37°C in 5% CO2 in air.
RESULTS
Production, purification, and partial characterization of streptin.
All attempts to detect streptin activity in liquid cultures of strain M25 were unsuccessful. BaCa-THB biphasic culture systems, however, yielded fluid-phase supernatant preparations that typically had a titer of 2 AU/ml. Cells from each 2 liters of the fluid phase, when extracted with 95% acidified methanol, typically yielded 3,200 AU of streptin. Removal of the methanol yielded crude streptin preparations (titer of 2,048 AU/ml) with inhibitory activity against all nine standard indicator strains. Purification of streptin was then achieved by HPLC by using a combination of C8 and C18 resins (Fig. 2). Three different forms of streptin were sequentially eluted from C8 resins by using 32% isocratic acetonitrile: streptin 1 (2,424 Da), streptin 1′ (2,442 Da), and streptin 2 (2,809 Da). Trace amounts of several other molecular species were also detected in mass spectrographs of purified streptin fractions, and these appear to correspond to various less-dehydrated forms of either streptin 1 (2,494, 2,476, and 2,460 Da) or streptin 2 (2,840 and 2,821 Da). N-terminal amino acid sequencing showed that the first six amino acid residues of both streptin 1 and streptin 1′ were VGXRYL, where “X” indicates no recognizable amino acid. Streptin 2, however, had three additional amino acid residues at the N terminus, its sequence being TPYVGXRYL. The amino acid composition analysis of streptin 1 matched almost exactly the numbers of each amino acid predicted by translation of the streptin structural gene (Table 1). Two lanthionine residues were detected, as was an unidentified peak that may correspond to 3-methyllanthionine.
FIG. 2.
(A) C8 reversed-phase HPLC elution profile of a partially purified streptin preparation that had been obtained by SepPak C18 chromatography of a 95% acidified methanol cell extract of strain M25. Protein material eluted in an acetonitrile gradient was detected by A214 measurement, and the inhibitory activity of each fraction was tested against indicator I1. Solid bar A represents a fraction further purified by C18HPLC (see panel B), and solid bar B represents a fraction further purified by C8 HPLC (see panel C). The unlabeled solid bar indicates fractions that contained inhibitory activity. (B) C18 elution profile of fraction A. Streptin 1 was detected in the fractions indicated by solid bar 1. The unlabeled solid bar represents fractions containing inhibitory activity. (C) C8 elution profile of fraction B. Streptin 1′ was detected in the fractions indicated by solid bar 1′. Streptin 2 was detected in the fraction labeled 2. The unlabeled solid bar represents fractions containing inhibitory activity.
TABLE 1.
Amino acid composition of streptin 1
| Amino acid residue | Estimated no. of residuesa (mol ratio) | Expected no. of residuesb |
|---|---|---|
| Asp/Asn | 1 (0.65) | 0 |
| Glu/Gln | 0 (0.31) | 0 |
| Ser | 0 (0.20) | 0 |
| Gly | 2 (1.97) | 2 |
| His | 0 (0) | 0 |
| Arg | 1 (0.82) | 1 |
| Thr | 0 (0.49) | 0 |
| Ala | 0 (0.48) | 0 |
| Pro | 1 (1.30) | 1 |
| Tyr | 1 (0.81) | 1 |
| Lanthionine | 2 (1.77) | 2 |
| Val | 3 (2.98) | 3 |
| Met | 0 (0) | 0 |
| Cys | 0 (0) | 0 |
| Ile | 0 (0) | 0 |
| Leu | 2 (1.82) | 2 |
| Phe | 1 (0.89) | 1 |
| Lys | 2 (2.12) | 2 |
| Trp | NDc | 1 |
| Unknownd | 2e (2.02) | 4f |
| Total | 18 | 20 |
The number of residues was estimated from the mole ratio (shown in parentheses).
Expected number of amino acid residues in the streptin propeptide resulting from translation of srtA. Each thioether ring is taken to be one amino acid.
ND, not determined.
This amino acid could not be determined.
This value is not accurate due to lack of knowledge about the nature of the unknown peak.
This value includes three dehydrated Thr and one 3-methyllanthionine.
Crude streptin preparations showed no loss of activity when stored for three months at either 4 or 20°C. These preparations were also stable in response to heating at 100°C for 10 min and were relatively more stable under acidic conditions (pH 3), retaining full activity over 24 h, than under alkaline conditions (pH 10), in which 50% of the activity was lost within 24 h. Streptin 1 and crude streptin preparations were inactivated upon treatment with trypsin.
Evaluation of the distribution and composition of the streptin locus in S. pyogenes.
The lantibiotic purified in the present study from strain M25 appears to be consistent with the predicted product of the streptin gene cluster described by Karaya et al. (16). The streptin structural gene srtA was used as a probe to test a variety of streptococci in dot and colony blot hybridizations. Of 58 tested S. pyogenes isolates, each representing a different M-type, 41 hybridized with the srtA probe (Table 2). Interestingly, only 10 of these strains appeared to express biologically active (inhibitory) streptin. None of 75 S. salivarius, 8 S. mutans, and 9 S. uberis strains tested were found to be positive for the streptin structural gene.
TABLE 2.
Distribution of srtA and production of P-type 777 inhibitory activity within S. pyogenes
| M-prototype S. pyogenes categorya | Type(s) |
|---|---|
| Positive for srtA and producing a P-type 777 pattern | 2, 11, 12, 25, 28, 60, 66, 67, 71, 76 |
| Positive for srtA but not giving a P-type 777 pattern | 1, 4, 5, 9, 13, 15, 18, 19, 23, 24, 26, 27, 29, 30, 31, 39, 41, 43, 46, 53, 55, 54, 58, 62, 64, 65, 72, 73, 79, 81 |
| Negative for srtA and not giving a P-type 777 pattern | 3, 6, 22, 33, 36, 38, 42, 47, 48, 51, 52, 57, 63, 69, 74, 75, 77, 78 |
As determined by colony blot analysis. All strains that appeared to be negative for srtA were screened for salA (known to be present in each of the strains) as a positive control of adequate DNA being present on the membrane.
The nucleotide sequence of srtA in 10 S. pyogenes isolates was found to be identical, regardless of whether these strains expressed streptin (strains 73220 [M-type 60], 71948 [M-type 11], M-12 [M-type 12], M25 [M-type 25], P5 [T-type 25], and 79009 [M-type 2]) or did not express streptin (strains 76068 [M-type 60], 74823 [M-type 11], and SF370 [M-type 1]). In each case the sequence was identical to that published for S. pyogenes strain BL-T (16).
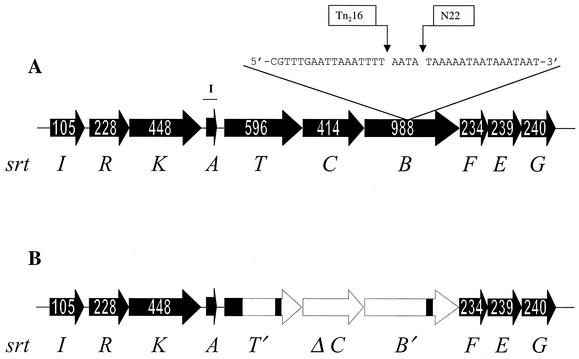
A direct comparison of the streptin loci in the P-type 777 (streptin-expressing) strains M25 and BL-T with that of the P-type 000 (streptin-nonexpressing) strain SF370 revealed that the strain SF370 locus contains three deletions within the srtT, srtC, and srtB region (Fig. 3).
FIG. 3.
Structures of the srt loci in S. pyogenes BL-T (A) and S. pyogenes SF370 (B). Open reading frames are depicted by solid arrows that show the numbers of amino acid residues in the deduced polypeptides. Unfilled regions represent areas of the streptin locus that are absent in strain SF370. Open reading frame srtA consists of a deduced polypeptide of 46 amino acids. The sites of insertion of Tn916 into srtB of strain M25 (the present study) and BL-T (16) are shown as Tn216 and N22, respectively. The position of the PCR-amplified segment of DNA used to probe for the presence of srtA is also shown (I).
PCR amplification of the entire streptin locus in a variety of S. pyogenes strains by using the primers srtIF and srtGR indicated that some other inhibitor-negative srtA-positive strains such as 76068 (M-type 60) also contained detectable deletions within the streptin locus. However, other srtA-positive, streptin-negative strains such as 148 (P-type 655) and 74823 (P-type 000) had srt amplicons that appeared to be the same size as in strain M25.
Northern analysis of srtA transcription and autoinduction.
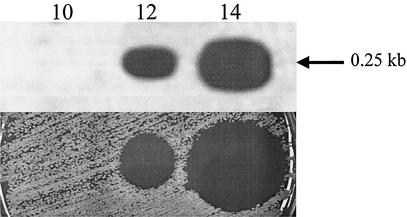
The levels of srtA mRNA were estimated in cells from 10 to 14 h strain M25 cultures (Fig. 4). No srtA transcript was detected in the mRNA extracted from 10-h cultures, but both the 12- and the 14-h cultures contained 0.25-kb srtA transcripts.
FIG. 4.
Northern analysis of srtA transcription in S. pyogenes strain M25 probed with a srtA fragment corresponding to bar I (see Fig. 3). S. pyogenes strain M25 was grown on BaCa at 30°C in air for 10 h (lane 10), 12 h (lane 12), and 14 h (lane 14). The srtA mRNA transcript (0.25 kb) is indicated by an arrow. The lanes contained equivalent amounts of total RNA (15 μg). Shown directly underneath each lane is the streptin inhibitory activity against indicator I1 produced by cultures of strain M25 under the corresponding incubation conditions. Aliquots (20 μl) of strain M25 from an 18-h THB culture were incubated on BaCa in adjacent positions for either 10, 12, or 14 h prior to removing the growth, sterilizing the agar surface with chloroform vapor, and then applying the indicator I1 lawn. After incubation, the level of streptin activity is indicated by the size of the inhibition zone.
Pure preparations of either streptin 1 or streptin 2 induced streptin production in strain M25 but failed to cross-induce the production of the heterologous lantibiotics SA-FF22, nisin Z, and salivaricin A in S. pyogenes strain FF22, L. lactis strain A5, and S. salivarius strain 20P3, respectively (Table 3).
TABLE 3.
Induction specificity of streptin, nisin Z, SA-FF22, and salivaricin A
| Producer strain | Inducing lantibiotica | P-type of producer strain | Induction of inhibitor production in straind
|
|||
|---|---|---|---|---|---|---|
| M25 | A5 | FF22 | 20P3 | |||
| M25 | Streptin 1b | 777 | + | − | − | − |
| M25 | Streptin 2b | 777 | + | − | − | − |
| A5 | Nisin Zc | 777 | − | + | − | − |
| FF22 | SA-FF22c | 436 | − | − | + | − |
| 20P3 | Salivaricin Ac | 677 | − | − | − | + |
A total of 10 μl of each lantibiotic (adjusted to a titer of 1 against indicator I1) was used as the test preparation against each producer strain in the plate induction assay.
Inducing preparations of streptin were HPLC purified and shown to consist of streptin 1 or streptin 2.
Inducing preparations were freeze-thaw extracts from 18-h, 37°C BaCa lawn cultures of the producer strain sterilized by heating them to 60°C for 30 min prior to use in the assay.
As indicated by an increased inhibitory zone against indicator I1 compared to that produced by the uninduced (control) culture.
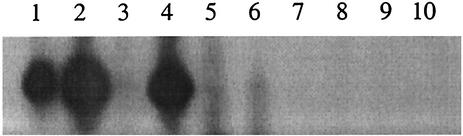
Northern analysis of srtA transcripts was carried out on strains M25, Tn216, SF370, 148, and M-57 (Fig. 5). Samples were obtained from 14-h cultures on BaCa that had either been presupplemented with a subinhibitory amount of streptin (induced culture) or not supplemented (control culture). A 0.25-kb srtA transcript was detected in the total mRNA extracted from streptin-induced strain Tn216 but appeared to be absent in the corresponding (uninduced) strain Tn216 mRNA. An srtA transcript was also evident in the total mRNA from both induced and uninduced strain M25 cultures. However, the induced strain M25 mRNA contained substantially increased levels of the transcript. The total mRNA from strain SF370 cultures contained only very low levels of the srtA transcript, irrespective of whether or not the cells had been grown on streptin-supplemented medium. Similarly, neither induced nor uninduced cultures of strains 148 or M-57 appeared to contain srtA transcripts.
FIG. 5.
Autoinduction of srtA transcription in S. pyogenes. Northern analysis of srtA mRNA transcripts from S. pyogenes grown at 30°C for 14 h on BaCa supplemented with subinhibitory amounts of crude streptin preparation (+ streptin) or not supplemented. Lane 1, S. pyogenes M25; lane 2, S. pyogenes M25 + streptin; lane 3, S. pyogenes Tn216; lane 4, S. pyogenes Tn216 + streptin; lane 5, S. pyogenes SF370; lane 6, S. pyogenes SF370 + streptin; lane 7, S. pyogenes 148; lane 8, S. pyogenes 148 + streptin; lane 9, S. pyogenes M-57; lane 10, S. pyogenes M-57 + streptin. Each lane contained 15 μg of total RNA.
Association between the production of streptin and of SLS.
Previously, Hynes and Tagg had derived nine inhibitor-negative derivatives of strain M25 by Tn916 mutagenesis and concluded that there was no apparent direct link between the production of the P-type 777 inhibitor and SLS in this strain (14). In contrast, Karaya et al. (16) suggested that the production of SLS and streptin may be interdependent in strain BL-T (16). Therefore, in the present study the nine inhibitor-negative mutants of strain M25 originally described by Hynes and Tagg (14) were reevaluated, and one of these (strain Tn216) was found to contain only a single Tn916 insert. This insertion was found to be within srtB (base pair position 7463) of the streptin locus, only 4 bp upstream of the Tn916 insertion site in the strain BL-T mutant described by Karaya et al. (base pair position 7467) (16). SLS production by strains M25 and Tn216 was compared, and no difference in the level of hemolysis was observed on Ba between the two strains. The hemolysis associated with strains of S. pyogenes is typically due to the action of both SLS and SLO. SLO-associated hemolysis is reduced in the presence of cholesterol. Similar levels of hemolysis were given by strains Tn216, M25, and Blackmore (SLS+ SLO−) on Ba plus cholesterol medium. In contrast, the hemolytic activity of strain C203U (SLS− SLO+) was much reduced (Fig. 6).
FIG. 6.
Hemolytic activity of four S. pyogenes strains on Ba plus cholesterol medium after 18 h of incubation at 37°C in 5% CO2 in air. Strains C203U (SLS− SLO+), Blackmore (SLS+ SLO−), M25 (streptin producer), and Tn216 (Tn916 inhibitor negative derivative of strain M25) are indicated.
DISCUSSION
In the present study, the agent responsible for production of P-type 777 inhibitory activity on blood agar medium by S. pyogenes strain M25 was shown to be the lantibiotic streptin. Our previous finding (14) that the inhibitory agent is not produced in liquid cultures of P-type 777 S. pyogenes was confirmed. Sufficient starting material for purification purposes could, however, be derived from cultures of strain M25 in a biphasic growth medium. Hynes had previously observed that, when relatively dilute suspensions of strain M25 cells were used as the inocula in deferred antagonism tests, there was greatly reduced production of inhibitory activity (12). These observations are consistent with streptin production being under quorum-sensing control. In the present study it was indeed demonstrated that streptin production by strain M25 on BaCa is specifically enhanced in the presence of biologically active (inhibitory) streptin. It seems likely that the formation of inducing levels of biologically active streptin must occur too late in conventional liquid cultures to effect significant amplification of production before cell metabolism ceases. In the dually incubated biphasic cultures, streptin produced during the initial growth of strain M25 on the agar surface presumably diffuses into the fluid phase of the secondary culture sufficiently quickly to effect induced lantibiotic production. Apparently, other factors are also involved, since it was also found (results not shown) that presupplementation of THB cultures of strain M25 with various concentrations of crude streptin did not lead to enhanced streptin production.
Washed cells from these biphasic cultures provided excellent starting material for the subsequent extraction and purification of cell-associated streptin. A number of other bacteriocins have been shown to exist in both cell-associated and extracellular forms in cultures (6, 15). Two markedly different molecular forms of streptin were detected in acidified 95% methanol extracts of the producer cells. The relatively stronger hydrophobic nature of streptin 2 compared to streptin 1 was shown by its delayed elution from C8 resins in acetonitrile gradients. Streptin 1 and streptin 2 appeared, however, to have similar inhibitory and autoinducing characteristics. Streptin 2 is thought to represent an incompletely processed form of the molecule. Streptin 1 lacks the residues TPY present at the N terminus of streptin 2 and was confirmed to be present in the cell extracts in either a relatively more (streptin 1, Mr 2,424) or less (streptin 1′, Mr 2,442) dehydrated form. Several other molecular species apparently corresponding to differing degrees of dehydration of streptin 1 and streptin 2 were also detected by mass spectroscopy but were not present in sufficient quantities or degrees of purification for N-terminal sequencing. The lantibiotic Pep5 has also been shown to exist in multiple Mr forms representing differing degrees of dehydration of its six serine or threonine residues (22). Interestingly, the incompletely dehydrated forms of Pep5 were only detected in lysates of the producer cells, with only the fully dehydrated peptide detected in the culture supernatant. Since the processing of lantibiotics appears commonly to be completed during the final stages of secretion through the cell envelope, it may be speculated that the various incompletely processed forms of streptin are relatively unlikely to be recovered from the extracellular fluids of streptin-producing cultures.
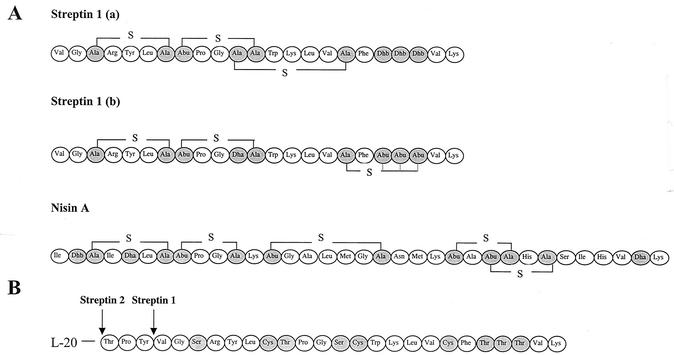
The strong homology between srtA and the corresponding nisin and subtilin structural genes indicates that streptin should also be classified as a class A1 lantibiotic. On this basis, a comparison of the nisin and streptin propeptides (Fig. 7) leads us to predict that the Ser residue encoded in position 3 of streptin 1 is probably involved in lanthionine ring formation with the Cys residue in position 7. The failure to detect any Cys in the streptin amino acid composition analysis indicates that all three Cys residues predicted by the srtA sequence are likely to be incorporated within either lanthionine or 3-methyllanthionine ring structures in the mature peptide. Streptin differs from nisin and subtilin in that it contains a Ser residue between the Gly and Cys residues of the otherwise highly conserved LCTPGC propeptide motif. One implication of this is that involvement of this Ser in lanthionine bridge formation would be expected to constrain the flexibility of the streptin molecule [Fig. 7, streptin 1 (a)]. This relatively flexible hinge region in other type A1 lantibiotics has been shown to be important for their membrane pore-forming capability (36). Thus, pore formation may not be the major mechanism of the killing action of streptin. Alternatively, a 3-methyllanthionine ring may form between Cys-17 and one of the Thr residues downstream at either position 19, 20, or 21 in streptin 1 [Fig. 7, streptin 1 (b)]. This would allow retention of a flexible hinge region in streptin 1 and facilitate pore formation as a potential mechanism of cell killing. It appears that the total activity of streptin is not dependent upon the complete modification of the propeptide since variant, incompletely modified forms such as streptin 1′ appear to retain inhibitory activity against M. luteus.
FIG. 7.
(A) Two possible bridging structures of the streptin propeptide, streptin 1 (a) and streptin 1 (b), based on the known nisin propeptide structure. Thioether bridges are indicated by “—S—”. Modified residues are shaded: Ala-S-Ala, lanthionine; Abu-S-Ala, 3-methyllanthionine; Dha, didehydroalanine; Dhb, didehydrobutyrine. In streptin 1 (b), 3-methyllanthionine could form between any one of the three dehydrobutyrine residues (as indicated by the dotted lines). (B) Schematic representation of the streptin 1 and streptin 2 prepeptides. L-20 indicates the leader peptide consisting of 20 amino acid residues. Labeled arrows indicate cleavage sites of the prepeptide to release either streptin 1 or streptin 2. Residues that are believed to be modified in the propeptide are shaded.
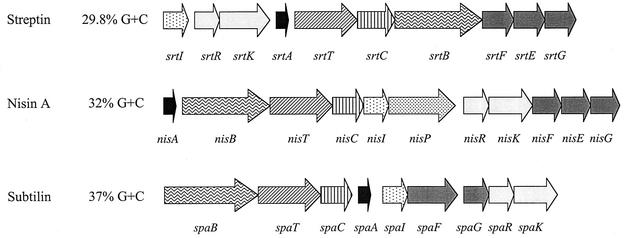
The arrangement of the streptin locus in strain M25 is identical to that reported by Karaya et al. (16) for S. pyogenes strain BL-T and is composed of 10 open reading frames encoding putative proteins involved in streptin immunity (SrtI) and regulation (SrtR and SrtK), followed by open reading frames for the streptin prepeptide (SrtA), transport (SrtT), prepeptide modification (SrtC and SrtB), and self-protection (SrtF, SrtE, and SrtG) (Fig. 8). Although these proteins are broadly similar to those described for nisin and subtilin biosynthesis, the arrangements of the individual open reading frames in the operons are quite different. The subtilin and streptin operons differ also from the nisin operon in that they do not encode a dedicated proteinase for processing of the lantibiotic prepeptide. It is most unusual for lantibiotic operons to not include a gene specifying a specific proteinase.
FIG. 8.
Organization of the streptin, nisin A, and subtilin gene clusters. Structural genes (not drawn to scale) are black, and genes with similar proposed functions have the same patterns. The GC content of each locus is presented as a percentage. Gene designations are according to de Vos et al. (7).
Interestingly, srtA appears to be highly conserved in 67% of the 58 S. pyogenes strains of different M-types tested. However, only 10 of these strains appear to produce biologically active streptin. There may be various reasons for the lack of streptin production in srtA-positive S. pyogenes. For example, S. pyogenes strains SF370 and 76068 have been demonstrated to have incomplete srt loci. The missing regions appear to be confined to srtT, srtC, and srtB. Inadequate expression of these genes is likely to result in the loss of active streptin production through an inability to modify the prepeptide and to export it through the cell membrane. Other srtA-positive streptin-negative strains such as S. pyogenes strains 148 and 74823 seem, however, to have intact streptin loci. Strain 148 does not appear to be capable of sensing and upregulating the expression of srtA in response to the presence of external streptin in the culture media. Therefore, the reason for the lack of streptin production in this strain may be a nonfunctional promoter sequence upstream of srtA. Interestingly, both the salivaricin A (salA) (35) and nisin (nisA) (19) structural genes have also been identified in strains that do not appear to produce the active lantibiotics. Upton et al. (35) have demonstrated that the lack of salivaricin A production in some salA-positive S. pyogenes is due to a deletion within the region of the sal locus containing the salT, salC, and salB open reading frames. nisA-positive nisin-nonproducing L. lactis strains have been identified that do not transcribe nisA, whereas others have a nisA transcript but do not produce active nisin (19).
The site of insertion of Tn916 within srtB of strain Tn216 was just 4 bp upstream of the corresponding Tn916 insertion site reported by Karaya et al. (16) in strain BL-T. Tn916 insertion “hot spots” have been identified to be regions that contain an A-rich sequence separated by about six bases from a T-rich sequence (23). The Tn916 insertion site within srtB conforms to this criterion. Northern analysis showed that strain Tn216, unlike the parent strain M25, failed to produce srtA transcripts unless exposed to preformed streptin. This finding supports the model of control of streptin production through a response regulator system similar to that controlling nisin production (17).
Application of the plate induction assay showed that both streptin 1 and streptin 2 are inducer molecules for streptin production in strain M25. Northern analysis also indicated enhanced production of streptin mRNA in induced cultures. In spite of this enhanced messenger formation, the production of biologically active streptin in THB cultures is poor, and so other factors are clearly involved in the effective expression of streptin. One such factor may be the need for the expression and activation of a host cell proteinase capable of processing the streptin prepeptide. In the case of subtilin, prepeptide processing is effected by B. subtilis host-encoded proteinases (27). Blocking of proteinase activity with the serine proteinase inhibitor phenylmethylsulfonyl fluoride led to the accumulation of biologically inactive subtilin precursor peptides with leader peptide moieties of differing lengths still attached (27). Incubation of these species with culture media of different subtilin-nonproducing B. subtilis strains yielded active subtilin. By analogy, our previous studies demonstrated that production of P-type 777 inhibitory activity by S. pyogenes appeared to be dependent on the concomitant expression of streptococcal proteinase (SpeB) by the producer strain (14). Evidence included the elimination of inhibitor production when cultures were grown under conditions inimical to proteinase production, such as low temperature and alkaline pH, or upon addition to the medium of substances known to have anti-proteinase activity such as glucose, iodoacetic acid, lincomycin, and trypan blue. Moreover, all of six proteinase-negative mutants of S. pyogenes strain M25 were found also to have become inhibitor negative (14). This apparent dependence of biologically active streptin production on functional SpeB implicates this molecule in the processing of the streptin prepeptide. Furthermore, SpeB has been shown previously not to degrade the mature form of streptin (14) but could be capable of effecting cleavage of the streptin leader. S. pyogenes proteinase activity (possibly SpeB) has also been shown to enhance the release of SLS (1), now known to be encoded by an operon closely resembling those characterizing the lantibiotics (20). Evidence for the role of proteinase in the release of SLS includes the interference with SLS production by protease inhibitors and reversal of this effect on addition of trypsin (1). Interestingly, the formation of both SLS and streptin occurs principally during the stationary-growth phase of S. pyogenes cultures, at a time when SpeB expression is also maximal (3).
In the study by Karaya et al. (16) it was noted that inactivation of the streptin locus by insertion of Tn916 into srtB appeared to be associated with a concomitant loss of SLS production in the producer strain, and it was speculated that some of the gene products from the streptin locus may also be essential for the production of SLS. In the present study, however, we have found that a single Tn916 insert in a position just 4 bp upstream of that in the Karaya et al. (16) study did not affect the production of SLS in strain M25. Further evidence for the lack of a direct association between the expression of streptin and of SLS is our current finding that 18 of 58 SLS-positive S. pyogenes strains of different M types are negative for srtA.
Acknowledgments
We thank M. Upton, K. Dierksen, A. Carne, C. Ronson, and G. Cook for helpful discussions and advice. We gratefully acknowledge the BLAST search facilities at the National Library of Medicine, Washington, D.C., and the University of Oklahoma Streptococcal (GAS) Genome Sequencing Project, funded by a USPHS/NIH grant to B. A. Roe, S. P. Linn, L. Song, X. Yuan, S. Clifton, R. E. McLaughlin, M. McShan, and J. Ferretti.
This work was supported by grant UO0605 from the Marsden Fund, Royal Society of New Zealand, and by the Health Research Council of New Zealand.
REFERENCES
- 1.Akao, T., K. Kobashi, and C. Y. Lai. 1983. The role of protease in streptolysin S formation. Arch. Biochem. Biophys. 223:556-561. [DOI] [PubMed] [Google Scholar]
- 2.Beall, B., R. Facklam, and T. Thompson. 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaussee, M. S., E. R. Phillips, and J. J. Ferretti. 1997. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect. Immun. 65:1956-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 5.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daba, H., C. Lacroix, J. Huang, R. E. Simard, and L. Lemieux. 1994. Simple method of purification and sequencing of a bacteriocin produced by Pediococcus acidilactici UL5. J. Appl. Bacteriol. 77:682-688. [DOI] [PubMed] [Google Scholar]
- 7.de Vos, W. M., O. P. Kuipers, J. R. van der Meer, and R. J. Siezen. 1995. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by gram-positive bacteria. Mol. Microbiol. 17:427-437. [DOI] [PubMed] [Google Scholar]
- 8.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flanagan, J., N. Collin, J. Timoney, T. Mitchell, J. A. Mumford, and N. Chanter. 1998. Characterization of the haemolytic activity of Streptococcus equi. Microb. Pathog. 24:211-221. [DOI] [PubMed] [Google Scholar]
- 10.Hubbard, M. J. 1995. Calbindin28kDa and calmodulin are hyperabundant in rat dental enamel cells Identification of protein phosphatase calcineurin as a calmodulin target and of a secretion-related role for calbindin28kDa. Eur. J. Biochm. 230:68-79. [DOI] [PubMed] [Google Scholar]
- 11.Hubbard, M. J., and N. J. McHugh. 1996. Mitochondrial ATP synthase F1-β- subunit is a calcium-binding protein. FEBS Lett. 391:323-329. [DOI] [PubMed] [Google Scholar]
- 12.Hynes, W. L. 1985. PhD thesis. University of Otago, Dunedin, New Zealand.
- 13.Hynes, W. L., and J. R. Tagg. 1985. Production of broad-spectrum bacteriocin-like activity by group A streptococci of particular M-types. Zentbl. Bakteriol. Mikrobiol. Hyg. A 259:155-164. [DOI] [PubMed] [Google Scholar]
- 14.Hynes, W. L., and J. R. Tagg. 1986. Proteinase-related broad-spectrum inhibitory activity among group A streptococci. J. Med. Microbiol. 22:257-264. [DOI] [PubMed] [Google Scholar]
- 15.Jack, R. W., and J. R. Tagg. 1992. Factors affecting production of the group A streptococcus bacteriocin SA-FF22. J. Med. Microbiol. 36:132-138. [DOI] [PubMed] [Google Scholar]
- 16.Karaya, K., T. Shimizu, and A. Taketo. 2001. New gene cluster for lantibiotic streptin possibly involved in streptolysin S formation. J. Biochem. 129:769-775. [DOI] [PubMed] [Google Scholar]
- 17.Kuipers, O. P., M. M. Beerthuyzen, P. G. de Ruyter, E. J. Luesink, and W. M. de Vos. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270:27299-27304. [DOI] [PubMed] [Google Scholar]
- 18.Lunsford, R. D. 1995. Recovery of RNA from oral streptococci. BioTechniques 18:412-414. [PubMed] [Google Scholar]
- 19.Moschetti, G., F. Villani, G. Blaiotta, A. Baldinelli, and S. Coppola. 1996. Presence of non-functional nisin genes in Lactococcus lactis subsp. lactis isolated from natural starters. FEMS Microbiol. Lett. 145:27-32. [DOI] [PubMed] [Google Scholar]
- 20.Nizet, V., B. Beall, D. J. Bast, V. Datta, L. Kilburn, D. E. Low, and J. C. D. Azavedo. 2000. Genetic locus for streptolysin S production by group A streptococcus. Infect. Immun. 68:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross, K. F., C. W. Ronson, and J. R. Tagg. 1993. Isolation and characterization of the lantibiotic salivaricin A and its structural gene salA from Streptococcus salivarius 20P3. Appl. Environ. Microbiol. 59:2014-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahl, H.-G., M. Reis, M. Eschbach, C. Szekat, A. G. Beck-Sickinger, J. Metzger, S. Stefanovic, and G. Jung. 1991. Isolation of Pep5 prepeptides in different stages of modification, p. 332-346. In G. Jung and H.-G. Sahl (ed.), Nisin and novel lantibiotics. Escom Publishers, Leiden, The Netherlands.
- 23.Scott, J. R., F. Bringel, D. Marra, G. Van Alstine, and C. K. Rudy. 1994. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol. Microbiol. 11:1099-1108. [DOI] [PubMed] [Google Scholar]
- 24.Shah, H. N., and S. E. Gharbia. 1992. Biochemical and chemical studies on strains designated Prevotella intermedia and proposal of a new pigmented species, Prevotella nigrescens sp. nov. Int. J. Syst. Bacteriol. 42:542-546. [DOI] [PubMed] [Google Scholar]
- 25.Simmonds, R. S., W. J. Simpson, and J. R. Tagg. 1997. Cloning and sequence analysis of zooA, a Streptococcus zooepidemicus gene encoding a bacteriocin-like inhibitory substance having a domain structure similar to that of lysostaphin. Gene 189:255-261. [DOI] [PubMed] [Google Scholar]
- 26.Simpson, W. J., and J. R. Tagg. 1983. M-type 57 group A streptococcus bacteriocin. Can. J. Microbiol. 29:1445-1451. [DOI] [PubMed] [Google Scholar]
- 27.Stein, T., and K. D. Entian. 2002. Maturation of the lantibiotic subtilin: matrix-assisted laser desorption/ionization time-of-flight mass spectrometry to monitor precursors and their proteolytic processing in crude bacterial cultures. Rapid Commun. Mass. Spectrom. 16:103-110. [DOI] [PubMed] [Google Scholar]
- 28.Tagg, J. R. 1984. Production of bacteriocin-like inhibitors by group A streptococci of nephritogenic M types. J. Clin. Microbiol. 19:884-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tagg, J. R., and L. V. Bannister. 1979. “Fingerprinting” beta-haemolytic streptococci by their production of and sensitivity to bacteriocine-like inhibitors. J. Med. Microbiol. 12:397-411. [DOI] [PubMed] [Google Scholar]
- 30.Tagg, J. R., R. S. D. Read, and A. R. McGiven. 1971. Bacteriocine production by group A streptococci. Pathology 3:277-278. [Google Scholar]
- 31.Tagg, J. R., and L. G. Vugler. 1986. Enhancement of the hemolytic activity of group B streptococci and streptolysin S-deficient group A streptococci on blood agar medium containing staphylococcal beta-lysin. J. Microbiol. Methods 5:215-220. [Google Scholar]
- 32.Tagg, J. R., and L. W. Wannamaker. 1978. Streptococcin A-FF22: nisin-like antibiotic substance produced by a group A streptococcus. Antimicrob. Agents Chemother. 14:31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Triglia, T., M. G. Peterson, and D. J. Kemp. 1988. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 16:8186. [DOI] [PMC free article] [PubMed]
- 34.Upton, M., P. E. Carter, M. Morgan, G. F. Edwards, and T. H. Pennington. 1995. Clonal structure of invasive Streptococcus pyogenes in northern Scotland. Epidemiol. Infect. 115:231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Upton, M., J. R. Tagg, P. Wescombe, and H. F. Jenkinson. 2001. Intra- and interspecies signaling between Streptococcus salivarius and Streptococcus pyogenes mediated by SalA and SalA1 lantibiotic peptides. J. Bacteriol. 183:3931-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiedemann, I., E. Breukink, C. van Kraaij, O. P. Kuipers, G. Bierbaum, B. de Kruijff, and H. G. Sahl. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772-1779. [DOI] [PubMed] [Google Scholar]