Abstract
A two-probe proximal chaperone detection system consisting of a species-specific capture probe for the microarray and a labeled, proximal chaperone probe for detection was recently described for direct detection of intact rRNAs from environmental samples on oligonucleotide arrays. In this study, we investigated the physical spacing and nucleotide mismatch tolerance between capture and proximal chaperone detector probes that are required to achieve species-specific 16S rRNA detection for the dissimilatory metal and sulfate reducer 16S rRNAs. Microarray specificity was deduced by analyzing signal intensities across replicate microarrays with a statistical analysis-of-variance model that accommodates well-to-well and slide-to-slide variations in microarray signal intensity. Chaperone detector probes located in immediate proximity to the capture probe resulted in detectable, nonspecific binding of nontarget rRNA, presumably due to base-stacking effects. Species-specific rRNA detection was achieved by using a 22-nt capture probe and a 15-nt detector probe separated by 10 to 14 nt along the primary sequence. Chaperone detector probes with up to three mismatched nucleotides still resulted in species-specific capture of 16S rRNAs. There was no obvious relationship between position or number of mismatches and within- or between-genus hybridization specificity. From these results, we conclude that relieving secondary structure is of principal concern for the successful capture and detection of 16S rRNAs on planar surfaces but that the sequence of the capture probe is more important than relieving secondary structure for achieving specific hybridization.
DNA microarrays are currently used for gene expression profiling (18, 25), DNA sequencing (24), disease screening (17), diagnostics (9, 29), and genotyping (14), usually within the context of clinical applications. The extension of microarray technology to the detection and analysis of 16S rRNAs in mixed microbial communities likewise holds tremendous potential for microbial community analysis, pathogen detection, and process monitoring in both basic and applied environmental sciences (7, 12, 27). The application of microarrays for unattended in-field or point-of-use environmental applications, however, frequently involves requirements to (i) detect many different microorganisms simultaneously, (ii) utilize a bioanalytical detection method that is conducive to automation and/or field deployment, (iii) monitor RNA as a qualitative indicator of microbial activity, and (iv) quantify RNA levels and/or the extent of microbial activity. Such requirements are especially pertinent for monitoring changes in microbial community composition and activity through time and space. Thus, continued use of microarray protocols that rely on PCR amplification represents a significant bottleneck for the routine application and deployment of microarrays in the field and highlights the need to develop sensitive and specific direct RNA detection methods for environmental samples.
There are several reports of direct rRNA detection on oligonucleotide microarrays. Guschin et al. (12) and Bavykin et al. (4), for example, used gel element microarrays to directly (and specifically) detect fragmented RNA from simple model microbial communities. A surface plasmon resonance sensor was used to detect intact rRNA binding to an immobilized probe (22), and Small et al. recently described an oligonucleotide array for the direct detection of intact 16S rRNA from unpurified soil extracts (26). Solution-phase capture of rRNA, such as within a gel element microarray, can progress with reasonable sensitivity even when probes are targeted toward 16S rRNA regions rich in secondary and tertiary structures (2, 12, 16). Capturing 16S rRNA with surface-immobilized oligonucleotides, however, is strongly influenced by secondary and tertiary structures, such that oligonucleotides are frequently targeted only toward accessible regions of the 16S rRNA target, such as bulges and hairpin loops (e.g., see reference 22).
To design 16S rRNA-targeted microarrays for microbial ecology, microbial community profiling, or other environmental monitoring applications where multiple species and genera may be encountered, however, probe sequences frequently must be developed to specifically target RNA-RNA duplexes and other inaccessible regions, such as the stem of a 16S rRNA stem-loop structure. To overcome secondary structure constraints yet still allow direct detection of rRNA without a PCR amplification step, Small et al. developed a two-probe proximal chaperone detection system that consists of a species-specific capture probe and a labeled detector probe that is mixed with the rRNA sample prior to microarray hybridization (26). Direct detection of intact rRNA required the capture and detector probes to be in close proximity to each other on the rRNA molecule. However, recent results with an expanded microarray showed cross-reactivity with nontarget probes, albeit at levels that were significantly lower than and statistically distinguishable from those obtained with perfectly matched probes. We hypothesize that the observed cross-reactivity of multispecies, multigenus rRNA detection on planar microarrays is due to the base-stacking contributions of the proximal oligonucleotide probes (5, 10, 15, 20, 21, 23). In order to test this hypothesis and to better understand how to improve the specificity of direct 16S rRNA microarray detection, we examined the physical spacing and nucleotide mismatch tolerance between capture and proximal chaperone detector probes that are required to achieve species-specific 16S rRNA detection with a dissimilatory metal and sulfate reducer oligonucleotide microarray of relevance to subsurface bioremediation and microbial ecology.
MATERIALS AND METHODS
Bacterial strains.
Cultures of Desulfovibrio desulfuricans, Geobacter chapellei, Geobacter sulfurreducens, and Shewanella putrefaciens were obtained from the U.S. Department of Energy Subsurface Microbial Culture Collection. Geobacter isolates were grown anaerobically in 100-ml serum bottles containing an 80% N2-20% CO2 gas headspace and (per liter) the following: 5 mg of tryptone, 3 mg of yeast extract, 1 mg of glucose, 420 mg of KH2PO4, 220 mg of K2HPO4, 200 mg of NH4Cl, 380 mg of KCl, 360 mg of NaCl, 40 mg of CaCl2 · 2H2O, 100 mg of MgSO4 · 7H2O, 1.8 g of NaHCO3, 500 mg of Na2CO3, 8.0 g of fumarate, 10 ml of mineral elixir [containing, per liter, 2.14 g of nitrilotriacetic acid, 100 mg of MnCl2 · 4H2O, 300 mg of FeSO4 · 7H2O, 170 mg of CoCl2 · 6H2O, 200 mg of ZnSO4 · 7H2O, 30 mg of CuCl2 · 2H2O, 5 mg of AlK(SO4)2 · 12H2O, 5 mg of H3BO3, 90 mg of Na2MoO4, 110 mg of NiSO4 · 6H2O, and 20 mg of Na2WO4 · 2H2O], 25 ml of 2 M lactate, and 1 ml of 1 mM Na2SeO3. After sterilization, 150 μl of a vitamin mixture (containing, per liter, 2 mg of biotin, 2 mg of folic acid, 10 mg of pyridoxine HCl, 5 mg of riboflavin, 5 mg of thiamine, 5 mg of nicotinic acid, 5 mg of pantothenic acid, 0.1 mg of cyanocobalamin, 5 mg of p-aminobenzoic acid, and 5 mg of thioctic acid) was added anaerobically to each serum bottle. Bottles were inoculated with 1 ml of a log-phase culture and grown in the dark at an ambient temperature for 2 weeks prior to RNA isolation.
Culture conditions for D. desulfuricans are described at the Subsurface Microbial Culture Collection website (http://caddis.esr.pdx.edu/smccw/) and in reference 6. Briefly, cells (10% inoculum) were cultivated in 100 ml of medium C (containing, per liter, 7.9 ml of 60% sodium lactate syrup, 4.5 g of Na2SO4, 0.06 g of CaCl2 · 2H2O, 0.3 g of sodium citrate, 1.0 g of NH4Cl, 0.5 g of KH2PO4, 2.0 g of MgSO4 · 7H2O, and 1.0 g of yeast extract [Difco] [pH 7.2]; degassed with N2 and sterilized by autoclaving). Sterilized C medium was supplemented with 0.8 ml of FeSO4 solution (containing, per 50 ml, 0.025 g of FeSO4 · 7H2O and 5 ml of 1 M H2SO4; degassed with N2 and filter sterilized) and 1 ml of reductant solution (containing, per 50 ml, 0.5 g of sodium thioglycolate and 0.5 g of ascorbic acid; degassed with N2 and filter sterilized). The complete medium was then anaerobically inoculated (10% [vol/vol]) with starter cultures. Cultures were incubated on a shaker platform in the dark at 30°C for 4 days prior to RNA extraction.
RNA extraction and fragmentation.
Cells were harvested by centrifugation, and total RNA was extracted with an RNAeasy extraction kit as described by the manufacturer (Qiagen, Valencia, Calif.). Total RNA was also fragmented by standard methods (3). Briefly, 4 μl of fragmentation buffer (200 mM Tris, 500 mM potassium acetate, 150 mM magnesium acetate [pH 8.4]; made with diethyl pyrocarbonate-treated water and filter sterilized) and 6 μg of total RNA were adjusted to a 20-μl total volume in microarray hybridization buffer (30% [vol/vol] formamide, 5× SSPE [0.75 M NaCl, 50 mM NaH2PO4, 5 mM EDTA] [pH 7.0], 2.5× Denhardt's solution). RNA was incubated for 30 min at 95°C, cooled on ice, amended with chaperone detector probes (see below), adjusted to a 105-μl total volume with hybridization buffer, and applied directly to replicate microarrays.
Probe design.
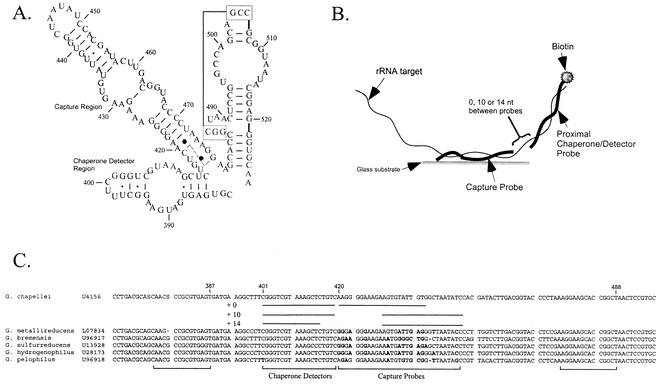
Oligonucleotide probes were designed based upon an alignment of dissimilatory metal and sulfate reducer 16S rRNA sequences deposited in GenBank. The alignment (and array) was targeted to full-length sequences for which an isolate was available in a public culture collection. Capture and chaperone detector probes were designed to examine the region of the 16S rRNAs of metal- and sulfate-reducing bacteria at about nucleotide (nt) 420 (G. chapellei numbering) (Fig. 1A). The region around nt 420 contains both variable and conserved sequences, a fact which allowed us to design species-specific capture probes and genus-level (or higher taxonomic rank) chaperone detector probes according to the scheme shown in Fig. 1B. Capture probes contained at least two mismatched nucleotides relative to all other capture probes on the array (Table 1). The locations of mismatched nucleotides in the capture and chaperone detector probes varied depending on the physical distance between the two probes, the length of the capture probe, and the target RNA sequences (Fig. 1B and C). Thus, capture probes immediately proximal to the chaperone detector probe (+0 capture probes) had mismatched nucleotides located primarily at the 5′ and 3′ ends, while capture probes located 10 and 14 nt away from the chaperone detector probe (+10 and +14 probes) had variable nucleotides in the center. All oligonucleotides were purchased from Sigma-Genosys (The Woodlands, Tex.). Unmodified capture probes were desalted, while biotinylated chaperone detector probes were purified by high-pressure liquid chromatography.
FIG. 1.
Metal and sulfate reducer 16S rRNA target region and detection strategy. (A) 16S rRNA secondary structure model for the target region based on the secondary structure models of Gutell (13) for D. desulfuricans. Numbering is based on the G. chapellei sequence (GenBank accession number U41561). (B) Conceptual representation of the proximal chaperone detector hybridization strategy. (C) Alignment of Geobacter targets showing the sequences and locations of capture and detector probes: +0, 0-nt separation between the 3′ end of the capture probe and the 5′ end of the detector probe; +10, 10-nt separation; +14, 14-nt separation.
TABLE 1.
Oligonucleotide capture and detector probes
| Probe | Name (designation)a | Sequence (5′-3′)b | Perfect match(es) | GenBank accession no. |
|---|---|---|---|---|
| Capture | S-S-Gbc.chap-0420-a-A-22 | CACAATACACTTCTTTCCCCTT | Geobacter chapellei | U41561 |
| S-S-Gbc.chap-0430-a-A-22 | GGATATTAGCCACAATACACTT | |||
| S-S-Gbc.sulf-0370-a-A-22 | TCTCAATCATTTCTTCCCTCCC | Geobacter sulfurreducens | U13928 | |
| S-S-Gbc.sulf-0380-a-A-22 | AGGTATTAGCTCTCAATCATTT | |||
| S-S-Gbc.met-0442-a-A-22 | CCTCAATCACTTCTTCCCTCCC | Geobacter metallireducens | L07834 | |
| S-S-Gbc.met-0452-a-A-22 | GGGTATTAACCCTCAATCACTT | |||
| S-S-Gbc.brem-0433-a-A-22 | CCAGCCCCATTTCTTCCCTTCT | Geobacter bremensis | U96917 | |
| S-S-Gbc.brem-0443-a-A-22 | TGGATATTAGCCAGCCCCATTT | |||
| S-S-Gbc.pel-0433-a-A-22 | CCGCACACATTTCTTTCCCTCT | Geobacter pelophilus | U96918 | |
| S-S-Gbc.pel-0443a-A-22 | CGGCTATTAACCGCACACATTT | |||
| S-S-Dsv.des-0448-a-A-22 | ACAACGTAGTTTCTTCCCTTCT | Desulfovibrio desulfuricans | M34113 | |
| S-S-Dsv.des-0458-a-A-22 | GCTGATTAGCACAACGTAGTTT | |||
| S-S-Dsv.gab-0395-a-A-22 | ATCCTCGGGGTTCTTCCCTCCT | Desulfovibrio gabonensis | U31080 | |
| S-S-Dsv.gab-0405-a-A-22 | CACTATTCGCATCCTCGGGGTT | |||
| S-S-Dsv.halo-0443-a-A-22 | CTCTAATGGTTTCTTCCCTCCT | Desulfovibrio halophilus | U48243 | |
| S-S-Dsv.halo-0453-a-A-22 | GCCTATTCGACTCTAATGGTTT | |||
| S-S-Dsv.grac-0400-a-A-22 | CCTCAAGGGTTTCTTCCCTTCT | Desulfovibrio gracilis | U53464 | |
| S-S-Dsv.grac-0410-a-A-22 | GCCTATTCGACCTCAAGGGTTT | |||
| S-S-Dsv.afr-0418-a-A-22 | ACCTTGGGGGTTCTTCCCTTCT | Desulfovibrio africanus | X99236 | |
| S-S-Dsv.afr-0428-a-A-22 | CCCTATTCGAACCTTGGGGGTT | |||
| S-S-S.putr-0425-a-A-22 | GCGTATTAAAGCTACACCCTTT | Shewanella putrefaciens | X82133 | |
| S-S-Dfm.geo-0513-a-A-22 | TCCTCCAGGGGTACCGTCATCC | Desulfotomaculum geothermicum | Y11567 | |
| S-S-Dfm.the-0523-a-A-22 | TCCTCGTTGGGTACCGTCACTT | Desulfotomaculum thermobenzoicum | L15628 | |
| S-S-Dsn.mag-0416-a-A-22 | TGGTATTAACATAAGACAGGTT | Desulfonema magnum | U45989 | |
| S-S-Dsbm.vac-0451-a-A-22 | ACTCTATTAAAGCATAATAATT | Desulfobacterium vacuolatum | M34408 | |
| S-S-Dsbm.nia-0429-a-A-22 | TGCTATTAACACAAAATAACTT | Desulfobacterium niacini | U51845 | |
| S-S-Pel.car-0388-a-A-22 | GGCCTATTCGACCACGATAGTT | Pelobacter carbinolicus | U23141 | |
| S-S-Dsb.hyd-0462-a-A-22 | TACTATTAATAGAAGCTAATTT | Desulfobacter hydrogenophilus | M34412 | |
| Detector | S-S-Gbc.chap-0401-a-A-19 (1) | GACAGAGCTTTACGACCCG-biotin | G. chapellei, G. metallireducens, G. bremensis | |
| S-S-Gbc.sulf-0351-a-A-19 (2) | GACAGGGCTTTACGACCCG-biotin | G. sulfurreducens | ||
| S-S-Gbc.pel-0414-a-A-19 (3) | GACAGAGCTTTACGACCCA-biotin | G. pelophilus | ||
| S-S-Dsv.des-0429-a-A-19 (4) | GACAGAGGTTTACGATCCG-biotin | D. desulfuricans | ||
| S-S-Dsv.gab-0376-a-A-19 (5) | GACAGAGGTTTACGACCCG-biotin | D. gabonensis, D. gracilis, D. africanus | ||
| S-S-Dsv.hal-0424-a-A-19 (6) | GACAGCGGTTTACGACCCG-biotin | D. halophilus | ||
| S-S-Dsv.bas-0381-a-A-19 (7) | GACAGAGGTTTACGACCCT-biotin | Desulfovibrio bastinii | U53462 | |
| S-S-Dsv.cal-0381-a-A-19 (8) | GACAGTAGTTTACGACCCG-biotin | Desulfovibrio caledoniensis | U53465 | |
| S-S-S.putr-0396-a-A-19 (9) | GAAAGTGCTTTACAACCCG-biotin | S. putrefaciens | ||
| S-S-Gbc.chap-0401-a-A-15 (10) | GAGCTTTACGACCCG-biotinc | G. chapellei, G. metallireducens, G. bremensis | ||
| S-S-Dsv.des-0429-a-A-15 (11) | GAGGTTTACGATCCG-biotinc | D. desulfuricans | ||
| S-G-Gbc-0474(G.chap)-a-A-15 (16) | Biotin-AGCCGGTGCTTCCTT | Geobacter | ||
| S-G-Gbc-0374(G.chap)-a-A-15 (17) | ACTCACGCGGCGTTG-biotin | Geobacter |
Oligonucleotide probes were named according to the guidelines of Alm et al. (1).
Bold italicized letters in detector probe sequences indicate mismatches relative to the G. chapellei target.
The use of shortened detector probes in combination with +10 capture probes resulted in the +14 probe separation distance described in Materials and Methods and shown in Fig. 1B.
Microarray design and fabrication.
The study objective was to compare the specificity of +0 capture probes to that of +10 and +14 capture probes for 10 bacterial species. The statistical design for accommodating this comparison had to accommodate signal variations across replicates, across wells, across hybridizations, and across slides in order to separate the probe effect from other factors affecting pixel intensity. The experimental design had to accommodate limitations in the allowable number of spots per array, in the number of arrays per slide, and in the application of treatments to wells. These constraints determined the layout of probes within an array and the application of target nucleic acids to the array.
Each planar array used in this study contained 60 individual spots printed in 10 rows and 6 columns. A species-specific +0 capture probe was replicated over the first three spots in a row; a species-specific +10 (or +14) capture probe was replicated over the next three spots. Each array featured a seventh row of two quality control oligonucleotide spots. Microarrays were fabricated as described in detail elsewhere (8). Briefly, 12-well, precleaned, Teflon-masked glass slides (Erie Scientific, Portsmouth, N.H.) were coated with epoxysilane (3-glycidoxyproplytrimethoxysilane) (Aldrich, Milwaukee, Wis.) for a minimum of 2 h, rinsed in methanol, and dried under compressed nitrogen. Probes were printed in triplicate at 80 to 90 μM in 0.01% sodium dodecyl sulfate-50 mM NaOH (pH 12.7) by using a Genetic Microsystems (now Affymetrix) 417 Arrayer after the array design. A biotin-labeled quality control oligonucleotide (5′-biotin-TTGTGGTGGTGTGGT-3′) was also printed in duplicate and acted both as a positive control for the signal development procedure and as a positional reference mark for imaging. One complete array was printed in each of the 12 wells, resulting in 12 replicate arrays per slide. After printing, slides were baked for 30 min in a 130°C vacuum oven and stored at room temperature.
Microarray hybridization and detection.
All hybridizations were performed with 2 μg of intact or fragmented total RNA in each microarray well. RNA was mixed with hybridization buffer and 1 μl of the appropriate detector probe (50 μmol) for a total of 35 μl per well. For each experiment, one bacterial RNA target was mixed with a detector probe and applied to one array. The same treatment was replicated in a second well on the same slide. The multifactorial experiment used two slides to accommodate the 10 bacterial species, giving four replicate arrays per test condition. Slides were incubated for 2 h at room temperature, the hybridization solution was aspirated from each well, and the slides were rinsed twice in 4× SSPE. The signal was developed by incubating the slides for 1 h at room temperature in AMDEX streptavidin-alkaline phosphatase (Amersham-Pharmacia) diluted 1:500 in 4× SSPE. Slides were then rinsed with 1× ELF wash buffer (ELF-97 mRNA in situ hybridization kit; Molecular Probes, Eugene, Oreg.) and 20 μl of ELF-97 substrate (1:100 in 1× ELF developing buffer C). The substrate solution was aspirated from the wells, and the slides were washed in a solution of 50 mM NaCl-0.1% Tween 20-100 mM Tris (pH 8) and then rinsed twice in double-distilled H2O. The remaining double-distilled H2O was removed by vacuum aspiration.
Extracting spot summary statistics.
To estimate the level of hybridization at each spot (i.e., spot intensity), we identified two sets of pixels: spot and spot neighborhood. However, estimated spot and spot neighborhood background signal intensities will vary from well to well and slide to slide because of nonhybridization effects such as pin-to-pin variability, spot shape and texture, wash or rinse residue, nonspecific ELF deposition, and slide illumination (see, e.g., Fig. 4B). Spot and spot neighborhood intensities were estimated from an image of fluorescent emissions by using an ArrayWoRx microarray scanner (Applied Precision, Issaquah, Wash.) with illumination at 360 nm and image capture at 535 nm. Spot estimation produced 1,440 sets of spot summary statistics, one for each of the 60 spots/well times 12 wells times 2 slides. The set of statistics for each array included mean spot and mean background intensities with sample variances as well as classification identifiers for spot position within an array, slide, hybridization, well, bacteria, probe, and replicate. The set of individual spot and spot neighborhood statistics was the basis for estimating the level of hybridization according to the response function y = log(spot intensity/background intensity).
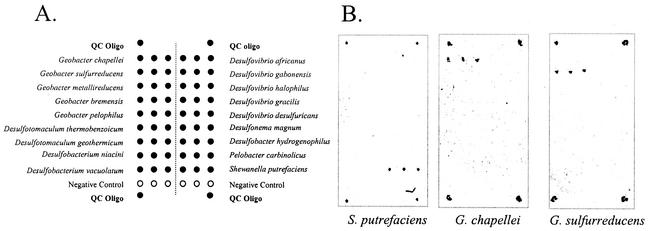
FIG. 4.
Multigenus microarray. (A) Print pattern for oligonucleotide (Oligo) probes. QC, quality control. (B) Hybridization results for 2 μg of intact RNA. Target RNA species are shown beneath the hybridization results.
Linear statistical modeling.
Although similar hybridization profiles within an rRNA-detector probe combination were observed, the signal intensities between replicate arrays varied considerably (see Results). Thus, an analysis-of-variance (ANOVA) model was developed to account for the observed variability between replicate arrays, so that we could statistically evaluate the effects of chaperone detector probe proximity and nucleotide mismatches on hybridization specificity. A comparison of signal intensities to determine probe effects was accomplished by using linear statistical modeling. For this application, the linear (ANOVA) statistical model included crossed, nested, fixed, and random terms (Tables 2 and 3). The model is a direct reflection of the experimental design, which featured random effects, such as slide; fixed effects, such as hybridization; crossed terms, such as slide crossed with hybridization; and nested terms, such as wells within slides and hybridization. Table 2 lists the factors with levels and types that were considered in this study. The full mixed-effects linear model is listed in Table 3. The model was analyzed by using the ANOVA capabilities in the JMP statistical analysis software package (SAS, Inc., Cary, N.C.).
TABLE 2.
Experimental factors influencing microarray spot signal intensities
| Factor | Identification | Codes | Type/effect |
|---|---|---|---|
| Slide | S | s = 1, 2 | Random |
| Hybridization | H | h = 1-6 | Fixed |
| Well | W | w = 1, 2 | Random |
| Bacteria | B | b = 1-10 | Fixed |
| Probe | P | p = 1, 2 | Fixed |
| Error (uncertainty) | E | e = 1-3 | Random |
TABLE 3.
ANOVA full mixed-effects linear model
| Terma | Effect | Degrees of freedomb |
|---|---|---|
| μ | Fixed | 1 |
| Ss | Random | 1 |
| Hh | Fixed | 5 |
| SHsh | Random | 5 (= 1 × 5) |
| Ww(sh) | Nested random | 12 (= 1 × 2 × 6) |
| Bb | Fixed | 9 |
| SBsb | Random | 9 (= 1 × 9) |
| HBhb | Crossed fixed | 45 (= 5 × 9) |
| SHBshb | Mixed random | 45 (= 1 × 5 × 9) |
| WBwb(sh) | Nested random | 108 (= 1 × 9 × 2 × 6) |
| Pp | Fixed | 1 |
| SPsp | Mixed random | 1 (= 1 × 1) |
| HPhp | Crossed fixed | 5 (= 5 × 1) |
| SHPshp | Mixed random | 5 (= 1 × 5 × 1) |
| WPwp(sh) | Nested random | 12 (= 1 × 1 × 2 × 6) |
| BPbp | Crossed fixed | 9 (= 9 × 1) |
| SBPsbp | Mixed random | 9 (= 1 × 9 × 1) |
| HBPhbp | Crossed fixed | 45 (= 5 × 9 × 1) |
| SHBPshbp | Mixed random | 45 (= 1 × 5 × 9 × 1) |
| WBPwbp(sh) | Nested random | 108 (= 1 × 9 × 1 × 2 × 6) |
| Ee(shwbp) | Random | 960 (= 2 × 2 × 6 × 2 × 10 × 2) |
| Totalc | 1,440 |
Subscripts (codes) indicate the interacting factors (nested and crossed) as described in Table 2.
The numbers in parentheses indicate the respective contribution of each code to the degrees of freedom calculation.
Yshwbpe = μ + Ss + Hh + SHsh + Ww(sh) + Bb + SBsb + HBhb + SHBshb + WBwb(sh) + Pp + SPsp + HPhp + SHPshp + WPwp(sh) + BPbp + SBPsbp + HBPhbp + SHBPshbp + WBPwbp(sh) + Ee(shwbp).
RESULTS
Probe proximity.
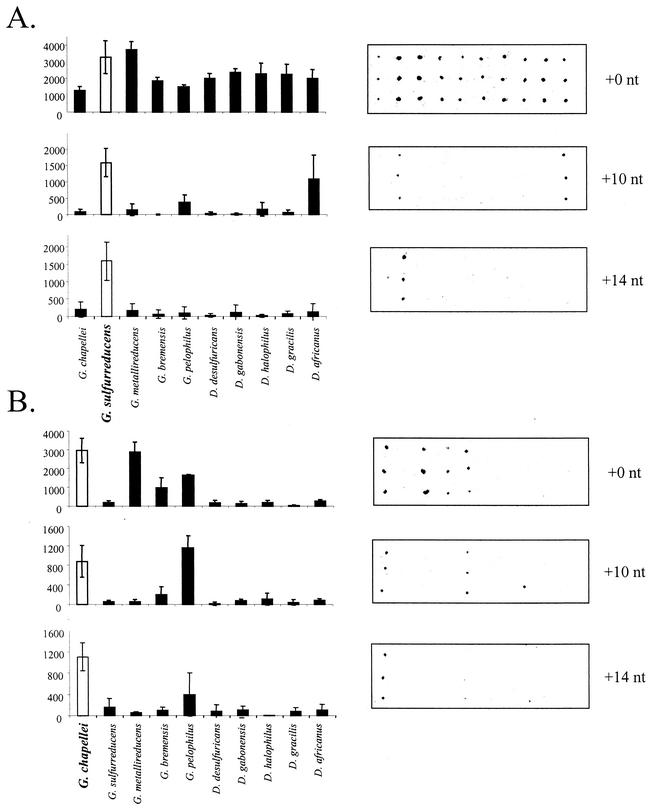
Initial experiments were performed by using a simple array targeting five species of Geobacter and five species of Desulfovibrio. The capture probes were situated either directly adjacent to (+0) or 10 nt away from (+10) the 3′ end of the detector probe, in keeping with the hybridization scheme shown in Fig. 1B. Capture and detector probes in immediate proximity to each other (+0) showed significant cross-hybridization to nontarget probes, regardless of the target RNA used for the experiment (Fig. 2). On the other hand, the +10 capture probes showed greatly improved species specificity. Signal intensities plotted in Fig. 2 are the averages and standard deviations from four independent hybridizations and illustrate the large standard deviations inherent in replicate microarray experiments. By applying the ANOVA model described above, we were able to assess whether there was a statistically significant difference in +10 signal intensities between the perfectly matched and mismatched capture probes.
FIG. 2.
Increased hybridization specificity with increased separation between the capture probe and the detector probe. Hybridizations were performed as described in Materials and Methods with 2 μg of intact G. chapellei RNA (A) or intact G. sulfurreducens RNA (B). Numbers on the y axes are mean fluorescence intensity (in arbitrary units). White columns indicate the perfectly matched probe for the target RNA. Error bars indicate standard deviations.
For the G. sulfurreducens target, not only was there a qualitative improvement in hybridization specificity between +0 and +10 capture probes, but the perfectly matched +10 probe yielded a significantly higher signal intensity than all the other +10 capture probes. It is unclear why G. sulfurreducens RNA (+10 sequence, AGGTATTAGCTCTCAATCATTT) hybridized to the D. africanus-specific capture probe (+10 sequence, CCCTATTCGAACCTTGGGGGTT) with a 10-nt separation, as there are only eight homologous nucleotides within the 22-bp target region and the results of a BLAST search indicated that there is no significant homology between the capture probe and the complete G. sulfurreducens RNA. The G. chapellei target showed significant cross-reaction with the Geobacter pelophilus-specific capture probe, even in the +10 and + 14 configurations (Fig. 2B). Nonetheless, improved hybridization specificity was observed with the +10 capture probes, and similar results were obtained for fragmented and nonfragmented D. desulfuricans and S. putrefaciens RNA targets (not shown). In all instances, intact RNA targets resulted in lower overall signal intensities on the arrays than did fragmented RNA targets.
Although the +10 capture probes were qualitatively and quantitatively more specific than the +0 capture probes, cross-hybridization to some nontarget +10 probes was still detectable at significant levels above background, including capture probes targeted toward a different genus (e.g., Desulfovibrio africanus-specific probe) (Fig. 2A). In an attempt to further reduce the incidence and intensity of nonspecific hybridization, especially at the level of genus cross-reactivity, a shorter detector probe (detector probe 10) was designed to extend the distance between the ends of the capture and detector probes to +14 nt. By shortening the detector probe to 15 nt and increasing the distance between the capture and detector probes, detection specificity was enhanced for both G. chapellei and G. sulfurreducens rRNAs (Fig. 2); identical results (albeit weaker signals) were obtained for intact RNA (data not shown). Cross-hybridization between G. sulfurreducens rRNA and the D. africanus-specific capture probe eliminated the observed cross-genus hybridization and gave a signal that was at least specific for genus-specific target RNA. Similar results were obtained for G. chapellei RNA (intact and fragmented) (Fig. 2B) and D. desulfuricans RNA (Fig. 3, detector probes 4 and 11, +10 capture probes).
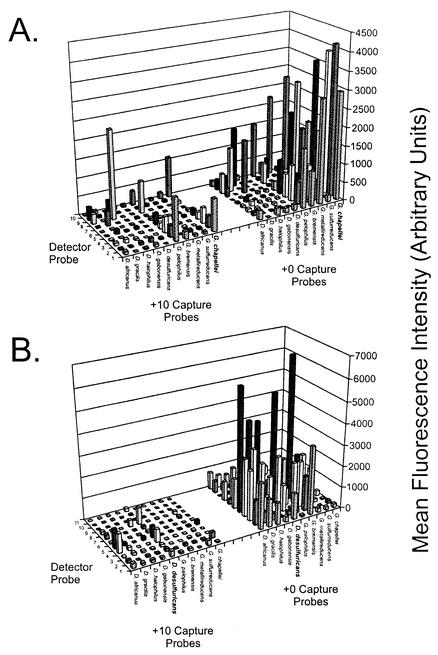
FIG. 3.
Effect of nucleotide mismatches between the detector probe and target rRNA. Detector probe sequences and perfect matches are listed in Table 1. Probes1 to 3 are specific for Geobacter species; probes 4 to 8 are specific for Desulfovibrio species; probe 9 is specific for S. putrefaciens; and probe 10 is identical to probe 1 and is specific for Geobacter species, except that it is shortened by 4 nt to achieve a +14 hybridization condition. Signals represent the average hybridization intensity from at least three replicate arrays (across two slide lots), with each array containing three replicate spots. Hybridizations were performed as described in Materials and Methods with 2 μg of fragmented G. chapellei target RNA (A) or fragmented D. desulfuricans target RNA (B). Similar results were obtained with intact RNA targets (not shown). The graphing software did not permit error bars to be plotted in the three-dimensional graphing format, and so they are not shown.
Secondary structure.
Separating the detector probe from the capture probe clearly improved hybridization specificity, presumably by eliminating or reducing the base-stacking energy in the duplex. However, the results also suggested that disrupting the secondary structure between nt 411 and 420 (G. chapellei numbering) (Fig. 1A) was important for achieving specific hybridization. In order to distinguish between base-stacking and secondary structure effects on hybridization specificity, two additional detector probes were synthesized to specifically disrupt the stem from nt 411 to 414 or the stems from nt 415 to 419 and nt 480 to 488 without contributing base-stacking energy to the duplex (Fig. 1C). Interestingly, detector probe 17 resulted in nondetection for both G. chapellei and G. sulfurreducens RNAs in either the intact or the fragmented condition. Detector probe 16, on the other hand, resulted in a detectable signal for both fragmented and intact RNA targets. The intact RNA resulted in a substantially higher signal intensity than the fragmented RNA; the +10 capture probes were again more specific than the +0 capture probes (data not shown). When detector probes 16 and 17 were combined, the results mirrored those obtained with detector probe 16 alone. An earlier study (26) showed that detector probes targeting the universal region at nt 519 likewise resulted in nondetection. In combination, these results suggest that disrupting the stem at nt 415 to 419 is critical for achieving a detectable hybridization signal with this particular suite of capture probes, regardless of base-stacking interactions.
Detector probe mismatch tolerance.
Having established that secondary structure is a primary obstacle to successful 16S rRNA detection, we investigated the extent to which mismatched nucleotides in the detector probe affect hybridization specificity on the array. As shown in Table 1, the detector probe sequences in this region of the 16S rRNA either can contain mismatched nucleotides within the genus or can be fairly conserved across genera. Either situation could result in nonspecific hybridization, detection, or erroneous interpretation of hybridization patterns from samples with unknown compositions. Further, specificity can be defined in terms of either (i) the detection of a specific target in a mixture of nontarget sequences or (ii) the cross-hybridization of a target to nontarget capture probes on the array. Both aspects of specificity were tested for three RNA targets and 11 detector probes with both +0 and +10 capture probes, as defined above.
The results for two of the three target RNAs are shown in Fig. 3. The G. chapellei target RNA (Fig. 3A) was detected with the +10 capture probes and all but detector probes 7 and 9, which share perfect identity with Desulfovibrio bastinii and S. putrefaciens, respectively. Hybridization signals resulting from non-Geobacter-specific detector probes were generally less intense than those resulting from Geobacter-specific detector probes, although they were easily detectable above background. We expected that mismatches in the middle of a detector probe would be more unstable than terminal mismatches, as has been shown for oligonucleotide array capture probes (11), and might therefore lead to nondetection. However, there was no obvious or predictable pattern to the microarray hybridization profile based solely on the nucleotide or position of mismatches between the detector probe and the target. For example, detector probe 7 (one central mismatch, one terminal mismatch) resulted in nondetection of the G. chapellei target, whereas detector probe 8 (three contiguous central mismatches) resulted in positive hybridization and detection of the G. chapellei target. Cross-hybridization to the G. pelophilus-specific capture probe was similar in relative magnitude to the results shown in Fig. 2A, regardless of the detector probe sequence. Interestingly, the use of Geobacter-specific detector probes with G. chapellei target RNA resulted in the lowest overall cross-hybridization to Desulfovibrio-specific capture probes (+0 and +10 variants), whereas Desulfovibrio-specific detector probes and G. chapellei target RNA sometimes resulted in nonspecific hybridization signals that exceeded those obtained with the G. chapellei-specific capture probe (e.g., detector probe 8 with +10 capture probes) (Fig. 3A). These same general trends were also observed for G. sulfurreducens target RNA (data not shown).
The results obtained with the D. desulfuricans target RNA were qualitatively similar to those obtained with the Geobacter target RNA. The only detector probe that failed to produce a detectable signal with the +10 capture probes was detector probe 9 (S. putrefaciens), which contained five mismatches with the D. desulfuricans target. The results again were generally consistent with the results shown in Fig. 2, in that regardless of the detector probe, cross-hybridization to nontarget capture probes was typically constrained to the genus. Cross-hybridization to Geobacter-specific capture probes was consistently less intense than that to within-genus capture probes, even when Geobacter-specific detector probes were used. In combination, these results indicate that the detector probe sequence is not the principal determinant for achieving hybridization specificity in the planar microarray format used here. They also indicate that a species-specific chaperone detector probe is not required in order to achieve positive, specific hybridization, a practical result that can greatly simplify the design and application of 16S rRNA arrays for microbial community profiling.
Multigenus array.
Given these results, we designed and fabricated a multigenus array to qualitatively assess the cross-hybridization of target RNA to more distantly related genera. Capture and detector probes were designed to conform to the +10 structural motif described above and were printed as shown in Fig. 4A. Two micrograms of intact RNA was hybridized to the array as described in Materials and Methods; the results for three RNA targets are shown in Fig. 4B. These preliminary results indicate that the chaperone detector probe strategy can result in specific (within-genus) hybridization with an expanded array that includes primary groups of metal- and sulfate-reducing bacteria.
DISCUSSION
In general, attempts to discriminate between specific hybridization and nonspecific hybridization on microarrays depend on a statistical comparison of signal intensities arising from perfectly matched and mismatched capture probes (28). Subtracting, normalizing, or statistically comparing signals from perfectly matched probes against signals from mismatched probes is meaningful when both the sequence of the target RNA (or organism) and the complete sample composition are known. However, the true abundance of viable but nonculturable organisms (or rRNA) in nature cannot be known a priori (such as microbial community profiling in environmental samples). Thus, perfectly matched/mismatched ratios have little meaning for the detection and characterization of rRNA targets from uncharacterized, mixed microbial communities with unknown sequence compositions. Indeed, recent results obtained with a very-high-density photolithographic phylochip were able to identify bacteria in concentrated aerosols only to the third level of phylogenetic rank as defined in the Ribosomal Database Project (19, 28).
For the analysis of unknown or mixed samples, then, any detectable signal (regardless of mean fluorescence intensity) cannot be ignored, as it may have biological or practical significance. Designing, applying, and interpreting microarrays in an environmental context are therefore much more complicated than would otherwise be anticipated given the rapid pace of microarray development and implementation in other contexts. Indeed, the results shown in Fig. 2 and 3 indicate that despite improved discrimination between target and nontarget sequences upon elimination of the base-stacking energy in the capture-detector probe contig, detectable cross-species hybridization is still observed. The question then turns to how 16S rRNA microarray data can be deconvolved into ecologically meaningful results.
Our approach to phylogenetic profiling is to capture and detect 16S rRNA directly on oligonucleotide arrays (26), as opposed to the more universal approach of amplifying 16S rDNA and analyzing the diversity of resulting amplification products (28). This philosophy of phylogenetic profiling brings with it unforeseen challenges and unpredictable hybridization behavior on oligonucleotide microarrays. The results shown in Fig. 2 and 3, for example, illustrate the inherent difficulty in achieving species-specific detection for 16S rRNA targets hybridized directly on an oligonucleotide array, even with multiple internal mismatches between the capture probe and the target (Fig. 1C). Despite the difficulty in achieving direct, species-specific capture and detection of rRNA and the occasional cross-genus hybridization signal, our results do indicate that the proximal chaperone detector strategy can lead to qualitative, genus-level rRNA hybridization specificity (Fig. 4). We conclude from these data that eliminating the base-stacking interaction between the capture and detector probes improves hybridization specificity, but the data are not conclusive with respect to the optimal spacing between the capture and detector probes either for this particular array or for 16S rRNA-targeted arrays in general. We anticipate that optimal probe spacing will be dependent upon the microbial targets of interest (i.e., beyond metal and sulfate reducers) and the specific 16S rRNA region within which useful capture probes can be designed (i.e., outside the region around nt 420). On the basis of the results presented here, however, we are designing an expanded +10 metal and sulfate reducer array in which species-specific capture probes are expected to cross-hybridize with relatives (and uncultured microorganisms) from within the same genus. In this manner, species-level capture probes can be used to examine near neighbors within the uncultured population, and microarray hybridization profiles and ecological parameters can be correlated at the genus level (not the species level).
Cross-species (and even cross-genus) hybridization also leads to the question of when a hybridization signal is significant. Assessing significance (of any kind) requires statistical models and methods that allow signals to be quantitatively compared within an array, across arrays, and between samples. Assessing the significance of different signal intensities within an array (i.e., between different species-specific capture probes) requires a better understanding of differential hybridization efficiency and the relationship between signal intensity and RNA copy number in the hybridization solution. Such studies will result in statistically defensible thresholds that can be applied to (validated) microarrays and determinants for judging the ecological significance of a given microarray probe response.
For the time being, however, we assume that a single RNA target will hybridize to any given probe with a defined and reproducible efficiency from one experiment to the next. The statistical model described here is therefore important because it defines an experimental design and provides a mathematical framework that does allow quantitative comparison of signal intensities from a single probe across arrays, slides, or samples. Thus, by analyzing RNA directly (no PCR) and using a statistical model of the type developed here, fluctuations in signal intensities (and, hence, microbial abundance and/or activity) for individual capture probes can be quantified, statistically compared, and correlated with ecological (or other) properties of interest. The next steps toward realizing this goal include expanding the metal and sulfate reducer array (species and genera), validating the probe set with a wider range of cultivated organisms, and applying the array and statistical methods to spatially and/or temporally correlated environmental samples.
Acknowledgments
This work was supported by the U.S. Department of Energy (DOE) under the Assessment Element of the NABIR Program. Argonne National Laboratory is operated for the U.S. DOE by the University of Chicago under contract W-31-109-ENG-38. Pacific Northwest National Laboratory is operated for the U.S. DOE by Battelle Memorial Institute under contract DE-AC06-76RLO 1830.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I. 1995. Fluorescently labelled, rRNA-targeted oligonucleotide probes in the study of microbial ecology. Mol. Ecol. 4:543-554. [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Bavykin, S. G., J. P. Akowski, V. M. Zakhariev, V. E. Barksy, A. N. Perov, and A. D. Mirzabekov. 2001. Portable system for microbial sample preparation and oligonucleotide microarray analysis. Appl. Environ. Microbiol. 67:922-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beattie, W. G., L. Meng, S. L. Turner, R. S. Varma, D. D. Dao, and K. L. Beattie. 1995. Hybridization of DNA targets to glass-tethered oligonucleotide probes. Mol. Biotechnol. 4:213-225. [DOI] [PubMed] [Google Scholar]
- 6.Boone, D. R., R. L. Johnson, and Y. Liu. 1989. Diffusion of the interspecies electron carriers H2 and formate in methanogenic ecosystems and its implications in the measurement of Km for H2 or formate uptake. Appl. Environ. Microbiol. 55:1735-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Call, D. R., F. J. Brockman, and D. P. Chandler. 2001. Genotyping Escherichia coli O157:H7 using multiplexed PCR and low-density microarrays. Int. J. Food Microbiol. 67:71-80. [DOI] [PubMed] [Google Scholar]
- 8.Call, D. R., D. P. Chandler, and F. J. Brockman. 2001. Fabrication of DNA microarrays using unmodified oligomer probes. BioTechniques 30:368-379. [DOI] [PubMed] [Google Scholar]
- 9.Drmanac, S., D. Kita, I. Labat, B. Hauser, C. Schmidt, J. D. Burczak, and R. Drmanac. 1998. Accurate sequencing by hybridization for DNA diagnostics and individual genomics. Nat. Biotechnol. 16:54-58. [DOI] [PubMed] [Google Scholar]
- 10.Gunderson, K. L., X. C. Huang, M. S. Morris, R. J. Lipshutz, D. J. Lockhart, and M. S. Chee. 1998. Mutation detection by ligation to complete n-mer DNA arrays. Genome Res. 8:1142-1153. [DOI] [PubMed] [Google Scholar]
- 11.Guo, Z. G., A. Guilfoyle, A. J. Thiel, R. Wang, and L. M. Smith. 1994. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res. 22:5456-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guttell, R. R. 1994. Collection of small subunit (16S and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 22:3502-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacia, J. G., and F. S. Collins. 1999. Mutational analysis using oligonucleotide microarrays. J. Med. Genet. 36:730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iannone, M. A., J. D. Taylor, J. Chen, M.-S. Li, P. Rivers, K. A. Slentz-Kesler, and M. P. Weiner. 2000. Multiplexed single nucleotide polymorphism genotyping by oligonucleotide ligation and flow cytometry. Cytometry 39:131-140. [PubMed] [Google Scholar]
- 16.Jacobsen, C. S. 1995. Microscale detection of specific bacterial DNA in soil with a magnetic capture-hybridization and PCR amplification assay. Appl. Environ. Microbiol. 61:3347-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan, J., L. H. Saal, M. L. Bittner, Y. Chen, J. M. Trent, and P. S. Meltzer. 1999. Expression profiling in cancer using cDNA microarrays. Electrophoresis 20:223-229. [DOI] [PubMed] [Google Scholar]
- 18.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Folletti, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 19.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maldonado-Rodriguez, R., M. Espinosa-Lara, A. Calixto-Suárez, W. G. Beattie, and K. L. Beattie. 1999. Hybridization of glass-tethered oligonucleotide probes to target strands preannealed with labeled auxiliary oligonucleotides. Mol. Biotechnol. 11:1-12. [DOI] [PubMed] [Google Scholar]
- 21.Maldonado-Rodriguez, R., M. Espinosa-Lara, P. Loyoloa-Abitia, W. G. Beattie, and K. L. Beattie. 1999. Mutation detection by stacking hybridization on genosensor arrays. Mol. Biotechnol. 11:13-25. [DOI] [PubMed] [Google Scholar]
- 22.Nelson, B. P., T. E. Grimsrud, M. R. Liles, R. M. Goodman, and R. M. Corn. 2001. Surface plasmon resonance imaging measurements of DNA and RNA hybridization adsorption onto DNA microarrays. Anal. Chem. 73:1-7. [DOI] [PubMed] [Google Scholar]
- 23.Parinov, S., V. Barsky, G. Yershov, E. Kirillov, E. Timofeev, A. Belgovskiy, and A. Mirzabekov. 1996. DNA sequencing by hybridization to microchip octa- and decanucleotides extended by stacked pentanucleotides. Nucleic Acids Res. 24:2998-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pease, A. C., D. Solas, E. J. Sullivan, M. T. Cronin, C. P. Holmes, and S. P. A. Fodor. 1994. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl. Acad. Sci. USA 91:5022-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schena, M., D. Shalon, R. Heller, A. Chai, P. O. Brown, and R. W. Davis. 1996. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93:10614-10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Small, J. A., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts using oligonuleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiro, A., M. Lowe, and D. Brown. 2000. A bead-based method for multiplexed identification and quantitation of DNA sequences using flow cytometry. Appl. Environ. Microbiol. 66:4258-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson, K. H., W. J. Wilson, J. L. Radosevich, T. Z. DeSantis, V. S. Viswanathan, T. A. Kuczmarski, and G. L. Andersen. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yershov, G., V. Barsky, A. Belgovskiy, E. Kirillov, E. Kreindlin, I. Ivanov, S. Parinov, D. Guschin, A. Drobishev, S. Dubiley, and A. Mirzabekov. 1996. DNA analysis and diagnostics on oligonucleotide microchips. Proc. Natl. Acad. Sci. USA 93:4913-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]