Abstract
Listeria monocytogenes is a gram-positive, facultative intracellular bacterium implicated in severe food-borne illness (listeriosis) in humans. The construction of well-defined gene replacements in the genome of L. monocytogenes has been instrumental to several genetic studies of the virulence and other attributes of the organism. Construction of such mutations by currently available procedures, however, tends to be labor intensive, and gene replacement mutants are sometimes difficult to recover due to lack of direct selection for the construct. In this study we describe the construction and use of plasmid vector pGF-EM, which can be conjugatively transferred from Escherichia coli S17-1 to L. monocytogenes and which provides the genetic means for direct selection of gene replacements.
Listeria monocytogenes is a gram-positive, facultative intracellular bacterium implicated in severe food-borne illness (listeriosis) in individuals who are at risk, especially pregnant women, neonates, the elderly, and the immunocompromised (4, 15). Although listeriosis cases are infrequent, the high mortality of the disease (commonly 20 to 30%) has precipitated the need for efficient monitoring of the organism in foods and for a better understanding of the pathogenic potential of the bacterium. Virulence studies have involved a number of genetic tools, including transposons and plasmid-mediated mutagenesis systems and animal as well as cell culture models (reviewed in references 8 and 15).
The construction of well-defined gene deletions is highly desirable for genetic studies of this pathogen. Unlike mutations mediated by transposon insertion or other insertion mutagenesis schemes, which can be accompanied by polar effects, deletions allow the precise evaluation of the loss of a specific genetic determinant.
The construction of in-frame deletions and their application in the genetic study of L. monocytogenes was originally described by Camilli et al. (3) with the temperature-sensitive shuttle plasmid pKSV7 (13), which can be introduced into L. monocytogenes by transformation of protoplasts (3) or by electroporation (10). Subsequent studies have also utilized pCON1, a plasmid with the genetic features of pKSV7 and, in addition, an origin of transfer element which allows it to be transferred from Escherichia coli into L. monocytogenes by conjugation (1).
To select for deletions and other allelic exchange mutants generated with these vectors, the recombinant plasmids with the cloned mutation of interest are first integrated into homologous regions of the L. monocytogenes genome following growth of the bacteria at the restrictive temperature (42°C) in the presence of an antibiotic (chloramphenicol), the resistance to which is vector borne. The strains containing the integration are repeatedly grown at permissive temperature (30°C) without chloramphenicol, and allelic exchanges mediated by a second crossover are identified by the loss of chloramphenicol resistance. However, identification of the final allelic exchange recombinants is often cumbersome because of the lack of positive selection. If the allelic exchange products harboring the mutation (e.g., deletion mutants) have any growth bias relative to the wild-type recombinants, which theoretically are expected to constitute 50% of the population, the desired mutants are often found to be a minute (1 to 2%) fraction of the chloramphenicol-sensitive recombinants and sometimes are not recovered at all. Thus, the need exists for plasmids with additional selection markers to facilitate the construction of such allelic exchange mutants. The construction and application of such a vector are described in this report.
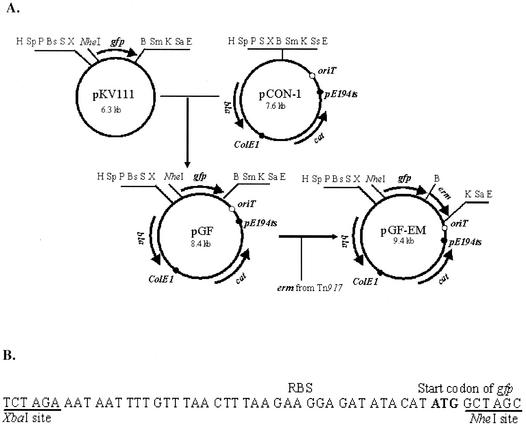
Plasmid pGF-EM was derived from plasmid pGF, a pCON1 derivative that has been generated in our laboratory for the purpose of generating green fluorescence protein (GFP) fusions in L. monocytogenes (9). Plasmid pGF was generated by cloning a promoterless gfp with a putative ribosomal binding site and flanking multiple cloning sites in pCON1 (Fig. 1A). On the pCON1 backbone, the plasmid harbors genes for ampicillin resistance (expressed in E. coli) and chloramphenicol resistance (expressed in Listeria). An erythromycin resistance gene from Tn917, erm, was cloned into pGF, to generate pGF-EM. To amplify erm, we used primers ermA and ermB (with BamHI and KpnI sites, respectively) (Table 1), based on the Tn917 sequence (11) and the Tn917 derivative pLTV3 (2) as a template. The amplified erm fragment was directionally cloned in BamHI- and KpnI-digested pGF downstream of gfp and in the same transcriptional orientation as the latter. The erythromycin resistance gene erm was amplified so as to lack both a promoter and a terminator in order to avoid influencing expression of downstream genes. The steps involved in the construction of the plasmid are diagrammatically shown in Fig. 1A. Figure 1B shows the sequence and multiple cloning sites immediately upstream of gfp in pGF-EM (as well as pGF). Especially useful is the NheI site immediately following the gfp start codon, as it is readily amenable to the construction of in-frame fusions to gfp.
FIG. 1.
Construction of pGF-EM. (A) Steps in the construction. The ca. 0.8-kb gfp fragment was excised from pKV111 (14) by HindIII and EcoRI digestion and ligated to HindIII- and EcoRI-digested pCON1 to generate pGF. ColE1, origin of replication function in E. coli; pE194ts, temperature-sensitive origin of replication functional in Listeria and other gram-positive bacteria (13); oriT, origin of transfer, allowing conjugative transfer from E. coli S17-1 to Listeria; cat, chloramphenicol resistance gene, expressed in Listeria; erm, erythromycin resistance gene; bla, beta- lactamase gene, conferring resistance to ampicillin, expressed in E. coli. Restriction enzyme sites: H, HindIII; Sp, SphI; P, PstI; Bs, BspMI; S, SalI; X, XbaI; B, BamHI; Sm, SmaI; K, KpnI; Sa, SacI; Ss, SspI; E, EcoRI. (B) DNA sequence between XbaI and NheI sites (present in pKV111, pGF, and pGF-EM). RBS, putative ribosomal binding site. The putative gfp start codon is indicated in bold. Restriction sites for NheI and XbaI are underlined.
TABLE 1.
Primers used in this study
| Primer | Sequence | Accession no. and locationa | Restriction siteb |
|---|---|---|---|
| P1 | 5′GCAAGCTTCGTAAGAGAAAAGCGAAACGGATGT3′ | AF467001, 2823-2850 | HindIII |
| P2 | 5′TGCGCTAGCTGTGAAGCGAACTTTCTTCTTGT3′ | AF467001, 3516-3493 | NheI |
| P3 | 5′CGGGTACCAATCAAAGAGGCTGGGCTTC3′ | AF467001, 5040-5059 | KpnI |
| P4 | 5′AAGAATTCGCAAAATGTGGTCGCAGTGCCA3′ | AF467001, 5565-5544 | EcoRI |
| ermA | 5′GTCGGATCCATTAAGAAGGAGGGATTCGTCA3′ | M11180, 489-510 | BamHI |
| ermB | 5′GTTGGTACCGAATTATTTCCTCCCGTTAAAT3′ | M11180, 1453-1432 | KpnI |
Location coordinates correspond to the primer sequence portion shown in italics.
Restriction site corresponds to the underlined portion of the primer sequence.
Application of pGF-EM for gene replacement construction.
To test the applicability of pGF-EM, it was used to construct a mutant with a deletion in a gene, being studied in our laboratory, a gene which encodes the putative FtsH of L. monocytogenes. In other bacteria, FitsH has been shown to be a stress-induced protease with important adaptive physiology roles (7). The identification of this gene (ftsH) in L. monocytogenes and its detailed functional characterization by mutagenesis and gene expression studies will be described elsewhere.
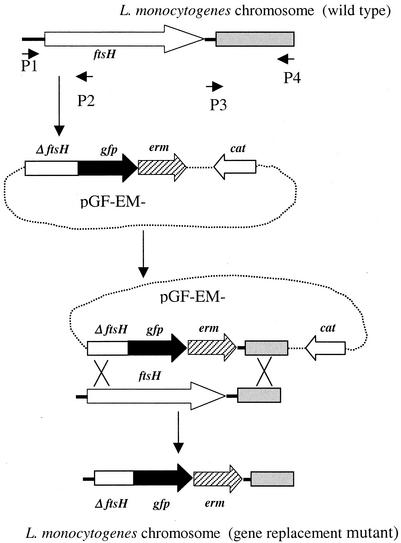
To construct an in-frame deletion in the putative ftsH, we used the steps outlined in Fig. 2. We first used primers P1 and P2 (Table 1 and Fig. 2) with L. monocytogenes strain 4b1 (10) DNA as a template to amplify a 694-bp fragment which includes 148 bp of the intergenic region immediately upstream of ftsH and the 5′ portion of ftsH (546 bp). The PCR product was digested by HindIII and NheI and ligated to HindIII- and NheI-digested pGF-EM to generate pGF-EM-FtsH1. We then used primers P3 and P4 (Table 1 and Fig. 2) to amplify a 501-bp fragment corresponding to the genomic region downstream of ftsH. The resulting PCR product was digested with KpnI and EcoRI and ligated to KpnI- and EcoRI-digested pGF-EM-FtsH1 to generate pGF-EM-FtsH2. The constructed deletion in pGF-EM-FtsH2 harbored the 5′ portion of ftsH fused in frame to gfp and lacked codons 181 to 687 of the ftsH coding sequence.
FIG. 2.
Application of pGF-EM in construction of gene replacement. A 5′ portion of ftsH was amplified with primers P1 and P2 and cloned into pGE-EM, yielding pGF-EM-FtsH1. ΔftsH indicates the truncated ftsH portion (open section in diagram). An additional genomic fragment (gray section in diagram) downstream of ftsH was amplified with primers P3 and P4 and cloned in pGF-EM-FtsH1, yielding pGF-EM-FtsH2, which was introduced into L. monocytogenes and used for construction of allelic exchange products. Primer positions and orientations are as indicated. Further details are provided in the text. Crosses indicate crossover events. Black arrows in the plasmid diagrams indicate gfp, cross-hatched arrows indicate the erythromycin resistance gene erm, and open arrows indicate the chloramphenicol resistance gene cat; arrows point in the direction of transcription.
For allelic exchange of this deletion into the genome of L. monocytogenes, the deletion-harboring plasmid pGF-EM-FtsH2 was transformed into E. coli S17-1 (12), which was then conjugated with L. monocytogenes strain 4b1 (streptomycin resistant) on agar plates (1). Transconjugants were selected on streptomycin (1,200 μg ml−1) and chloramphenicol (5 μg ml−1) at the permissive temperature (30°C). Integrants were selected at the restrictive temperature (42°C) on the basis of their chloramphenicol resistance and were subsequently grown repeatedly at the permissive temperature (30°C) to select for the second homologous recombination event, which would result in excision of the vector sequences and loss of chloramphenicol resistance. Of 192 erythromycin-resistant colonies screened for chloramphenicol resistance, 127 (66%) were found to be chloramphenicol sensitive. Of these, four were checked by PCR with primers P1 and ermB (Table 1) to confirm the presence of the deletion. All four were found to harbor the ftsH deletion. In addition, Southern blots with the entire ftsH as well as with the second (deleted) half of ftsH as probes confirmed that all four strains harbored the expected deletion (data not shown).
One-step selection for allele replacement mutants.
The construction of the ftsH deletion described above involved two major steps: first, selection for integration of plasmid containing the fusion construct into the bacterial chromosome, and second, repeated passage at the permissive temperature and screening for the excision of plasmid. The presence of erm on pGF-EM, however, suggests the possibility of directly selecting for the recombinants in a single step, without the need to first isolate integrants of the plasmid into the chromosome.
Following conjugative transfer of pGF-EM-FtsH2, which harbors the ftsH deletion into L. monocytogenes as described above, the culture was transferred five to seven times in the presence of erythromycin following overnight growth in stationary cultures at the restrictive temperature (42°C). During each transfer, the cultures were allowed to grow for 24 h in brain heart infusion broth (Difco) without shaking. Under these conditions we observed a substantial enrichment (90%) of the culture for erythromycin-resistant, chloramphenicol-sensitive recombinants. PCR analysis of four randomly chosen colonies (done as described above) confirmed that in these bacteria the wild-type ftsH gene was indeed replaced by the deleted version. The deletion was confirmed by Southern blotting, as described above (data not shown). Thus, allelic replacements can be obtained by positive selection and in a single step, obviating the need to first bring about the integration of the plasmid into the chromosome. Overall, mutants were constructed in a substantially shorter time and with higher efficiency (90 versus 66%) than by the two-step method.
The most attractive feature of pGF-EM is the presence of a drug resistance gene (erm) that allows direct selection for the allelic exchange products. In the absence of such selection, the desired products are often a small fraction of the chloramphenicol-sensitive population. In our experience with deletions of several genes in L. monocytogenes, yields were often ca. 2% or lower, and substantial time and expense were dedicated to screening derivatives that proved to harbor the wild-type sequence. The construction of pGF-EM is such that in the two-step method only the allelic exchange products would harbor the erm gene whereas the remainder (second homologous recombination products with the wild-type sequence) would be erythromycin sensitive. In the one-step process, the combination of restrictive temperature and erythromycin selects for double-crossover events that integrate the cloned deletion into the chromosome and result in the desired allelic exchange mutants.
An additional useful feature of pGF-EM is the presence of the promoterless gfp gene and multiple cloning sites, especially the NheI site mentioned earlier, which facilitates cloning in frame with gfp. Thus, if the promoter region of the gene that harbors the deletion is retained, the resulting allelic exchange is actually a “deletion-fusion” construct that can provide indications of the transcriptional level of the gene of interest. In addition, the construction of chromosomal translational fusions of the entire coding sequence of the gene of interest with gfp can be facilitated by pGF-EM. The usefulness of GFP fusions for genetic studies of L. monocytogenes has been demonstrated (5). The presence of GFP in constructs generated by pGF-EM can be monitored by fluorescence as well as by Western blotting analyses with commercially available antibodies (e.g., anti-GFP monoclonal antibody mAb11E5; QbioGene). The gfp gene used in these studies was originally derived from pQBI63 (Qbiogene) and is a red-shifted mutant in which amino acid residues Phe64, Ser65, and Ile168 of the GFP from Aequorea victoria were changed to Leu64, Cys65, and Thr168, respectively. This red-shifted GFP was chosen for these studies because of its reported stability, high resistance to photobleaching, and good expression in other bacterial systems (N. Ruby, personal communication). The presence of the gfp and erm genes in the constructs does not seem to affect cellular fitness under laboratory conditions. There were no noticeable differences between insertion mutants harboring these genes (obtained following the first crossover) and the parental wild-type strain in terms of colony size, cell shape, and growth rate (data not shown).
The recent availability of the genome sequence data of two strains of L. monocytogenes (6; http://www.tigr.org) will greatly facilitate the extensive functional analysis of the organism's genetic repertoire. The plasmid described here will be useful in such studies not only with L. monocytogenes but also with the closely related but nonpathogenic species Listeria innocua, whose genome has also been recently deciphered (6), and is expected to contribute to several studies of the virulence and adaptive physiology of the organism, currently pursued by ourselves and others. In addition, since the plasmid is a pKSV7 derivative, it can be used for genetic analysis of Bacillus subtilis, in which pKSV7 is known to replicate (13), and possibly other gram-positive bacteria as well.
Acknowledgments
We thank Ned Ruby and Karen Visick for pKV111 and Dan Portnoy for pKSV7 and pCON1.
Funding for this work was partially provided by the International Life Sciences Institute-North America and by USDA grant NRI 99-35201-8183.
REFERENCES
- 1.Behari, J., and P. Youngman. 1998. Regulation of hly expression in Listeria monocytogenes by carbon sources and pH occurs through separate mechanisms mediated by PrfA. Infect. Immun. 66:3635-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camilli, A., D. A. Portnoy, and P. Youngman. 1990. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J. Bacteriol. 172:3738-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camilli, A., L. G. Tilney, and D. A. Portnoy. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freitag, N. E., and K. E. Jacobs. 1999. Examination of Listeria monocytogenes intracellular gene expression by using the green fluorescent protein of Aequorea victoria. Infect. Immun. 67:1844-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, et al. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 7.Hlavacek, O., and L. Vachova. 2002. ATP-dependent proteinases in bacteria. Folia Microbiol. 47:203-212. [DOI] [PubMed] [Google Scholar]
- 8.Kathariou, S. 2000. Pathogenesis determinants of Listeria monocytogenes, p. 295-314. In J. W. Cary, J. E. Linz, and D. Bhatnagar (ed.), Microbial foodborne diseases, mechanisms of pathogenesis and toxin synthesis. Technomics Publ. Co., Inc., Lancaster, Pa.
- 9.Li, G. 2001. Molecular mechanisms underlying the involvement of the ltrB locus in cold tolerance of Listeria monocytogenes. PhD dissertation, University of Hawaii, Honolulu.
- 10.Promadej, N., F. Fiedler, P. Cossart, S. Dramsi, and S. Kathariou. 1999. Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel, serogroup-specific gene. J. Bacteriol. 181:418-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw, J. H., and D. B. Clewell. 1985. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 164:782-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 13.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 14.Stabb, E. V., K. L. Visick, D. S. Millikan, A. A. Corcoran, L. Gilson, S. V. Nyholm, M. McFall-Ngai, and E. G. Ruby. 2000. The Vibrio fischeri-Euprymna scolopes symbiosis: a model marine animal-bacteria interaction, p. 269-275. In N. Saxena (ed.), Recent advances in marine science and technology. PACON International, Honolulu, Hawaii.
- 15.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 4:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]