Abstract
Arsenate [As(V); HAsO42−] respiration by bacteria is poorly understood at the molecular level largely due to a paucity of genetically tractable organisms with this metabolic capability. We report here the isolation of a new As(V)-respiring strain (ANA-3) that is phylogenetically related to members of the genus Shewanella and that also provides a useful model system with which to explore the molecular basis of As(V) respiration. This gram-negative strain stoichiometrically couples the oxidation of lactate to acetate with the reduction of As(V) to arsenite [As(III); HAsO2]. The generation time and lactate molar growth yield (Ylactate) are 2.8 h and 10.0 g of cells mol of lactate−1, respectively, when it is grown anaerobically on lactate and As(V). ANA-3 uses a wide variety of terminal electron acceptors, including oxygen, soluble ferric iron, oxides of iron and manganese, nitrate, fumarate, the humic acid functional analog 2,6-anthraquinone disulfonate, and thiosulfate. ANA-3 also reduces As(V) to As(III) in the presence of oxygen and resists high concentrations of As(III) (up to 10 mM) when grown under either aerobic or anaerobic conditions. ANA-3 possesses an ars operon (arsDABC) that allows it to resist high levels of As(III); this operon also confers resistance to the As-sensitive strains Shewanella oneidensis MR-1 and Escherichia coli AW3110. When the gene encoding the As(III) efflux pump, arsB, is inactivated in ANA-3 by a polar mutation that also eliminates the expression of arsC, which encodes an As(V) reductase, the resulting As(III)-sensitive strain still respires As(V); however, the generation time and the Ylactate value are two- and threefold lower, respectively, than those of the wild type. These results suggest that ArsB and ArsC may be useful for As(V)-respiring bacteria in environments where As concentrations are high, but that neither is required for respiration.
The contamination of groundwaters and surface waters with arsenic (As) is a major concern to public health in countries such as Bangladesh, China, Taiwan, Argentina, Chile, and the United States (40). Elevated As concentrations typically derive from the weathering of As-bearing minerals and/or from geothermal sources (2, 58). It is now known that a variety of microorganisms, including members of the Eukarya, Archaea, and Bacteria, influence As geochemistry in many locales throughout the world by virtue of their metabolism (31, 35, 53). These metabolic processes include oxidation (49, 58), reduction (1, 13), and methylation reactions (5) that strongly affect (and in some cases, control) As speciation in the environment. One process that is particularly intriguing is microbial respiration of arsenate [As(V); HAsO4−2]. In the absence of oxygen, microorganisms can gain energy by coupling the oxidation of organic material to As(V) reduction, resulting in the production of the highly toxic As compound, arsenite [As(III); HAsO2]. As(V)-respiring organisms can affect water quality by catalyzing the mobilization of As(III) from sediments (1), as well as affect the biogeochemical cycles of other elements. For example, a significant proportion (∼14%) of organic carbon remineralization to CO2 within the hyper-saline waters of Mono Lake, Calif., has been linked to the activity of As(V)-respiring microorganisms (42).
To date, numerous phylogenetically diverse bacteria have been isolated that use As(V) as a terminal electron acceptor for respiratory growth, suggesting that this metabolic process may be ancient in origin (35, 53). As(V)-respiring organisms have been isolated from various sites including: a Superfund site contaminated with As (1), a seleniferous freshwater marsh in Nevada (41), an Australian goldfield (27), mud from a reed bed in Australia (28), a freshwater lake in Massachusetts (37), an alkaline hypersaline lake in California (6), geothermal pools within Yellowstone National Park (15, 19), an As-contaminated lake in Idaho (39), bovine rumen fluid, hamster feces, and termite hind guts (17). All of these As(V)-respiring strains are obligate anaerobes.
Until now, only three studies have investigated the molecular basis of As(V) respiration (23, 28, 35; D. K. Newman, C. W. Saltikov, E. Afkar, S. Tiwari, B. W. Kail, R. S. Oremland, F. M. M. Morel, and J. F. Stolz, unpublished data). Although the enzymology of this process is emerging, biochemical approaches alone will not be sufficient to determine how As(V)-respiration is regulated and/or functionally integrated with other cellular pathways that traffic in As. Of specific interest is the relationship between pathways that control As(V) respiration and those that control As resistance, given that an inescapable consequence of As(V) respiration is the buildup of toxic As(III). Arsenic detoxification by the products of the ars genes has been studied in great detail (reviewed in references 31 and 44), and the ars genes have been found in many organisms (50). The ars operon encodes a multisubunit As(III) efflux pump comprising a transmembrane oxyanion conducting channel, ArsB, that often associates with an ATPase subunit, ArsA. In addition, As(V) resistance is conferred by a small 16-kDa cytoplasmic As(V) reductase, ArsC, that reduces As(V) to As(III). Regulation of the ars operon is controlled by the As(III)-sensitive trans-acting repressor, ArsR, and the inducer-independent trans-acting repressor, ArsD. The ars operon functions to lower the intracellular As concentration, which permits survival in environments with high concentrations of As. Although it seems logical that the ars genes might be present and functional in As(V)-reducing bacteria, this has not been directly proven to date.
As an entry into exploring the relationship between As(V) respiration and As resistance at the molecular level, we report here the isolation and characterization of an As(V)-respiring facultative anaerobe. The focus of this study concerns whether the arsB and arsC genes are required for As(V) respiration.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in the present study are listed in Table 1. E. coli strains were grown in Luria-Bertani (LB) Miller medium (Difco). The growth conditions for various Shewanella strains are described below.
TABLE 1.
Strains and plasmids used in this study
| Bacterial strain or plasmid | Genotype or markers and characteristics and usesa | Source or reference |
|---|---|---|
| E. coli | ||
| DH10β | Host for E. coli cloning; F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 Δ(codB-lac1)3 deoR recA1 endA1 araD139 Δ(ara-leu)7697 galU galK λ−rpsL (Strr) | Life Technology |
| UQ950 | E. coli DH5α λpir host for cloning; F− Δ(argF-lac)169 φ80dlacZ58(ΔM15) glnV44(AS) rfbD1 gyrA96 (Nalr) recA1 endA1 spoT1 thi-1 hsdR17 deoR λpir+ | D. Lies, Caltech |
| β2155 | Donor for bacterial conjugation; F′ lacZ58(ΔM15) lac1q traD36 proA+B+/λ−(?) thrB1004 pro thi rpsL (Strr) hsdS lacZ58(ΔM15) ΔdapA::erm (Ermr) pir+::RP4-2-Tc::Mu (Kmr) | 12 |
| AW3110 | W3110 Δars::cam | 9 |
| Shewanella spp. | ||
| ANA-3 | Isolated from an As-treated wooden pier piling in a brackish estuary (Eel Pond, Woods Hole, Mass.), contains arsDABC, respires on As(V) resistant to >5 mM arsenite | This study |
| ARSB1 | arsB mutant derived from ANA-3; arsB::Kan(EZ::TN<KAN-2>) | This study |
| S. oneidensis MR-1 | Manganese-reducing strain from Oneida Lake, N.Y., sediments, type strain | 32 |
| S. algae OK-1 | ATCC 51192, type strain, isolated from red algae | 51 |
| S. amazonensis SB2B | Isolated from Amazon water, type strain | 55 |
| S. baltica 63 | NCTC 10735, Japan, isolated from oil brine, type strain | 60 |
| Shewanella sp. strain MR-4 | Isolated from the Black Sea water column, type strain | 33 |
| S. pealeana ANG-SQ1 | Isolated from accessory nidamental gland of a squid, psychrotolerant, type strain | 25 |
| S. frigidimarina ACAM 591 | Isolated from Antarctic Sea ice, type strain | 7 |
| S. putrefaciens 95 | ATCC 8071, isolated from spoiled butter with surface taint, type strain | 46 |
| S. woodyi MS32 | Isolated from seawater detritus, Alboran Sea, type strain | 29 |
| 184 | ATCC 8073, isolated from spoiled butter with surface taint | 46 |
| CL 256/73 | NCTC 12093, isolate from human cerebrospinal fluid, type strain | 18 |
| CN32 | Isolated from anaerobic subsurface core sample, New Mexico | 14, 59 |
| Plasmids and vectors | ||
| pLAFR5 | 21.5-kb broad-host-range cosmid cloning vector; Tcr, lacZ | 22 |
| EZ::TN<KAN-2> | 1.2-kb transposon used for in vitro mutagenesis | Epicentre |
| pSALT1 | pLAFR5-based 45-kb As(III)r cosmid from ANA-3 genomic DNA library; contains arsDABC, Tcr, confers resistance to As(III) and As(V) | This study |
| pSALT1-B10 | As(III)-sensitive pSALT1; arsB::kan(EZ::TN<KAN-2>), Kmr Tcr | This study |
| pSMV8 | 9.1-kb mobilizable suicide vector; oriR6K mobRP4, Gmr | D. Lies, Caltech |
| parsB::kan | pSMV8 with Kmr Tn-interrupted arsB gene from pSALT1-B10 | This study |
Strr, streptomycin resistance; Nalr, nalidixic acid resistance; Tcr, tetracycline resistance; Gmr, gentamicin resistance.
Isolation of ANA-3.
ANA-3 was isolated in 1998 from an As-treated wooden pier located in a brackish estuary (Eel Pond [Woods Hole, Mass.]). Arsenate reducers were enriched by placing a 1-by-3-cm strip of the wood sample into a defined minimal medium (pH 7.2; NaHCO3 [1.9 g/liter], KH2PO4 [0.2 g/liter], NH4Cl [0.25 g/liter], KCl [0.5 g/liter], CaCl2 · 2H2O [0.1 g/liter], NaCl [1.0 g/liter], MgCl2 · 6H2O [0.4 g/liter], 1 ml of SL10 trace elements/liter, and 1 ml of vitamin solution [37]/liter) amended with lactate (10 mM), arsenate (5 mM), and sulfide (1 mM) and grown under anaerobic conditions by using the Hungate technique (30). As(V) reduction was visually observed by the formation of the yellow mineral As2S3 from the reaction of As(III) and S2− (36). The sample was subcultured into fresh medium once As(V) reduction had occurred. Subculturing was repeated three times before finally plating on LB agar aerobically. A single colony was inoculated back into the defined minimal medium, and after several days a yellow precipitate developed. This isolate was named strain ANA-3.
ANA-3 was routinely grown in either LB medium or a minimal medium (pH 7.2) containing the following: 0.225 g of K2HPO4/liter, 0.225 KH2PO4/liter, 0.46 g of NaCl, 0.225 g of (NH4)2SO4/liter, g of 0.117 MgSO4 · 7H2O/liter, 2.24 g of sodium lactate/liter, 10 mM Na2HAsO4 or 20 mM NaNO3, 4.2 g of NaHCO3/liter, SL10 trace elements (1 ml), and vitamins (10 ml) (41). The medium was boiled under a stream of N2-CO2 (4:1), dispensed anaerobically into bottles flushed with the mixed gas, and autoclave sterilized. Sterile anaerobic sodium bicarbonate solution was added by injection after the tubes had cooled.
Phylogenetic analysis.
Ten nanograms of purified genomic DNA (21) from liquid cultures was used as the template for PCR amplification. Universal 16S ribosomal DNA (rDNA) primers (Bact 11 and 1492) were used to amplify the 1.5-kb 16S rDNA fragment according to established protocols (46). Amplicons were sequenced directly after purification on Qiagen columns (Qiagen, Valencia, Calif.). The PCR product was sequenced by using the dideoxy chain termination method with the Sequenase DNA sequencing kit (U.S. Biochemical Corp., Cleveland, Ohio) and an ABI 373A automated sequencer (Perkin-Elmer Corp., Foster City, Calif.). The phylogenetic relationships of organisms covered in the present study were determined by comparison of individual 16S rDNA sequences to other existing sequences in the public database (GenBank [http://www.ncbi.nlm.nih.gov/]). Sequence alignments were obtained online from the Ribosomal Database Project (RDP [http://rdp.cme.msu.edu/html/]). Evolutionary trees were constructed by using PAUP∗, version 4.0b10 (54), with the optimality criterion set to distance (minimum evolution). The Kimura two-parameter model was used to estimate pairwise distances. Phylogenetic trees were inferred by neighbor-joining and tree bisection-reconnection branch-swapping algorithms. After a heuristic search was performed, bootstrap analysis was done with 1,000 replications. The final tree was assembled in Dendromaker (http://www.cib.nig.ac.jp/dda/timanish/dendromaker/home.html) and with Adobe Illustrator (Adobe Systems, Inc.). The GenBank nucleotide accession number for strain ANA-3 is AF136392.
Electron donors and acceptors.
Various electron donors listed in Table 2 were screened for the ability to support growth on As(V) as the sole terminal electron acceptor. Lactate was used as the electron donor and sole carbon source for testing the electron acceptors listed in Table 2. Fumarate and As(V) reduction were determined by high-pressure liquid chromatography analysis (described below). The humic acid functional analog 2,6-anthraquinone disulfonate (AQDS) reduction was determined spectrophotometrically by monitoring absorption at 450 nm (38). Thiosulfate reduction was confirmed by colorimetric detection of hydrogen sulfide by the methylene blue method (10). Nitrate reduction was determined by monitoring the formation of nitrite by colorimetry with Griess reagent (sulfanilamide and N-naphthylethylenediamine in HCl) (52). Mineral reduction was monitored by observing the change in color of the iron oxide (from rust to dark brown) or the transformation of the manganese oxide from black to white. Minerals were prepared as described by Lovley and Phillips (26). Growth was inferred either by monitoring increases in CFU (per milliliter) or by visually inspecting increases in turbidity compared to controls without electron donor or acceptor.
TABLE 2.
Growth characteristics of Shewanella sp. strain ANA-3
| Electron donor or fermentation substrate (concn [mM]) | Physiological parameter
|
||
|---|---|---|---|
| Growtha | TEAb (concn [mM]) | Reduction of TEA | |
| Electron donorsc | Oxygen | + | |
| Acetate (10) | − (+) | Arsenate (10) | + |
| Citrate (10) | − | Fumarate (20) | + |
| Ethanol (10) | − | Selenate (5) | − |
| Formate (10) | − | Nitrate (5) | + |
| Fumarate (10) | − | MnO2 (20) | + |
| Glucose (10) | − | Fe(OH)3 (50) | + |
| Glycerol (5) | − | AQDS (5) | + |
| Lactate (10) | + (+) | Sulfate (10) | − |
| Malate (10) | − | Thiosulfate (5) | + |
| Propionate (10) | − | Sulfite (5) | − |
| Pyruvate (10) | + (+) | DMSO (10) | − |
| Succinate (10) | − | ||
| Fermentation | |||
| Lactate (10) | − | ||
| Pyruvate (10) | − | ||
+, Growth; −, no growth; parentheses indicate aerobic growth; no parenthesis indicates that aerobic growth on the substrate was not tested.
20 mM lactate was used as the electron donor. TEA, terminal electron acceptor; DMSO, dimethyl sulfoxide.
5 mM arsenate was used as the terminal electron acceptor.
Construction of the genomic library.
Genomic DNA was prepared according to standard methods (3), partially digested with Sau3AI, and size fractionated on a 10 to 40% sucrose gradient (48). DNA fragments of 20 to 30 kb were ligated to the cosmid vector pLAFR5 previously digested with ScaI/BamHI. After being packaged into phage by using Gigapack Gold XL (Stratagene, La Jolla, Calif.), the cosmid library was transduced into Escherichia coli β2155 (12), mated en masse into E. coli AW3110 (9), and plated onto LB agar containing tetracycline (15 μg/ml) and 5 mM sodium meta-arsenite (Sigma). Cosmid DNA from an As-resistant clone was isolated and transferred back into As(III)-sensitive AW3110 by electroporation to confirm that it conferred the As(III)-resistant phenotype. This cosmid was designated pSALT1. The region of pSALT1 that conferred As(III) resistance was mapped by in vitro transposon mutagenesis by using the EZ::TN<KAN-2> system (Epicentre) according to the manufacturer's instructions. Randomly mutagenized pSALT1 was electroporated into AW3110, and clones were screened for sensitivity to 5 mM As(III). After the flanking region of the transposon of an As(III)-sensitive clone (pSALT1-B10) was sequenced by using EZ::TN<KAN-2> primers (supplied by Epicentre), the nucleotide sequence was analyzed by BLAST searching (http://www.ncbi.nlm.nih.gov/BLAST/). A 5-kb region was sequenced by primer walking upstream and downstream of the initial sequence. The nucleotide sequence was assembled by using AssemblyLIGN (Accelrys) and submitted to the National Center for Biotechnology Information (accession no. AY161137).
Construction of arsB insertion mutation.
An arsB gene replacement mutant was constructed from ANA-3 by exchanging the wild-type allele for the mutant allele of pSALT1-B10. PCR was used to generate a fragment with SpeI ends (underlined) from the mutated cosmid pSALT1-B10 by using the primers TNARSBF (GGACTAGTATGGGACGATTGATTAGGATGG) and TNARSBR (GGACTAGTGGTCGTGGCCGTTTACTCTTTA). The resulting 2.8-kb fragment contained the 1.2-kb kanamycin-resistant (Kmr) transposon flanked by ∼800 bp of arsB on either side and was cloned into the SpeI site of the mobilizable suicide vector pSMV8 to generate parsB::kan. The mutation was introduced into ANA-3 by conjugation from the E. coli donor strain β2155 containing parsB::kan. Overnight cultures of the donor (800 μl) and ANA-3 (200 μl) were centrifuged together, resuspended in ∼40 μl, and spotted onto LB agar containing 300 μM diaminopimelic acid. The mating reaction was incubated at 30°C for 6 h prior to plating onto LB medium plus kanamycin (50 μg/ml) without diaminopimelic acid. After overnight incubation at 30°C, 12 Kmr colonies were picked and tested for sensitivity to gentamicin (to indicate loss of pSMV8) and then analyzed for recombination of the mutant allele by PCR with primers TNARSBF and TNARSBR.
RT-PCR analysis.
Overnight cultures of ANA-3 and ARSB1 grown in LB medium were diluted 1/25 into LB medium amended with 1 mM As(V). After incubation at 30°C for 4 h, 1 ml was used to isolate total RNA by using the Trizol reagent (Invitrogen). Crude RNA samples were DNase treated and cleaned up by using the Qiagen RNeasy Mini kit. Reverse transcription (RT) was performed with primers 16S-1492-R1 (GGTTACCTTGTTACGACTT), ARSA-R1 (GGCTTAATCGTTCAATACCAAT), and ARSC-R1 (TCACTACTTCACCGTCTTCCTT) and 1 μg of DNase-treated RNA. Control reactions consisted of (i) primer without RT and (ii) RT without primer. RT reactions were diluted 1/50 into sterile nuclease-free water, followed by PCR analysis with the corresponding reverse primers used in the RT reactions and the following forward primers: 16S-8-F1 (AGAGTTTGATCCTGGCTCAG), ARSA-F1 (GCTAGAAGAGGATTTACGCTCA), or ARSC-F1 (CCAACCATTATCCTCTACCTTG). PCR products were analyzed on 1% agarose gels.
Arsenate respiration experiments.
Overnight cultures grown anaerobically on 20 mM lactate and 20 mM fumarate were used as the inocula for experiments to check for respiratory growth on 10 mM As(V). Cultures were centrifuged and rinsed twice in anaerobic minimal medium [without lactate and As(V)] and resuspended at ∼108 cells/ml. Washed cells were inoculated into 100 ml of low-phosphate (∼0.3 mM) minimal medium amended with arsenate (10 mM) and lactate (20 mM) at ∼106 cells/ml and incubated anaerobically at 30°C without shaking. Control experiments with or without As(V) and/or lactate were also done to determine whether ANA-3 could grow in the absence of either a terminal electron acceptor or electron donor. Cultures were sampled periodically and analyzed for cell density by staining formaldehyde-fixed cells with 1 μg DAPI (4′,6′-diamidino-2-phenylindole)/ml, followed by filtration onto polycarbonate Nuclepore (Millipore Corp.) membranes (0.2 μm [pore size]). Stained cells were enumerated by epifluorescence microscopy on a Zeiss Axioplan (Carl Ziess MicroImaging, Inc.). Arsenic compounds [As(V) and As(III)], lactate, and acetate were quantified by high-pressure liquid chromatography (Waters) by using a Hamilton PRP-X300 column in series with a Bio-Rad Aminex HPX-87H column heated to 50°C. A mobile phase of phosphoric acid (30 mM) was set to 0.7 ml/min. Compounds were detected by UV at 210 nm.
Other Shewanella species were tested for the ability to respire As(V) by inoculating ∼106 cells/ml into anaerobic LML medium (4) containing 5 mM As(V) as the electron acceptor and lactate as the carbon source and electron donor. Cultures were sampled before and after 24 h of incubation, and As(V) was measured by using the molybdenum blue assay (20).
Resistance to As(III).
A microtiter plate assay was developed to determine aerobic As(III) sensitivity for various strains listed in Table 1. Overnight LB medium-grown cultures were diluted 100-fold into fresh LB medium amended with increasing As(III) concentrations. A total of 150 μl of each arsenic concentration was pipetted in quadruplicate into a 96-well microtiter dish and then incubated at 30°C and at 100 rpm for 24 h. Growth was monitored by measuring the optical density at 630 nm (OD630) before and after the incubation period in a Dynex Opsys microplate reader (Dynex Technologies).
Anaerobic As(III) resistance in ANA-3 and the arsB mutant of ANA-3 (ARSB1) was tested by inoculating 1/100 of anaerobic starter cultures grown in 20 mM lactate and 20 mM fumarate into anaerobic Hungate tubes containing minimal medium supplemented with lactate (20 mM), fumarate (20 mM), and increasing As(III) concentrations. The OD600 was monitored periodically for 68 h. The maximum OD values reached during the incubation period were used for determining the resistance profiles on increasing As(III) concentrations.
RESULTS
Enrichment and isolation.
By targeting the tidal interface of an old marine pier piling whose wood had been treated with an As-based preservative, we hypothesized that we would be able to enrich for a facultative anaerobe that could respire As(V). Our goal was to isolate an organism that we could develop into a model system for studying As(V) respiration and arsenic resistance. After several passages of the enrichment culture, followed by plating on LB agar, only coral-pink colonies were observed on the plates, and one was inoculated into anaerobic As(V) medium. Growth and As(V) reduction were observed within several days. This rod-shaped, 0.5-by-2.5-μm strain was designated ANA-3.
16S rDNA phylogeny.
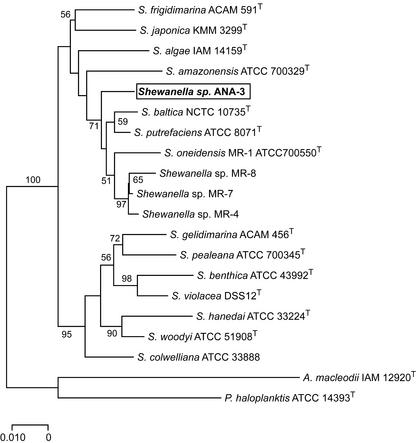
Phylogenetic analyses of the 16S rDNA sequence demonstrated that ANA-3 belongs to the Proteobacteria, gamma subdivision, genus Shewanella. Similarities among the 16S rDNA nucleotide sequences between ANA-3 and other Shewanella species in the RDP database are between 93 and 98%. A sequence variation of 1.7% was found between ANA-3 and S. putrefaciens ATCC 8071, and it was 2.8% between ANA-3 and S. oneidensis strain MR-1 (whose genome has been completely sequenced [16]). Of the strains we included in our analysis, we noticed the greatest sequence variation (6.3%) between ANA-3 and S. hanedai. A phylogenetic tree of the 16S rDNA sequences of strains from various sources (56) is shown in Fig. 1. ANA-3 is most closely related to S. putrefaciens based on the percent 16S rDNA similarity but does not cluster tightly with S. putrefaciens in the phylogenetic tree.
FIG. 1.
Phylogenetic relationships among 16S rDNA sequences from Shewanella strains. Shewanella sp. strain ANA-3 is boxed and in boldface type. The phylogenetic tree was constructed according to the distance criterion. The scale represents the number of substitutions per site. The percentage of 1,000 bootstrap replicates that supported the branching order is shown near the relevant nodes. Nodes without bootstrap values occurred <50%. Outgroups included Alteromonas macleodii (X82145) and Pseudoalteromonas haloplanktis (X67024). GenBank accession numbers for Shewanella species are given parenthetically as follows: S. figidimarina (U85903), S. japonica (Af145921), S. algae (U91546), S. amazonensis (AF005248), Shewanella sp. strain ANA-3 (AF136392), S. baltica (AJ000214), S. putrefaciens (U91550), S. oneidensis (AF005251), Shewanella sp. strain MR-8 (AF005254), Shewanella sp. strain MR-7 (AF005253), Shewanella sp. strain MR-4 (AF005252), S. gelidimarina (U85907), S. pealeana (AF011335), S. benthica (X82131), S. violacea (D21225), S. hanedai (U91590), S. woodyi (AF003549), and S. colwelliana (AF170794).
Arsenate respiration.
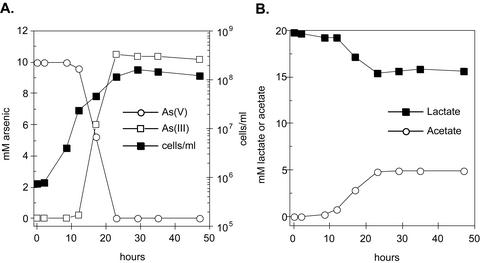
To confirm that ANA-3 was capable of respiring As(V), we characterized the growth of ANA-3 when As(V) served as the sole terminal electron acceptor. Figure 2A shows a time course for As(V) respiration and growth. Cell density increased by several logs to a maximum of 1.6 × 108 cells/ml. Controls without either electron donor (lactate) or acceptor [As(V)] exhibited neither As(V) reduction nor cell growth (data not shown). The early stationary phase was reached by ∼23 h. The generation time for ANA-3 was ∼2.8 h. After 23 h the initial 10 mM concentration of As(V) was completely reduced to 10 mM As(III). Concurrently, 4.4 mM of lactate was oxidized to acetate (Fig. 2B). We observed a lactate molar growth yield (Ylactate) of 10.2 g of cells/mol of lactate with ANA-3, assuming a cell dry weight of 2.8 × 10−13 g/cell (34). The oxidation of lactate and reduction of As(V) represents close to a 2:1 stoichiometric conversion of As(V) to As(III) and lactate to acetate as expected for the following reaction:
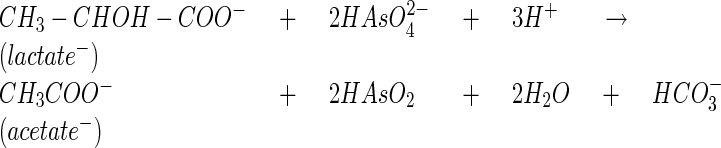 |
where ΔG° = −287.6 kJ/mol of lactate (−71.7 kJ/mol electron).
FIG. 2.
(A) Respiratory arsenate reduction and growth of Shewanella sp. strain ANA-3 on lactate as the electron donor. (B) Oxidation of lactate and accumulation of acetate during respiration on arsenate. Data are representative of triplicate cultures.
In comparison, we tested the other Shewanella species listed in Table 1 for the ability to respire As(V), but none of them were able to do so. Arsenate thus does not appear to be a common electron acceptor for Shewanella species.
Other growth characteristics.
ANA-3 respired on a variety of electron acceptors, including metal oxides of iron and manganese (Table 2). Among the carbon sources tested, lactate and pyruvate were the only electron donors that could support growth on As(V) (Table 2). Fermentation was not observed with either of these substrates, although ANA-3 was capable of metabolizing cysteine, evolving H2S (data not shown). We observed the formation of As2S3 in anaerobic As(V)-reducing cultures of ANA-3 when cysteine was included in the medium as a reducing agent. The precipitation of As2S3 by As(V) reducing microorganisms has been described elsewhere (36). ANA-3 completely reduced As(V) when grown aerobically in LB medium supplemented with 5 mM As(V) and could grow in the presence of 10 mM As(III).
Identification of the ANA-3 ars operon.
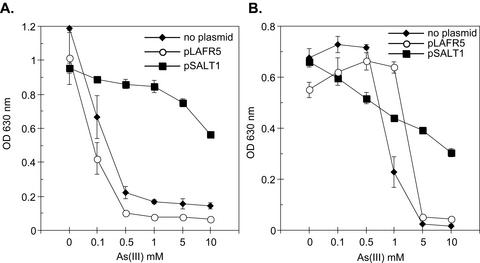
When ANA-3 was grown aerobically in LB medium, cell densities of ∼5 × 109 cells/ml were reached in overnight cultures. Similar cell densities were also observed in aerobically incubated LB medium-grown cultures supplemented with 5 mM As(III) or As(V). Given ANA-3's ability to resist the toxicity of As(V) and As(III) when grown in LB medium, we hypothesized that it might contain an ars operon. To test this, we identified a region of DNA from an ANA-3 genomic library that conferred high-level resistance to As(III) on other bacteria. Resistance to As(III) up to 10 mM was observed when the cosmid (pSALT1) containing this region was transformed into As(III)-sensitive strains of E. coli AW3110 (Fig. 3A) and S. oneidensis MR-1 (Fig. 3B). The arsB gene was shown to be essential for As(III) resistance. The ars deletion E. coli strain AW3110 harboring the mutagenized cosmid pSALT1-B10 no longer grew on LB agar plates containing 5 mM As(III). The genes on pSALT1 conferred As(III) resistance under aerobic conditions but were not sufficient to confer the ability to respire As(V), however, since pSALT1 was unable to promote growth on As(V) in addition to As(V) reduction when transformed into S. oneidensis strain MR-1 (data not shown).
FIG. 3.
The Asr cosmid pSALT1 confers As(III) resistance to E. coli AW3110 (A) and S. oneidensis MR-1 (B). Strains were grown aerobically in LB medium with the specified As(III) concentrations. Tetracycline was added at 15 μg/ml to strains harboring pSALT1 or the cosmid vector pLAFR5. The initial OD630 was <0.05 on average in all experiments. Values and error bars represent the averages and standard deviations of quadruplicate samples, respectively, after 24 h of incubation.
Molecular analysis of pSALT1 revealed the presence of four genes, arsDABC (Fig. 4) but no arsR homolog within 5 kb upstream of arsD and 1 kb downstream of arsC. The lack of an arsR gene upstream of arsD was intriguing, since ArsR is a repressor for the expression of the ars operon and the arsR gene is commonly found immediately upstream of the arsDABC gene cluster (44). The arsR gene in ANA-3 may be distantly located from the arsDABC cluster or it is possible that this ars operon may be regulated in a different way. The ArsD, ArsA, ArsB, and ArsC of ANA-3 are predicted to be similar to those found on the E. coli plasmid R773 (Table 3) but only exhibit low amino acid sequence similarity to homologs found in the S. oneidensis MR-1 genome (Table 3). BLAST searching the GenBank database with the putative arsenic resistance proteins in the S. oneidensis MR-1 genome suggests that their closest relatives are found in Pseudomonas aeruginosa PAO1 (e.g., ArsR and ArsCs) and Pyrococcus furiosus DSM 3638 (e.g., ACR3) (Table 4). No ArsB-like homologs were found in the S. oneidensis MR-1 genomic database.
FIG. 4.
(A) Map of the ANA-3 ars operon and location of the arsB mutation in ARSB1. The mutation in arsB with the EZ::TN<KAN-2> transposon is indicated by the black 1.2-kb size box. Numbers under the genes indicate the percent identity (similarity) to the corresponding R773 ars operon homolog. (B) Gel picture showing the results of PCR with primers TNARSBF and TNARSBR with genomic DNA of the ARSB1 strain (lane 2), pSALT1-B10 cosmid (lane 3), wild-type ANA-3 (lane 4), and a reagent negative control (lane 5). Lane 1 contains a 1-kb ladder. (C) RT-PCR analysis for the expression of arsC, arsA, and 16S rDNA genes after ANA-3 and ARSB1 were grown for 4 h in the presence of 1 mM As(V). Lanes 1 to 3 and lanes 4 to 6 correspond to ANA-3 and ARSB1, respectively. Lanes also correspond to RT with primer (lanes 1 and 4), no RT added (lanes 2 and 5), RT without primer (lanes 3 and 6), ANA-3 genomic DNA (PCR-positive control) (lane 7), water (PCR-negative control) (lane 8), and DNA ladders in kilobases (lane 9).
TABLE 3.
Percent amino acid identity and similarity between the ars homologs of Shewanella sp. ANA-3 and those of S. oneidensis MR-1 and E. coli R773 arsenic resistance plasmid
TABLE 4.
Percent amino acid identity and similarity of the predicted arsenic resistance proteins of S. oneidensis MR-1 to the closest known proteins in the GenBank database
| Putative protein | TIGR accession no. | Closest match in GenBank |
|---|---|---|
| ArsR | SO0532 | 58% identity and 70% similarity to Pseudomonas aeruginosa PAO1 ArsR; NP_250967 |
| ACR3 | SO0534 | 49% identity and 62% similarity to Pyrococcus furiosus DSM 3638 ACR3; NP_578281 |
| ArsC | SO0533 | 50% identity and 62% similarity to Pseudomonas aeruginosa PAO1 ArsC; NP_250969 |
| ArsC | SO2871 | 58% identity and 74% similarity to Pseudomonas aeruginosa PAO1 ArsC; NP_249641 |
ANA-3 can be maintained on LB medium for multiple generations in the absence of As selection without losing its arsenic resistance. Because the cosmid library was generated from total genomic DNA and electrophoresis of the genomic DNA on a 0.7% agarose gel did not show any distinguishable plasmid bands, this suggests that the ars genes are either chromosomally encoded or on a highly stable megaplasmid. In addition, Southern blot analysis of ANA-3 genomic DNA (hybridized with an ANA-3 arsB) probe detected the presence of only one copy of arsB.
Mutagenesis of arsB and the effects on As(V) respiration and As(III) resistance.
Although the ars system located on pSALT1 is not sufficient to confer respiratory As(V) reduction in S. oneidensis MR-1, it might still be necessary for growth on As(V), especially if high concentrations of As(III) are generated inside the cytoplasm. Therefore, we constructed a mutation in the arsB homolog in ANA-3 to determine whether arsB is also required for respiratory growth on As(V). Figure 4A shows the position of the transposon insertion in arsB introduced into ANA-3 to generate the strain ARSB1. ARSB1 was unable to reduce As(V) to As(III) under aerobic conditions (data not shown). We suspected that the mutation in arsB was polar to the downstream gene arsC, predicted to encode a small 17-kDa cytosolic As(V) reductase. To test this, we used RT-PCR to assay for the presence of arsC-specific message in RNA extracted from cells grown in the presence of 1 mM As(V). There was no detectable amount of arsC message in the arsB mutant strain ARSB1, unlike in wild-type ANA-3 grown under the same conditions (Fig. 4C). Controls for expression of the arsA and 16S rDNA genes were positive in both ANA-3 and ARSB1, confirming that the absence of arsC-message was due to a polar affect of the arsB mutation.
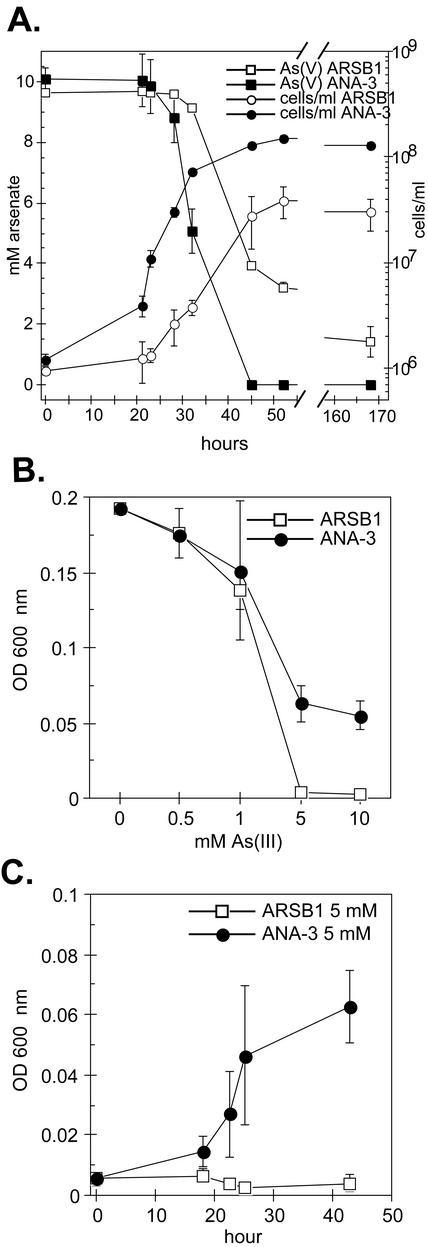
When we tested ARSB1 and ANA-3 for their ability to respire on As(V) in low-phosphate medium (∼0.3 mM Pi), a significant difference in the phenotype of ARSB1 was observed compared to the wild-type ANA-3 (Fig. 5A). The generation time and Ylactate value for ARSB1 were 5.3 h and 3.6 g of cells/mol, respectively, ∼2-fold longer and ∼3-fold less than for the wild type. The cell density of ARSB1 reached a maximum of 3.8 × 107 cells/ml, ca. 75% lower than wild-type ANA-3, when 7 mM of As(V) had been reduced to As(III). Lower phosphate concentrations did not appear to affect the growth of wild-type ANA-3 on As(V), evident in the Ylactate (9.9 g of cells/mol) and generation time (2.8 h), which are similar to the values obtained when ANA-3 is grown in higher-phosphate medium (Fig. 2). When ARSB1 and ANA-3 were grown in higher-phosphate medium (∼3 mM) and 5 mM As(V), no differences in As(V) respiration rates or growth rates were observed (data not shown).
FIG. 5.
Anaerobic As(V) respiration with lactate as the sole carbon source and electron donor (A), resistance profile to increasing As(III) concentrations (B), and time course for growth in 5 mM As(III) (C) for ANA-3 and ARSB1. In panels B and C, both strains were grown anaerobically on lactate and fumarate with As(III) added at the specified concentrations, and the initial OD600 values were similar to that of the blank medium (i.e., 0.005). The values and error bars in all three panels represent the averages and ranges of duplicate samples, respectively.
When ARSB1 and wild-type ANA-3 were grown anaerobically on lactate and fumarate in the presence of increasing As(III) (Fig. 5B and C), the growth of ARSB1 was completely inhibited in 5 mM As(III) (Fig. 5C). However, ARSB1 could grow in 1 mM As(III) similar to the wild type (Fig. 5B). At 2.5 mM As(III) concentrations, the growth of the other Shewanella species listed in Table 1 was also inhibited.
DISCUSSION
The primary objective of the present study was to isolate and characterize a bacterium that would be useful for dissecting the molecular basis of respiratory As(V) reduction. Many As(V) reducers have been described physiologically, yet little progress has been made in identifying the gene(s) involved in As(V) respiration and the biochemical details of their protein products. This stems in part from the fact that the previous isolates are all strict anaerobes that have short lifetimes on the bench. Although it is possible to successfully establish genetic systems and do biochemical work in strict anaerobes (e.g., Geobacter metallireducans and Desulfovibrio desulfuricans) (11, 57), because ANA-3 can grow overnight aerobically on LB medium, exhibits robust anaerobic growth in minimal medium, and plates easily, it provides an attractive model system for molecular studies. Moreover, many of the existing genetic tools developed for S. oneidensis strain MR-1 can be adapted for use in ANA-3, and because ANA-3 is so closely related to S. oneidensis MR-1 the recent completion of the genome sequence for S. oneidensis MR-1 (16) aids its genetic analysis. To demonstrate the utility of ANA-3 as a model genetic system for studying As(V)-respiration, we selectively disrupted genes involved in arsenic resistance (the ars genes) and studied their impact on As(V) respiration.
An initial hint that ANA-3 contained an ars operon came from its robust growth on high concentrations of As(V). When grown on 5 mM As(V) with lactate in excess, ANA-3's molar growth yield on lactate (∼10 g of cells/mol of lactate) was twice that previously reported for the As(V)-respiring strains Sulfurospirillum barnesii SES-3 (5.3 g of cells/mol of lactate) and Desulfotomaculum auripigmentum OREX-4 (5.6 g of cells/mol of lactate) (24, 37). At 10 mM As(V), ANA-3 grew as well as it did at 5 mM As(V), whereas the growth of SES-3 and OREX-4 was significantly impaired. This suggested that ANA-3's resistance to high concentrations of arsenic might be due to the presence of a high-level ars detoxification system, including an arsA gene, since expression of this system confers resistance to high concentrations of As (31). Additional physiological evidence in support of this was provided by the fact that ANA-3 could reduce As(V) when grown aerobically and exhibited resistance to 10 mM As(III).
Several new ars operons have recently been identified by using the As(III)-sensitive E. coli ars deletion strain AW3110. For example, the ars operons of Pseudomonas fluorescens MSP3 and Thiobacillus ferrooxidans were shown to confer resistance up to 2 mM As(III) when expressed in AW3110 (8, 43). To determine whether ANA-3 contained an ars operon as predicted, we tested whether DNA from ANA-3 could confer As(III) resistance to AW3110 and to the As(III)-sensitive S. oneidensis strain MR-1. Positive identification and sequencing of a cosmid that functionally rescued strains AW3110 and MR-1 in the presence of high As(III) concentrations confirmed the presence of four open reading frames with striking homology to arsD, arsA, arsB, and arsC of the E. coli R773 (Table 3). Although many genomes of sequenced microorganisms possess an As(III) efflux pump [namely, a homolog of the ArsB or ACR3, both encoding membrane-bound As(III) efflux channels], high-level resistance normally requires the addition of a large 63-kDa ATPase subunit, ArsA (44). The presence of an arsA homolog in the ANA-3 ars operon thus may explain ANA-3's resistance to 10 mM As(III).
ANA-3 is the first respiratory As(V) reducer shown to have an ars gene cluster, but identification of the ars genes in an As(V)-respiring organism is not surprising. Although Macy et al. (28) could not detect ars genes in the As(V) reducer Desulfomicrobium strain Ben-RB when using an E. coli R773 arsC probe, this could be due to sequence differences between the Ben-RB ars operon and the E. coli R773 operon. Indeed, this would be expected given the sequence diversity of the ars operon among different genera of As-resistant bacteria (47) and the fact that Ben-RB is phylogenetically distant from E. coli, whereas ANA-3 is more closely related. As more genetic work on different As(V)-respiring strains is performed, it will be interesting to see whether ANA-3 is exceptional with respect to its possession of an arsA-containing ars operon or representative of many As(V) respirers. How ANA-3 acquired this operon is an intriguing open question.
Knowing that ANA-3 possessed an ars operon, our next question became whether the ars detoxification system was required by ANA-3 to respire As(V). This was interesting for two reasons. First, we were curious as to whether the ars detoxification system conferred an advantage to cells respiring As(V). Second, we wanted to determine whether ArsC could account for As(V) reduction under conditions of As(V) respiration. The construction of an arsB mutant that was polar onto arsC (strain ARSB1) enabled us to consider both of these issues. ARSB1 did not grow in medium amended with >5 mM As(III), nor did ARSB1 reduce As(V) in LB medium-As(V) cultures. Interestingly, when respiring on As(V) in low-phosphate medium, ARSB1 reduced 7 to 8 mM As(V) at a slower rate compared to the wild type, achieving 25% lower cell density. These observations, along with the fact that no arsC-specific mRNA was detected by RT-PCR in ARSB1, suggest that there is an additional As(V) reductase that is used for As(V) respiration and that ArsC is not used for respiratory As(V) reduction.
There are several possible explanations for the As(III) resistance phenotype in ARSB1: (i) the presence of a duplicate ars operon compensates for the loss of this copy of arsB and arsC; (ii) the expression of an additional arsenic detoxification pathway during As(V) respiration compensates for the loss of this copy of arsB and arsC; and/or (iii) the enzyme used to reduce As(V) to As(III) during respiration resides in the periplasm and has a high affinity for As(V)—thus, the loss of this copy of arsB and arsC does not seriously affect the cell. Because Southern blot analysis with an arsB gene probe revealed only one hybridizing band within the genomic DNA of ANA-3, we believe the first explanation is unlikely. Although we do not yet have any direct evidence that either supports or rejects the second explanation, it seems possible, based on positive identification of homologs in the S. oneidensis MR-1 genome to genes involved in other As resistance systems (Table 4), that ANA-3 may also possess an additional As resistance system. However, because the arsB mutant could not reduce As(V) aerobically, it appears that a functional ArsC homolog is not made.
Regardless of whether an additional As resistance pathway exists and is expressed when ANA-3 is respiring As(V), we favor the third explanation. We base this position upon the fact that we recently identified a respiratory As(V) reductase whose coding sequence motifs suggest that it resides in the periplasm of ANA-3 (C. W. Saltikov and D. K. Newman, unpublished data). If this enzyme is able to scavenge As(V) faster than As(V) can enter the cytoplasm through inorganic phosphate (Pi) transporters (45) when the As(V)/Pi ratio is low, we would expect the need for a cytosolic As efflux system to be minimal. Conversely, when the As(V)/Pi ratio is high, we would expect more As(V) to enter the cell. Preliminary evidence in support of this interpretation is that the ARSB1 mutant exhibits a growth defect relative to the wild type when the As(V)/Pi ratio is high. Although more work is required to confirm this interpretation, including kinetic analyses of As(V) binding and/or turnover rates in the presence of various Pi, we favor it as a working hypothesis.
In summary, our results show that the ArsB efflux system is not required for ANA-3 to respire on As(V) and that ArsC is not required for ANA-3 to reduce As(V) to As(III) when respiring As(V). Nevertheless, the presence of the ars operon does appear to provide ANA-3 with additional protection against the toxicity of As(III) when respiring high concentrations of As(V). Whether or not As(V)-respiring microorganisms inhabiting natural systems require high-level As detoxification systems remains to be determined.
Acknowledgments
We thank Joanna Levitt and Anabel Anton for help with the isolation of ANA-3 during the 1998 MBL Microbial Diversity Course, Doug Lies and members of the Newman lab for valuable discussions, Angela Snow for laboratory assistance, and B. P. Rosen for providing E. coli strain AW3110.
Funding was provided by grants from the Luce Foundation and the Packard Foundation to D.K.N. and by a National Science Foundation Postdoctoral Fellowship in Microbial Biology to C.W.S.
REFERENCES
- 1.Ahmann, D., L. R. Krumholz, H. F. Hemond, D. R. Lovley, and F. M. Morel. 1997. Microbial mobilization of arsenic from sediments of the Aberjona watershed. Environ. Sci. Technol. 31:2923-2930. [Google Scholar]
- 2.Armienta, M. A., G. Villasenor, R. Rodriguez, L. K. Ongley, and H. Mango. 2001. The role of arsenic-bearing rocks in groundwater pollution at Zimapan Valley, Mexico. Environ. Geol. 40:571-581. [Google Scholar]
- 3.Ausubel, F. M. 1992. Short protocols in molecular biology, 2nd ed.: a compendium of methods from current protocols in molecular biology. John Wiley & Sons, Inc./Green Publishing Associates, New York, N.Y.
- 4.Beliaev, A. S., and D. A. Saffarini. 1998. Shewanella Putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J. Bacteriol. 180:6292-6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bentley, R., and T. G. Chasteen. 2002. Microbial methylation of metalloids: arsenic, antimony, and bismuth. Microbiol. Mol. Biol. Rev. 66:250-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum, J. S., A. B. Bindi, J. Buzzelli, J. F. Stolz, and R. S. Oremland. 1998. Bacillus arsenicoselenatis, sp. nov., and Bacillus selenitireducens, sp. nov.: two haloalkaliphiles from Mono Lake, California, that respire oxyanions of selenium and arsenic. Arch. Microbiol. 171:19-30. [DOI] [PubMed] [Google Scholar]
- 7.Bowman, J. P., S. A. Mccammon, D. S. Nichols, J. H. Skerratt, S. M. Rea, P. D. Nichols, and T. A. Mcmeekin. 1997. Shewanella gelidimarina sp. nov. and Shewanella frigidimarina sp. nov., novel Antarctic species with the ability to produce eicosapentaenoic acid (20:5 Omega 3) and grow anaerobically by dissimilatory Fe(III) reduction. Int. J. Syst. Bacteriol. 47:1040-1047. [DOI] [PubMed] [Google Scholar]
- 8.Butcher, B. G., S. M. Deane, and D. E. Rawlings. 2000. The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli. Appl. Environ. Microbiol. 66:1826-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlin, A., W. Shi, S. Dey, and B. P. Rosen. 1995. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J. Bacteriol. 177:981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cline, E. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 11.Coppi, M. V., C. Leang, S. J. Sandler, and D. R. Lovley. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67:3180-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dehio, C., and M. Meyer. 1997. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179:538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowdle, P. R., A. M. Laverman, and R. S. Oremland. 1996. Bacterial dissimilatory reduction of arsenic(V) to arsenic(III) in anoxic sediments. Appl. Environ. Microbiol. 62:1664-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fredrickson, J. K., J. M. Zachara, D. W. Kennedy, H. L. Dong, T. C. Onstott, N. W. Hinman, and S. M. Li. 1998. Biogenic iron mineralization accompanying the dissimilatory reduction of hydrous ferric oxide by a groundwater bacterium. Geochim. Cosmochim. Acta 62:3239-3257. [Google Scholar]
- 15.Gihring, T. M., and J. F. Banfield. 2001. Arsenite oxidation and arsenate respiration by a new Thermus isolate. FEMS Microbiol. Lett. 204:335-340. [DOI] [PubMed] [Google Scholar]
- 16.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, Peterson, J. D., L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nature 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 17.Herbel, M. J., J. S. Blum, S. E. Hoeft, S. M. Cohen, L. L. Arnold, J. Lisak, J. F. Stolz, and R. S. Oremland. 2002. Dissimilatory arsenate reductase activity and arsenate-respiring bacteria in bovine rumen fluid, hamster feces, and the termite hindgut. FEMS Microbiol. Ecol. 41:59-67. [DOI] [PubMed] [Google Scholar]
- 18.Holmes, B., S. P. Lapage, and H. Malnick. 1975. Strains of Pseudomonas-Putrefaciens from clinical material. J. Clin. Pathol. 28:149-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber, R., M. Sacher, A. Vollmann, H. Huber, and D. Rose. 2000. Respiration of arsenate and selenate by hyperthermophilic Archaea. Syst. Appl. Microbiol. 23:305-314. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, D. L., and M. E. Q. Pilson. 1972. Spectrophotometric determination of arsenite, arsenate, and phosphate in natural waters. Anal. Chim. Acta 58:289-299. [Google Scholar]
- 21.Johnson, J. L. 1981. Genetic characterization, p. 450-472. In P. Gerhardt, R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. B. Phillips (ed.), Manual of methods for general bacteriology. American Society for Microbiology, Washington, D.C.
- 22.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 23.Krafft, T., and J. M. Macy. 1998. Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur. J. Biochem. 255:647-653. [DOI] [PubMed] [Google Scholar]
- 24.Laverman, A. M., J. S. Blum, J. K. Schaefer, E. J. P. Phillips, D. R. Lovely, and R. S. Oremland. 1995. Growth of strain SES-3 with arsenate and other diverse electron acceptors. Appl. Environ. Microbiol. 61:3556-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonardo, M. R., D. P. Moser, E. Barbieri, C. A. Brantner, B. J. Macgregor, B. J. Paster, E. Stackebrandt, and K. H. Nealson. 1999. Shewanella pealeana sp. nov., a member of the microbial community associated with the accessory nidamental gland of the squid Loligo pealei. Int. J. Syst. Bacteriol. 49:1341-1351. [DOI] [PubMed] [Google Scholar]
- 26.Lovley, D. R., and E. J. P. Phillips. 1988. Novel mode of microbial energy-metabolism-organic-carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macy, J. M., K. Nunan, K. D. Hagen, D. R. Dixon, P. J. Harbour, M. Cahill, and L. I. Sly. 1996. Chrysiogenes arsenatis gen. nov., sp. nov., a new arsenate-respiring bacterium isolated from gold mine wastewater. Int. J. Syst. Bacteriol. 46:1153-1157. [DOI] [PubMed] [Google Scholar]
- 28.Macy, J. M., J. M. Santini, B. V. Pauling, A. H. O'Neill, and L. I. Sly. 2000. Two new arsenate/sulfate-reducing bacteria: mechanisms of arsenate reduction. Arch. Microbiol. 173:49-57. [DOI] [PubMed] [Google Scholar]
- 29.Makemson, J. C., N. R. Fulayfil, W. Landry, L. M. Vanert, C. F. Wimpee, E. A. Widder, and J. F. Case. 1997. Shewanella woodyi sp. nov., an exclusively respiratory luminous bacterium isolated from the Alboran Sea. Int. J. Syst. Bacteriol. 47:1034-1039. [DOI] [PubMed] [Google Scholar]
- 30.Miller, T. L., and M. J. Wolin. 1974. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl. Environ. Microbiol. 27:985-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukhopadhyay, R., B. P. Rosen, L. Phung, and S. Silver. 2002. Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol. Rev. 26:311-321. [DOI] [PubMed] [Google Scholar]
- 32.Myers, C. R., and K. H. Nealson. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole electron-acceptor. Science 240:1319-1321. [DOI] [PubMed] [Google Scholar]
- 33.Nealson, K. H., C. R. Myers, and B. B. Wimpee. 1991. Isolation and identification of manganese-reducing bacteria and estimates of microbial Mn(IV)-reducing potential in the Black Sea. Deep-Sea Res. 38(Suppl. 2):S907-S920. [Google Scholar]
- 34.Neidhardt, F. C., and H. E. Umbarger. 1991. Chemical composition of Escherichia coli, p. 13-16. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 35.Newman, D. K., D. Ahmann, and F. M. M. Morel. 1998. A brief review of microbial arsenate respiration. Geomicrobiol. J. 15:255-268. [Google Scholar]
- 36.Newman, D. K., T. J. Beveridge, and F. M. M. Morel. 1997. Precipitation of arsenic trisulfide by Desulfotomaculum auripigmentum. Appl. Environ. Microbiol. 63:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman, D. K., E. K. Kennedy, J. D. Coates, D. Ahmann, D. J. Ellis, D. R. Lovley, and F. M. Morel. 1997. Dissimilatory arsenate and sulfate reduction in Desulfotomaculum auripigmentum sp. nov. Arch. Microbiol. 168:380-388. [DOI] [PubMed] [Google Scholar]
- 38.Newman, D. K., and R. Kolter. 2000. A role for excreted quinones in extracellular electron transfer. Nature 405:94-97. [DOI] [PubMed] [Google Scholar]
- 39.Niggemyer, A., S. Spring, E. Stackebrandt, and R. F. Rosenzweig. 2001. Isolation and characterization of a novel As(V)-reducing bacterium: implications for arsenic mobilization and the genus Desulfitobacterium. Appl. Environ. Microbiol. 67:5568-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nriagu, J. 2002. Arsenic poisoning through the ages, p. 21-22. In W. T. Frankenberger (ed.), Environmental chemistry of arsenic. Marcel Dekker, Inc., New York, N.Y.
- 41.Oremland, R. S., J. S. Blum, C. W. Culbertson, P. T. Visscher, L. G. Miller, P. R. Dowdle, and F. E. Strohmaier. 1994. Isolation, growth, and metabolism of an obligately anaerobic, selenate-respiring bacterium, strain SES-3. Appl. Environ. Microbiol. 60:3011-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oremland, R. S., P. R. Dowdle, S. Hoeft, J. O. Sharp, J. K. Schaefer, L. G. Miller, J. S. Blum, R. L. Smith, N. S. Bloom, and D. Wallschlaeger. 2000. Bacterial dissimilatory reduction of arsenate and sulfate in meromictic Mono Lake, California. Geochim. Cosmochim. Acta 64:3073-3084. [Google Scholar]
- 43.Prithivirajsingh, S., S. K. Mishra, and A. Mahadevan. 2001. Detection and analysis of chromosomal arsenic resistance in Pseudomonas fluorescens strain MSP3. Biochem. Biophys. Res. Commun. 280:1393-1401. [DOI] [PubMed] [Google Scholar]
- 44.Rosen, B. P. 1999. Families of arsenic transporters. Trends. Microbiol. 7:207-212. [DOI] [PubMed] [Google Scholar]
- 45.Rosenberg, H., L. M. Russell, P. A. Jacomb, and K. Chegwidden. 1982. Phosphate exchange in the pit transport system in Escherichia coli. J. Bacteriol. 149:123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruimy, R., V. Breittmayer, P. Elbaze, B. Lafay, O. Boussemart, M. Gauthier, and R. Christen. 1994. Phylogenetic analysis and assessment of the genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas deduced from small-subunit ribosomal-RNA sequences. Int. J. Syst. Bacteriol. 44:416-426. [DOI] [PubMed] [Google Scholar]
- 47.Saltikov, C. W., and B. H. Olson. 2002. Homology of Escherichia coli R773 arsA, arsB, and arsC genes in arsenic-resistant bacteria isolated from raw sewage and arsenic-enriched creek waters. Appl. Environ. Microbiol. 68:280-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Santini, J. M., L. I. Sly, A. M. Wen, D. Comrie, P. De Wulf-Durand, and J. M. Macy. 2002. New arsenite-oxidizing bacteria isolated from Australian gold mining environments: phylogenetic relationships. Geomicrobiol. J. 19:67-76. [Google Scholar]
- 50.Silver, S., L. T. Phung, and B. P. Rosen. 2002. Arsenic metabolism: resistance, reduction, and oxidation, p. 254. In W. T. Frankenberger (ed.), Environmental chemistry of arsenic. Marcel Dekker, Inc., New York, N.Y.
- 51.Simidu, U., K. Kitatsukamoto, T. Yasumoto, and M. Yotsu. 1990. Taxonomy of four marine bacterial strains that produce tetrodotoxin. Int. J. Syst. Bacteriol. 40:331-336. [DOI] [PubMed] [Google Scholar]
- 52.Smibert, R. M., and N. Krieg. 1994. Phenotypic characterization, p. 607-654. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 53.Stolz, J. F., and R. S. Oremland. 1999. Bacterial respiration of arsenic and selenium. FEMS Microbiol. Rev. 23:615-627. [DOI] [PubMed] [Google Scholar]
- 54.Swofford, D. L. 1999. PAUP: phylogenetic analysis using parsimony (and other methods), version 4.0.b10. Sinauer Associates, Sunderland, Mass.
- 55.Venkateswaran, K., M. E. Dollhopf, R. Aller, E. Stackebrandt, and K. H. Nealson. 1998. Shewanella amazonensis sp. nov., a novel metal-reducing facultative anaerobe from Amazonian shelf muds. Int. J. Syst. Bacteriol. 48:965-972. [DOI] [PubMed] [Google Scholar]
- 56.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705-724. [DOI] [PubMed] [Google Scholar]
- 57.Wall, J. D., T. Murnan, J. Argyle, R. S. English, and B. J. Rapp-Giles. 1996. Transposon mutagenesis in Desulfovibrio desulfuricans: development of a random mutagenesis tool from Tn7. Appl. Environ. Microbiol. 62:3762-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilkie, J. A., and J. G. Hering. 1998. Rapid oxidation of geothermal arsenic(III) in streamwaters of the Eastern Sierra Nevada. Environ. Sci. Technol. 32:657-662. [Google Scholar]
- 59.Zachara, J. M., J. K. Fredrickson, S. M. Li, D. W. Kennedy, S. C. Smith, and P. L. Gassman. 1998. Bacterial reduction of crystalline Fe3+ oxides in single phase suspensions and subsurface materials. Am. Mineral. 83:1426-1443. [Google Scholar]
- 60.Ziemke, F., M. G. Hofle, J. Lalucat, and R. Rossello-Mora. 1998. Reclassification of Shewanella putrefaciens Owen's genomic group II as Shewanella baltica sp. nov. Int. J. Syst. Bacteriol. 48:179-186. [DOI] [PubMed] [Google Scholar]