Abstract
We have analyzed the diversity of microbial genomes represented in a library of metagenomic DNA from soil. A total of 24,400 bacterial artificial chromosome (BAC) clones were screened for 16S rRNA genes. The sequences obtained from BAC clones were compared with a collection generated by direct PCR amplification and cloning of 16S rRNA genes from the same soil. The results indicated that the BAC library had substantially lower representation of bacteria among the Bacillus, α-Proteobacteria, and CFB groups; greater representation among the β- and γ-Proteobacteria, and OP10 divisions; and no rRNA genes from the domains Eukaryota and Archaea. In addition to rRNA genes recovered from the bacterial divisions Proteobacteria, Verrucomicrobia, Firmicutes, Cytophagales, and OP11, we identified many rRNA genes from the BAC library affiliated with the bacterial division Acidobacterium; all of these sequences were affiliated with subdivisions that lack cultured representatives. The complete sequence of one BAC clone derived from a member of the Acidobacterium division revealed a complete rRNA operon and 20 other open reading frames, including predicted gene products involved in cell division, cell cycling, folic acid biosynthesis, substrate metabolism, amino acid uptake, DNA repair, and transcriptional regulation. This study is the first step in using genomics to reveal the physiology of as-yet-uncultured members of the Acidobacterium division.
For many decades, microbiologists have been intrigued by the observation that the vast majority of microorganisms in natural environments elude laboratory cultivation (34). In fact, in soils around the world, typically less than 1% of the cells observed by direct counting are recovered by standard cultivation methods (2, 36-38). The vast majority of microorganisms in natural environments were largely intractable to scientific inquiry until the advent of molecular phylogenetic analysis, a method that uses sequence heterogeneity within the 16S rRNA gene for inference of evolutionary relationships (27, 42). PCR amplification of 16S rRNA genes from natural environments frequently reveals sequences that are highly divergent from known cultured phyla, even among bacterial divisions well represented by cultured isolates (12, 15, 41). Many of the highly divergent 16S rRNA gene sequences together comprise entirely new prokaryotic lineages, forming newly recognized divisions (14-16). One of the greatest challenges facing microbiologists today is to understand the complex ecological roles and interactions of as-yet-uncultured microorganisms affiliated with these novel phylogenetic groups.
Of the bacterial divisions revealed by rRNA gene sequence data, the division Acidobacterium is ubiquitous in soils and sediments, yet it has few cultured representatives (4, 20, 22, 30). Based upon directly recovered rRNA sequences, eight subdivisions of the Acidobacterium division have been proposed, only three of which have cultured representatives (4, 15, 23, 24, 30). Typically, most Acidobacterium-related rRNA gene sequences recovered from soil either are in subdivisions without cultured representatives or are related to Acidobacterium capsulatum, a gram-negative bacterium isolated from an acidic mineral environment (18). Recently, the first Acidobacterium cultured isolates were obtained from a nonsaturated soil (17, 30). All of these cultured isolates collectively represent only a small fraction of the Acidobacterium phylum identified by rRNA gene sequences. By using information acquired from both cultured isolates and directly cloned genomic DNA, we gain the tools necessary to extend our limited knowledge of the Acidobacterium division beyond the phylogenetic context.
The work of DeLong and colleagues has demonstrated the utility of capturing community DNA in the form of bacterial artificial chromosome (BAC) libraries, and by linking phylogenetic and functional information within specific BAC clones, gaining new perspectives on the microbial ecology of natural environments (5, 8, 35, 39). Genomic DNA from marine picoplankton was cloned into a BAC vector; screening of the resulting E. coli transformants for clones containing an rRNA gene led Beja et al. (5, 6) to the discovery of a 140-kb BAC clone that contained both an rRNA operon (SAR86 clade) and an open reading frame (ORF) with high homology to bacteriorhodopsins. Bacteriorhodopsins are light-driven proton pumps capable of harnessing solar energy (5); these data along with results of other molecular analyses suggested that the uncultured SAR86 bacterium utilizes a novel mode of solar energy generation (7, 9). This strategy has also recently been used by Quaiser et al. to identify a fosmid clone derived from an uncultured soil crenarchaeote, revealing physiologically relevant functional genes adjacent to an rRNA operon (28).
Through a phylogenetic analysis of our soil metagenomic libraries, we can better characterize potential biases involved in metagenomic library construction (e.g., cell lysis, restriction digestion, and cloning) and also acquire molecular tools useful for microbial ecology studies. Here we report the results of a census of rRNA genes within a previously constructed BAC library (29) and present the first genomic sequence isolated from an uncultured member of the Acidobacterium division.
MATERIALS AND METHODS
BAC library construction.
The BAC library was constructed in Escherichia coli strain DH10B as previously described (29). Briefly, soil cores were collected from an undisturbed and uncultivated site at the West Madison Agricultural Research Station (WMARS), and high-molecular-weight soil DNA was extracted directly from the sample by successive freeze-thaw cycles and phenol extraction followed by isopropanol precipitation (29). Two rounds of preparative pulsed-field electrophoresis gels were used to isolate sufficiently pure and large genomic DNA that had been partially endonuclease-digested with HindIII, which was then ligated into the vector pBeloBAC11 and transformed into E. coli. Soil library 2 (SL2) contains 24,400 clones with an approximate average insert size of 42 kb; in total, this represents over a gigabase of genomic DNA, or approximately 200 genome equivalents, assuming an average genome size of 4 Mbp. Clones were individually stored in freezing medium (Luria-Bertani with 20% glycerol) at −80°C in 96-well plates. In addition, clone pools (12× and 144×) were prepared to provide for more-rapid analysis.
DNA isolation from soil.
At the same time that high-molecular-weight genomic DNA was extracted from soil for the construction of SL2, we also used a bead-beating method (Bio101, Inc., La Jolla, Calif.) to isolate genomic DNA (10). The genomic DNA isolated via a Bio101 kit is generally less than 20 kb in size, yet the harsh lysis conditions ensure that the genomic DNA is broadly representative of the soil microbial community (11). Samples were stored at −20°C until further analysis.
BAC DNA isolation from SL2.
BAC clone DNA was isolated using the R.E.A.L. prep 96 plasmid kit (Promega, Madison, Wis.), with some modifications. BAC clones in 12× format were replica plated into 96-well polypropylene plates (Fisher Scientific, Pittsburgh, Pa.) containing 200 μl of 2× yeast-tryptone medium with 12.5 μg of chloramphenicol/ml and incubated overnight at 37°C. The resulting stationary-phase cultures were then used to inoculate 1 ml of 2× yeast-tryptone medium within deep 96-well plates (Promega), sealed with a sterilized AirPore strip (Qiagen, Valencia, Calif.), and incubated for 12 to 16 h at 37°C with vigorous shaking. Cells were concentrated by centrifugation at 2,700 × g in an Eppendorf 5810R tabletop centrifuge cooled to 4°C. The cell pellets were resuspended in 300 μl of resuspension solution and vortexed until thoroughly in suspension. Cells were then lysed by the addition of 300 μl of lysis solution and incubated for 5 min. After lysis, 400 μl of neutralization solution was added, and the solution was very gently mixed by gentle side-to-side movement to avoid chromosomal shearing. The white precipitate was then removed by passing the solution through the SV96 clearing plate, and isopropanol (0.7× volume) was added to precipitate the DNA. After washing with 70% ethanol, the BAC DNA was air dried and then resuspended in 100 μl of T10E1. The DNA yield and purity were assessed for representative samples by agarose gel electrophoresis. Yields of 0.2 to 0.5 μg of BAC DNA per well of a 96-well plate were typically achieved by this method. BAC DNA from clones containing rRNA genes was restriction digested with HinfI or NotI, and then the samples were subjected to pulsed-field gel electrophoresis (Chef Mapper; Bio-Rad Corp., Hercules, Calif.) at 6 V/cm with a 3- to 15-s switch time from an included angle of 120° for 10 h.
Digestion of chromosomal DNA.
The majority of host chromosomal DNA was removed during the neutralization step of the BAC DNA preparation, but even trace quantities of host DNA may lead to PCR amplification of the host (i.e., E. coli) 16S rRNA gene. Therefore, we reduced the contribution of host DNA to subsequent PCRs by using plasmid-safe, ATP-dependent DNase (Epicentre Technologies, Inc., Madison, Wis.) to digest nicked chromosomal DNA at 37°C for 2 h. The digests were then passed over a Sephadex G-50 column matrix contained within a low nucleic acid-binding filtration plate (0.65-μm-pore-size DVPP filter; Millipore, Bedford, Mass.), and BAC DNA was eluted by centrifugation at 2,000 × g for 1 min in an Eppendorf centrifuge.
PCR amplification of rRNA genes.
The 16S rRNA genes from Bio101-extracted DNA or SL2 DNA were PCR amplified under the same conditions, by using approximately 100 ng of template DNA, 1 U of Taq polymerase, 200 μM deoxynucleoside triphosphates, and 200 nM concentrations of each of the primers 27F (broadly conserved in bacteria) (26), 23FPL (broadly conserved in eukaryotes and archaea) (3), and 1492R (broadly conserved across most phyla) (26) in a 50-μl PCR volume. The reaction was performed with 3 min of denaturation at 95°C and 30 cycles of 95°C for 1 min, 55°C annealing for 90 s, and 72°C extension for 150 s and followed by 7 min of extension at 72°C. All reactions were carried out in a Robocycler 96 (Stratagene, La Jolla, Calif.) with 50 μl of mineral oil added to each tube. Representative reactions were analyzed by agarose gel electrophoresis to insure efficient PCR. Other primer sets specific to the domains Eukaryota (519R) (21) and Archaea (133F) (33) were also used in this study to test for the presence of representatives of these domains within SL2.
E. coli rRNA gene template-specific termination.
In some PCRs, terminally modified oligonucleotides specific to the E. coli 16S rRNA gene were included to inhibit amplification from the host chromosomal template (12a). The terminal modifications included a 5′ acridine group that increases the affinity of the oligonucleotide to its cognate template and a 3′ phosphoramidite spacer that inhibits PCR extension, resulting in a set of E. coli rRNA terminators that selectively inhibit PCR amplification from targeted rRNA genes. This method, which is analogous to the peptide nucleic acid PCR clamping method of von Wintzingerode et al. (40), has proven effective during PCRs with mixtures of DNA templates from E. coli and other species and was used previously to screen a smaller soil library, soil library 1 (29). It is possible that inclusion of terminator oligonucleotides within a PCR could inhibit PCR amplification of non-E. coli rRNA genes contained within a metagenomic library. For this reason, we only used terminator oligonucleotides after the initial screening to PCR amplify rRNA genes from respective BAC clones without any host ribosomal DNA (rDNA) contamination.
16S rDNA clone libraries.
Multiple 16S rDNA clone libraries were prepared by using as a template either genomic DNA extracted directly from soil (bead beat method) or BAC DNA isolated from all of the SL2 clones pooled into one tube (following growth in 12× format), with both universal primer sets and a primer set specific to the Acidobacterium division (4). PCR products were first digested with HinfI and then analyzed by agarose gel electrophoresis to determine at a gross level the heterogeneity of each PCR product. PCR products prepared from soil DNA and SL2 template, with and without terminator oligonucleotides, were cloned into the pGEM-T cloning vector (Promega), and transformants were selected on appropriate media. For each rDNA library, clones were arrayed in 96-well plates containing 1 ml of Luria-Bertani per well and plasmid DNA was isolated for subsequent sequencing.
Phylogenetic analysis.
The first 500 bp of each rRNA gene clone was sequenced by using the bacterial primers 27F and 519R (21). Partial rRNA gene sequences were assembled with SeqMan (DNAStar, Madison, Wis.) to generate a consensus sequence. All sequences were checked for chimeras with Chimera Check, version 2.7 (25), and compared to available databases by use of the Basic Local Alignment Search Tool (BLAST) (1) to determine approximate phylogenetic affiliation. Sequences were aligned to a data set of 6,883 bacterial sequences (courtesy of Phillip Hugenholtz, http://rdp.cme.msu.edu/html/alignments.html) by using the ARB software package (http://www.arb-home.de/) and refined manually to remove regions of ambiguous homology. Alignments used for phylogenetic analysis were minimized by the Lane mask (21) for bacterial data or an Acidobacterium filter (phylum-specific 50% filter by base frequency) prepared in ARB. Phylogenetic trees for near full-length sequences (>1,400 nucleotides [nt]) were inferred within the ARB package by using evolutionary distance (neighbor-joining algorithms with Felsenstein correction) and the PHYLIP program for maximum parsimony (J. Felsenstein, Department of Genetics, University of Washington, Seattle). Partial sequences (<1,400 nt) were inserted into trees without branch arrangement of full-length sequences by using the parsimony insertion tool of ARB. The robustness of the tree topology was tested by bootstrap resampling with multiple out-groups.
RFLP analysis.
After each 96-well PCR, the resultant products were digested by using HinfI and AluI in separate reactions (enzymes were chosen for their abilities to differentiate between E. coli and non-E. coli restriction fragment length polymorphism [RFLP] patterns, based on in silica analysis), and the restriction fragments were analyzed on an agarose gel. E. coli rRNA gene PCR products were likewise digested and analyzed as a negative control. After ethidium bromide staining of the agarose gel, restriction fragment (RF) patterns were analyzed with an IS-1000 digital imaging system (Alpha Innotech, San Leandro, Calif.).
BAC insert sequencing.
A BAC clone (P17F9) containing an rRNA gene affiliated with the Acidobacterium division was chosen for full-insert sequencing. Since sequencing directly from a BAC clone is difficult and does not generate interpretable sequence information for long stretches of DNA, we chose to generate a shotgun library of the BAC clone within a multicopy plasmid. First, a large-scale BAC DNA preparation was performed by extracting vector DNA from a 1-liter culture using with a large construct kit (Qiagen). After exonuclease digestion and cleanup over the supplied anion-exchange column, the BAC DNA was subjected to partial Sau3A digestion and the 2- to 3-kb-sized fragments were isolated from a low-melting-point agarose gel. The restriction fragments were ligated with a BamHI-digested pUC19 vector by using T4 DNA ligase, and E. coli transformants were selected on appropriate media. BigDye sequencing reactions were conducted by using M13 forward and reverse primers, respectively, and analyzed on an ABI 377 automated sequencer at the University of Wisconsin's Biotechnology Center. A contiguous insert sequence was assembled by utilizing SeqMan. ORFs were identified with Glimmer, an algorithm developed at The Institute for Genomic Research for ORF identification (32), and all predicted ORFS were compared to the database of nonredundant genomic sequences at GenBank by the BLASTn and BLASTx algorithms.
Nucleotide sequence accession numbers.
Partial 16S rRNA sequences represented in Fig. 1a have been previously assigned GenBank accession numbers (10). These previously reported sequences, together with the 16S rRNA gene sequences reported (see Fig. 1b and 3, accession nos. AY214601 through AY214917), are listed with their respective GenBank accession numbers at http://www.plantpath.wisc.edu/goodman/addinfo/SL2.html. The annotated sequence of the BAC clone P17F9 has been assigned the accession number AY214600 in the GenBank database.
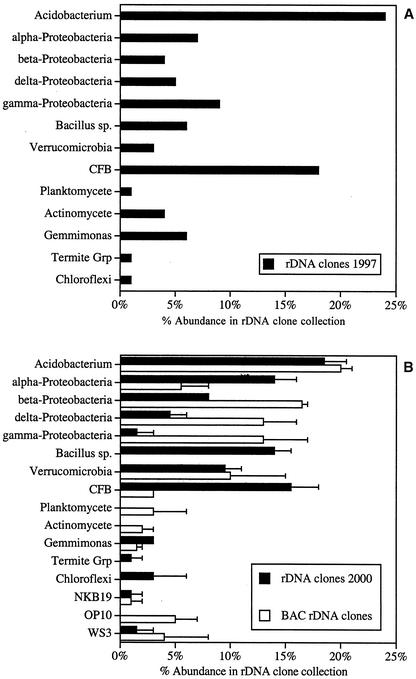
FIG. 1.
Phylogenetic distribution of 16S rRNA genes amplified from DNA recovered from soil and a soil metagenomic library. Shown are phylogenetic distributions for rDNA clones derived from soil isolated at the WMARS in the year 1997 (n = 124) (A) or in the year 2000 (n = 130) (B). Panel B also shows the phylogenetic distribution of rRNA genes amplified from the BAC SL2 (n = 132), which was constructed from the same soil sample used for the rDNA clone library in the year 2000. Each rRNA gene sequence was aligned with a large data set of 16S rRNA gene sequences, and maximum-parsimony analysis indicated a phylogenetic affiliation. The percentage of rDNA clones within each phylogenetic group is indicated. Groups that lack a bar (e.g., the Bacillus group in panel B) indicate the absence of clones affiliated with a particular phylogenetic group. Error bars represent the standard errors for the average percentages of abundance of each phylogenetic group within replicate rDNA clone libraries, each prepared from the same DNA template.
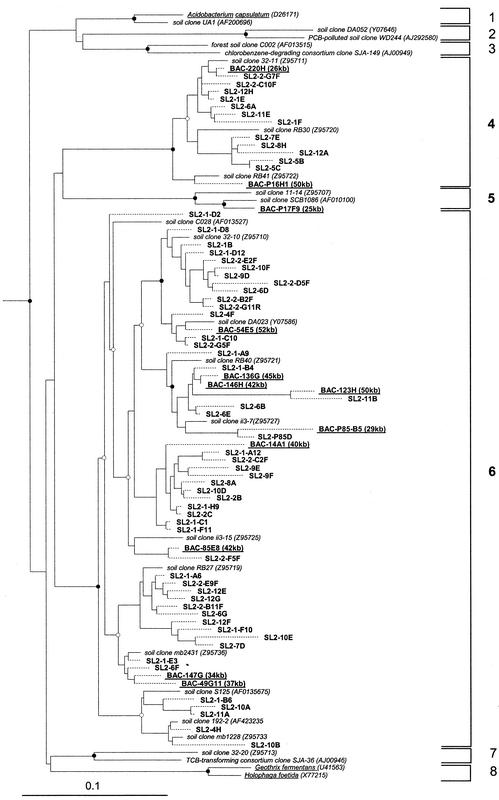
FIG. 3.
Phylogenetic dendrogram representing the analysis of recovered rRNA sequences affiliated with the Acidobacterium division. 16S rRNA sequences published in GenBank affiliated with the Acidobacterium division (4, 15, 24) were used to analyze all 16S rRNA gene sequences recovered from SL2 as described in the text. Published sequences are designated in italics; clones sequenced in this particular study are in boldface. Underlined sequences in bold indicate that a BAC clone has been identified that contains the respective rRNA gene, and the number in parentheses represents the approximate insert size. Published Acidobacterium subdivisions are bracketed on the right (15). Partial length sequences (<1,400 nt) are indicated by dashed lines. Branch points supported (bootstrap values of >75%) by parsimony analyses are indicated by solid circles; open circles represent those marginally supported (bootstrap values of 50 to 75%) by parsimony. Branch points without circles are unresolved (bootstrap values of <50%) by different analyses. Bacterial out-groups (data not shown) used for the analyses were E. coli (J01695) and Agrobacterium tumerfaciens (M11223). The bar represents 0.1 changes per nucleotide.
RESULTS
16S rRNA gene survey of WMARS soil and SL2.
A bacterial division or group level affiliation for each 16S rDNA clone was assigned based on the phylogenetic analysis performed with a data set of 6,883 rRNA gene sequences (Fig. 1). In addition to the rDNA clone libraries prepared from the soil microbial DNA used for SL2, we also included data from an rDNA clone collection prepared in a previous study at the same location in the year 1997 (9), designated rDNA clones 1997 (Fig. 1). These two independently isolated soil samples from the same location revealed similar division level representation, with the majority of the rRNA genes affiliated with the Proteobacteria, Acidobacterium, CFB, Bacillus, and Verrucomicrobia groups, cumulatively representing 76% of the year 1997 clones and 86% of the clones prepared in the year 2000. In contrast, the replicate rDNA clone libraries derived from SL2 (entire BAC library pooled into a single template) revealed a significantly altered community composition, with the most pronounced difference being the marked apparent underrepresentation in the clones affiliated with the Bacillus group compared to the rDNA clone library constructed from bead-beaten soil DNA. These data revealed that the rDNA clone library derived from SL2 contained a lower representation of the Bacillus, α-Proteobacteria, and CFB groups. The data also suggest that other phylogenetic groups, most notably the β- and γ-Proteobacteria and OP10 divisions, are more highly represented within the SL2-derived rDNA clone library than in the PCR-generated rDNA clone libraries from soil (Fig. 1). We also found rRNA gene sequences from soil samples collected in 1997 that affiliated with the divisions OS-K (one sequence), OP1 (one sequence), OP3 (two sequences), and TM7 (two sequences), as well as two rRNA sequences from the first bead-beaten library that affiliated with the Firmicutes division but not with the Bacillus group. In the SL2-1 clone bank, generated with the presence of terminator oligonucleotides, there were two E. coli rRNA gene sequences, whereas the SL2-2 clone bank, which did not have terminator oligonucleotides added during PCR, contained four E. coli rRNA gene sequences. No PCR products were detected when primer sets specific to the domains Eukaryota or Archaea were used, despite multiple attempts with the same DNA templates used successfully for amplification of bacterial rRNA genes.
Identification of BAC clones containing 16S rRNA genes.
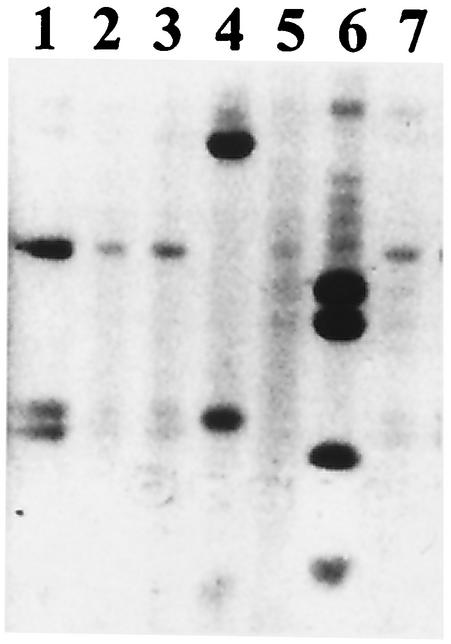
SL2 was screened in a 12× pool format to identify BAC clones containing a 16S rRNA gene. Results from screening of the 12× pools were highly variable depending upon the quality of the BAC DNA template. Exonuclease digestion, while largely effective at reducing chromosomal contamination of BAC preps, also eliminated any PCR product from most wells, making it difficult to distinguish between a false negative due to PCR error and a true negative. Therefore, we screened SL2 with and without exonuclease digestion. When chromosomal DNA was partially removed by exonuclease digestion, the rDNA PCR product derived from a BAC template was very evident by RFLP analysis (Fig. 2, lanes 4 and 6), with only faint bands observed that corresponded to an E. coli rDNA gene RFLP pattern (Fig. 2, lanes 2 and 3). In many experiments, the degree of host chromosomal contamination was variable, even after exonuclease digestion. Only when exonuclease treatment was performed and E. coli rRNA gene-specific terminator oligonucleotides were included in the PCR was the E. coli RFLP pattern completely absent by visual inspection (data not shown). Although we did not add terminator oligonucleotides to the initial PCR screening due to concerns about selective inhibition of rRNA gene amplification from microorganisms related to the E. coli host, we did employ these terminator oligonucleotides to prepare homogenous rRNA gene products for cloning after their initial identification. None of the BAC clones containing rRNA genes were observed to have any loss of rRNA gene product amplification when the terminator oligonucleotides were included in the reaction mixture (data not shown).
FIG. 2.
RFLP analysis of rRNA PCR products from metagenomic library clones. Depicted is an RF pattern for the E. coli negative control (lane 1) and 6 different BAC pools, each of which contains a DNA template from 12 different BAC clones. Lanes 4 and 6 reveal BACs containing rRNA genes, whereas lanes 2, 3, 5, and 7 reveal only a faint RF pattern presumably derived from E. coli 16S rRNA genes.
In SL2, we identified 28 BAC clones that carried rRNA genes. The results from this screening of SL2 mirrored the results from direct amplification from the entire pooled library, with the majority of the BAC clones identified from the Acidobacterium (10 clones) and Proteobacteria (7 clones) divisions. In addition, there were three clones from the Verrucomicrobia division, two clones from the WS3 division, one clone from the CFB division, one clone from the OP10 division, one clone from the Nitrospirae division, and one clone from the Bacillus group. The latter clone was 92% identical to the 16S rRNA gene (764 bp sequenced) of Bacillus licheniformis strain KL-164.
Phylogenetic analysis of Acidobacterium rRNA genes.
Each of the 12 Acidobacterium rRNA genes linked to a specific BAC clone (including 2 from soil library 1), as well as rRNA genes from the Acidobacterium division recovered from direct amplification from SL2, were aligned with a large data set of 16S rRNA gene sequences affiliated with the Acidobacterium division (121 in total), and a phylogenetic analysis was performed on the aligned sequences (Fig. 3). The vast majority of 16S rRNA gene sequences, 58 of 73 Acidobacterium-affiliated sequences recovered from SL2, were affiliated with subdivision 6 (15), which does not have any cultured representatives (Fig. 3). Fourteen of the remaining clones from SL2 affiliated with subdivision 4, and only one rRNA, contained within BAC clone P17F9 (see annotated sequence below), was recovered from subdivision 5. Neither of these has a cultured representative. The rRNA sequences recovered from the rDNA clone collection of 1997 were affiliated with subdivisions 4 (10 sequences), 5 (3 sequences), 6 (9 sequences), and 7 (2 sequences), and 8 sequences failed to place within any of the recognized subdivisions based on maximum-parsimony analysis (data not shown). None of the rDNA clones from any of the collections appeared related to the deeply branching subgroup 8, containing the cultured isolates Holophoga foetida and Geothrix fermentans.
Partial genomic sequence from an Acidobacterium taxon.
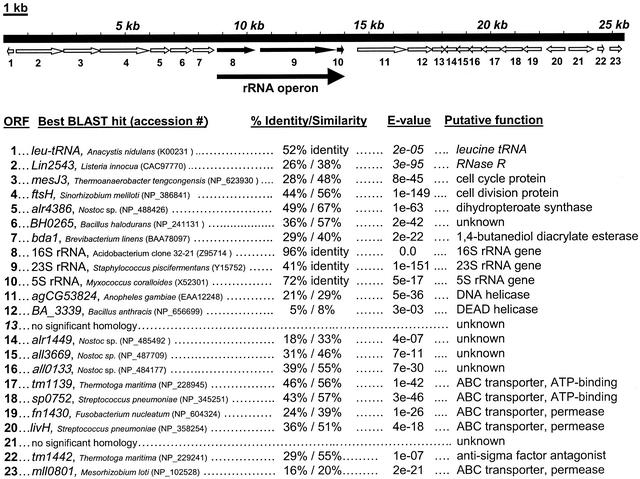
We fully sequenced the insert sequence from BAC clone P17F9, which contains an rRNA gene affiliated with the Acidobacterium subdivision 5, with 84% identity to the 16S rRNA gene of A. capsulatum (Fig. 3). This partial genome sequence has an average of 55.2% G+C content, similar to the 60% G+C content of A. capsulatum (19). Glimmer analysis of the insert DNA revealed 20 predicted ORFs as well as a complete rRNA operon (Fig. 4). BLAST analysis of each predicted ORF revealed predicted gene products involved in cell cycling (mesJ3, ORF 3), cell division (ftsH, ORF 4), folic acid biosynthesis (alr4386, ORF 5), DNA excision repair (agCG53824, ORF 11), transcriptional regulation (tm1442, ORF 19), and a high-affinity ABC transporter for branched-chain amino acids (ORFs 17, 18, 19, and 20), among other predicted gene products (Fig. 4). We also identified a putative gene product with 40% similarity to a novel 1,4-butanediol diacrylate esterase from Brevibacterium linens (31), located immediately upstream of the rRNA operon (ORF 7). Curiously, while ORFs 19 and 20, both putative permeases involved in ABC transporter function, have significant similarity to one another (50% amino acid identity), ORF 23 appears to be only a partial reading frame of 573 bp, lacking both N- and C-terminal sequences, with only 38% amino acid identity to ORF 19. Out of the 23 ORFs predicted by the Glimmer algorithm, two ORFs did not have any significant homology to entries in the GenBank database and four ORFs had significant homology to proteins of unknown function (Fig. 4).
FIG. 4.
Annotated sequence for the BAC clone P17F9. The 25.4-kb insert from P17F9 was shotgun subcloned, and sequences recovered from subclones were assembled into a contiguous sequence at 3.3× coverage. The Glimmer program was employed to search for ORFs, and each identified ORF was compared to the GenBank database of nonredundant sequences by using BLAST algorithms. For each ORF, the percent identity or similarity of the gene or gene product to its nearest relative in the GenBank database is listed along with the E value and putative function of the gene product.
DISCUSSION
This phylogenetic analysis of the SL2 metagenomic library suggests that the methodology employed for library construction resulted in a biased representation of the soil bacterial microflora. Since the same primer sets and PCR conditions were employed in the survey of both the BAC library and the soil microbial DNA, it is likely that the low representation of the Bacillus group reflects an incomplete lysis of bacterial spores or vegetative cells during the isolation of high-molecular-weight genomic DNA and is not a PCR artifact. Our discovery of a BAC clone derived from a Bacillus sp. indicates that some Bacillus spp. did contribute genomic DNA to the metagenomic library, although possibly not in proportion to their natural abundance. In contrast, the higher relative representation of other phylogenetic groups, such as the β- and γ-Proteobacteria and OP10 divisions, may be a consequence of avid cell lysis and release of their respective genomic DNAs during library construction.
The survey reported here is not exhaustive, nor is it a quantitative measurement of rRNA gene relative abundance in WMARS soil or the SL2 metagenomic library. The estimated number of 16S rRNA genes within SL2 depends upon a number of unknown variables. If SL2 contains 200 genomic equivalents based upon an average genome size of 4 Mbp, then the minimum number of rRNA genes within the library would be 200. Based upon recent work suggesting that the number of rRNA operon copies per genome is directly proportional to growth rate in culture (19) and that the vast majority of soil microorganisms have adapted to an oligotrophic existence, we speculate that, on average, the microorganisms represented within a soil metagenomic library have less than two copies of the rRNA operon per genome. Since we have discovered 197 unique rRNA gene sequences from SL2, without observing redundant sequences, we would expect that the total number of rRNA gene sequences in SL2 greatly exceeds 200. The potential biases affecting the identification of BAC clones containing rRNA genes include preferential amplification from the host rDNA template, an inability to differentiate between host and nonhost RFLP patterns, and potential toxicity of heterologously expressed rRNA gene operons. While the latter bias is considered unlikely due to maintenance of BACs in single copy, our laboratory has previously observed decreased cell growth in E. coli expressing an archaeal rRNA gene from a medium-copy vector (33) (H. Simon, J. Dodsworth, and R. Goodman, personal communication).
There are several ways in which screening for rRNA genes within a metagenomic library might be improved from the methods described here. First, by using an inducible-copy BAC (or fosmid) vector for metagenomic library construction, the ratio of cloned rRNA genes to host chromosomal contamination within a BAC DNA preparation would be increased, resulting in less PCR amplification from a host template. Of course, heterologous rRNA operon toxicity must be experimentally determined to not contribute to the loss of BAC clones upon copy induction, at least for the targeted groups, for this strategy to be used. Even with single-copy BAC libraries, it would be significantly easier to screen only for those BAC clones derived from a phylogenetic group of interest, using group-specific probes (35), thereby avoiding host background. Also, by arraying metagenomic libraries on glass slides or other substrates, a more-rapid sequence-based screening for BAC clones containing a specific gene may be accomplished.
While apparently not representative of the soil microfloral diversity from which it was constructed, SL2 is a rich source of genomic sequences from the Acidobacterium division. Of all the rRNA genes recovered from SL2 that are affiliated with the Acidobacterium division, none are affiliated with subdivisions that contain a cultured member (15). By linking the phylogenetic and functional information contained within large insert BAC clones, we may reveal the first glimpses of the physiological capabilities of these uncultured taxa. Indeed, the putative carboxylesterase found within the Acidobacterium clone P17F9 is related to a B. linens enzyme known to catalyze the conversion of insoluble butanediol diacrylate (or derivatives) to a hydrolyzed, soluble form that could potentially be used as a carbon source by the uncultured Acidobacterium taxa whose partial genome we recovered. From this small BAC clone we have already identified genes whose putative functions suggest testable hypotheses regarding ecological roles of this uncultured bacterium.
The analysis of phylogenetically linked genomic sequences from metagenomic libraries provides a variety of molecular tools facilitating both molecular ecology studies and natural product discovery. Since each BAC clone isolated from this study likely contains a complete rRNA operon, the rRNA genes and intergenic sequences allow a finer resolution of phylogenetic analysis, permitting studies of the distribution and natural abundance of uncultured taxa along environmental gradients and within soil microhabitats. Better knowledge of environmental distribution may suggest strategies for capturing the genomic sequences of particular phylogenetic groups. The molecular data acquired from BAC clones in our collection, as well as those from Christa Schleper’s group (T. Ochsenreiter, A. Quaiser, G. Raddatz, S. Schuster, and C. Schleper, Abstr. 10th Int. Conf. Microb. Genomes, p. 56, 2002), will broaden our understanding of the physiology of, and ecological niches inhabited by, members of the Acidobacterium division (13). Furthermore, the molecular data may suggest cultivation strategies for certain phyla by revealing substrate preferences. By using a combination of such experimental approaches, the data acquired from a metagenomic library will open new routes for exploration of the soil microbial world.
Acknowledgments
We thank the other members of the Goodman and Handelsman laboratories for support and critical analysis of this work.
This research was funded by grants from the David and Lucille Packard Foundation and the McKnight Foundation. M.R.L. was supported by NIH NRSA fellowship GM20623-02 and NSF DEB grant 0213048.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable Archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beja, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. [DOI] [PubMed] [Google Scholar]
- 6.Beja, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. [DOI] [PubMed] [Google Scholar]
- 7.Beja, O., E. N. Spudich, J. L. Spudich, M. Leclerc, E. F. DeLong. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786-789. [DOI] [PubMed] [Google Scholar]
- 8.Beja, O., E. V. Koonin, L. Aravind, L. T. Taylor, H. Seitz, J. L. Stein, D. C. Bensen, R. A. Feldman, R. V. Swanson, and E. F. DeLong. 2002. Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Appl. Environ. Microbiol. 68:335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beja, O., M. T. Suzuki, J. F. Heidelberg, W. C. Nelson, C. M. Preston, T. Hamada, J. A. Eisen, C. M. Fraser, and E. F. DeLong. 2002. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630-633. [DOI] [PubMed] [Google Scholar]
- 10.Bintrim, S. B., T. J. Donohue, J. Handelsman, G. P. Roberts, and R. M. Goodman. 1997. Molecular phylogeny of Archaea from soil. Proc. Natl. Acad. Sci. USA 94:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgmann, H., M. Pesaro, F. Widmer, and J. Zeyer. 2001. A strategy for optimizing quality and quantity of DNA extracted from soil. J. Microbiol. Methods 45:7-20. [DOI] [PubMed] [Google Scholar]
- 12.Dojka, M. A., J. K. Harris, and N. R. Pace. 2000. Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria. Appl. Environ. Microbiol. 66:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Goodman, R. M., and M. R. Liles. April2001. Template specific termination in a polymerase chain reaction. U.S. Patent 6,248,567.
- 13.Handelsman, J., M. R. Rondon, S. F. Brady, J. Clardy, and R. M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:R245-R249. [DOI] [PubMed] [Google Scholar]
- 14.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 15.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kishimoto, N., Y. Kosako, and T. Tano. 1991. Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr. Microbiol. 22:1-7. [DOI] [PubMed] [Google Scholar]
- 19.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analysis. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liesack, W., F. Bak, J. U. Kreft, and E. Stackebrandt. 1994. Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch. Microbiol. 162:85-90. [DOI] [PubMed] [Google Scholar]
- 23.Lonergan, D. J., H. L. Jenter, J. D. Coates, E. J. P. Phillips, T. M. Schmidt, and D. R. Lovley. 1996. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J. Bacteriol. 178:2402-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 25.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medlin, L., H. J. Elwood, S. Stickel, and M. L. Sogin. 1988. The characterization of enzymatically amplified eukaryotic 16S-like ribosomal RNA-coding regions. Gene 71:491-500. [DOI] [PubMed] [Google Scholar]
- 27.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 28.Quaiser, A., T. Ochsenreiter, H.-P. Klenk, A. Kletzin, A. H. Treusch, G. Meurer, J. Eck, C. W. Sensen, and C. Schleper. 2002. First insight into the genome of an uncultivated crenarchaeote from soil. Environ. Microbiol. 4:603-611. [DOI] [PubMed] [Google Scholar]
- 29.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 31.Sakai, Y., J. Ishikawa, S. Fukasaka, H. Yurimoto, R. Mitsui, H. Yanase, and N. Kato. 1999. A new carboxylesterase from Brevibacterium linens IFO 12171 responsible for the conversion of 1,4-butanediol diacrylate to 4-hydroxybutyl acrylate: purification, characterization, gene cloning, and gene expression in Escherichia coli. Biosci. Biotech. Biochem. 63:688-697. [DOI] [PubMed] [Google Scholar]
- 32.Salzberg, S. R., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon, H. M., J. A. Dodsworth, and R. M. Goodman. 2000. Crenarchaeota colonize terrestrial plant roots. Env. Microbiol. 2:495-505. [DOI] [PubMed] [Google Scholar]
- 34.Staley, J. T., and A. Konopka. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321-346. [DOI] [PubMed] [Google Scholar]
- 35.Stein, J. L., T. L. Marsh, K. Y. Wu, T. Shizuya, and E. F. DeLong. 1996. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J. Bacteriol. 178:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torsvik, V., J. Goksøyr, F. L. Daae, R. Sørheim, J. Michalsen, and K. Salte. 1994. Use of DNA analysis to determine the diversity of microbial communities, p. 39-48. In K. Ritz, J. Dighton, and K. E. Giller (ed.), Beyond the biomass. John Wiley and Sons, Chichester, England.
- 38.Torsvik, V., R. Sorheim, and J. Goksoyr. 1996. Total bacterial diversity in soil and sediment communities-a review. J. Ind. Microbiol. 17:170-178. [Google Scholar]
- 39.Vergin, K. L., E. Urbach, J. L. Stein, E. F. DeLong, B. D. Lanoil, and S. J. Giovannoni. 1998. Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl. Environ. Microbiol. 64:3075-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Wintzingerode, F., O. Landt, A. Ehrlich, and U. B. Gobel. 2000. Peptide nucleic acid-mediated PCR clamping as a useful supplement in the determination of microbial diversity. Appl. Environ. Microbiol. 66:549-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 42.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]