Abstract
We examined the population of unicellular cyanobacteria (Synechococcus) in the upper 3-mm vertical interval of a 68°C region of a microbial mat in a hot spring effluent channel (Yellowstone National Park, Wyoming). Fluorescence microscopy and microsensor measurements of O2 and oxygenic photosynthesis demonstrated the existence of physiologically distinct Synechococcus populations at different depths along a light gradient quantified by scalar irradiance microprobes. Molecular methods were used to evaluate whether physiologically distinct populations could be correlated with genetically distinct populations over the vertical interval. We were unable to identify patterns in genetic variation in Synechococcus 16S rRNA sequences that correlate with different vertically distributed populations. However, patterns of variation at the internal transcribed spacer locus separating 16S and 23S rRNA genes suggested the existence of closely related but genetically distinct populations corresponding to different functional populations occurring at different depths.
Developments in genetic-sequence-based identification methods and microenvironmental-analysis techniques have significantly enhanced the ability to describe the ecology of bacterial populations in situ (2). Our research has focused on the ecology of phototrophic bacteria in alkaline hot spring microbial mats, where cellular morphologies are of limited use in distinguishing populations. The original descriptions of these mats were relatively simple: a green surface layer containing unicellular cyanobacteria (Synechococcus) overlying an orange layer containing green nonsulfur (Chloroflexus-like) bacteria. Cultivation-independent genetic-sequence-based analysis of 16S rRNA gene segments has been used to sample the diversity and distribution of Synechococcus populations in these communities (33). These studies have shown that numerous genetically distinct Synechococcus populations appear to be adapted to and organized along temperature-defined niches due to thermal gradients in hot spring effluent channels (7, 9). These results were consistent with earlier studies of cultivated Synechococcus that were not genetically characterized (23), as well as recent studies in which isolates were genetically characterized (16). In nearly all instances, two or three Synechococcus sequences were detected at a given temperature by 16S rRNA analysis (7-9, 25).
We investigated the possibility that multiple 16S rRNA sequences at the same temperature site represented Synechococcus populations that coexist by adapting to different light environments within the mat. We analyzed Synechococcus populations over the vertical interval at a 60°C site (25). Here, the degree of 16S rRNA sequence variation between populations is relatively high (e.g., 3.8% [9]), so that members of two phylogenetically well-supported clades, designated Synechococcus OS type A and OS type B (33), cooccur. The results showed an organized pattern of vertical sequence distributions consistent with the idea of light-adapted populations (25). This idea is also consistent with sun- and shade-adapted strains of filamentous thermophilic cyanobacteria (30, 31). Our study also revealed that the Synechococcus layer contained a yellow-green upper layer and a distinct, dark-green subsurface layer of more brightly autofluorescent Synechococcus cells. Similar layers were evident at most temperature sites in the mat.
We hypothesized that cells in the subsurface layer might represent low-light-adapted populations, while those at the surface were high-light adapted. However, in the earlier studies, the light gradients within the mat were not analyzed. Also, because 16S rRNA sequence divergence among Synechococcus populations found at higher temperatures is lower (e.g., <1% within the A cluster [9]), we hypothesized that it might be necessary to use higher-resolution genetic markers to observe vertical population structure at higher-temperature sites. Here, we evaluate these hypotheses.
MATERIALS AND METHODS
Study site.
We studied a microbial mat growing at ∼68°C at the head of the effluent channel of Mushroom Spring, an alkaline hot pool in the Lower Geyser Basin of Yellowstone National Park, Wyoming. This mat and the neighboring Octopus Spring mat that we refer to below have been described previously (25, 33).
Sample collection.
Samples for denaturing gradient gel electrophoresis (DGGE) analysis were collected on 22 August 1999 using a no. 4 cork borer as described previously (25). DGGE patterns were generated for microbial populations over the top 3.5 mm of the mat by separate analysis of 100-μm-thick vertical intervals subsampled parallel to the mat surface using a cryotome as previously described (25). Samples for clone library construction were collected on 17 July 2001 and treated in the field as follows. A section of mat ∼3 cm in diameter and 0.5 cm thick was removed with a weighing spatula and placed in a sterile petri dish. Using a clean razor blade, a slice of this mat was placed on its side, exposing the vertical aspect. Using a dissecting microscope and a sterile no. 27 gauge syringe needle, material from the upper surface of the mat slice was collected and transferred into a clean 1.5-ml conical plastic screw-cap tube containing 50 μl of sterile water. Material from a subsurface, dark-green layer (sampled from ∼2 mm below the mat surface) was obtained similarly. The tubes were frozen on dry ice and stored at −20°C.
Samples for microscopic autofluorescence analysis of surface and subsurface Synechococcus cells were collected on 31 August 2001 using a needle and dissecting microscope as described above, except that these fresh, intact mat samples were processed in the laboratory and were not frozen prior to microscopic analysis. Wet mounts of cell suspensions were prepared on slides coated with Gelrite (Kelco, San Diego, Calif.) to reduce drifting of cells. The slides were coated by dipping them into molten Gelrite, quickly wiping the underside, and placing them on a flat surface to dry. Images of surface and subsurface Synechococcus cells were acquired under equivalent lighting conditions using both phase-contrast and epifluorescence microscopy on an Axioplan microscope equipped with a NEOFLUAR objective, a standard filter set (green excitation, 546 nm; red emission, >590 nm) and an AxioCam digital camera (all from Carl Zeiss Inc., Oberkochen, Germany).
Nucleic acid extraction.
DNA was extracted using slight modifications of a method described by Moré et al. (18). A rice grain-size sample of mat was placed in a 1.5-ml centrifuge tube with 800 μl of phosphate buffer (Na2HPO4-NaH2PO4; 120 mM; pH 8) and 260 μl of sodium dodecyl sulfate solution (10% sodium dodecyl sulfate, 0.1 M NaCl, 0.5 M Tris-HCl, pH 7.5). Zirconium beads (0.1-mm diameter; Biospec Products, Bartlesville, Okla.) were added to half the height of the tube. The samples were shaken for 45 s at the maximum setting (6.5) in a FastPrep bead beater (Savant Instruments Inc., Farmingdale, N.Y.), cooled on ice, and then centrifuged for 1 min. Approximately 600 μl of the supernatant fluid was transferred to a new tube, and proteins were precipitated with 0.4 volume of 10 M ammonium acetate for 5 min on ice. After centrifugation for 10 min, ∼650 μl of the supernatant fluid was transferred to a clean tube, and nucleic acids were precipitated with 0.7 volume of isopropanol for 5 min at room temperature. The samples were centrifuged for 30 min at 4°C, and the pellets were washed once with 70% ethanol, air dried, resuspended in 30 μl of molecular-grade water (Sigma, St. Louis, Mo.), and stored frozen. All centrifugations were at 14,000 rpm (16,000 × g) in a tabletop microcentrifuge.
DGGE genetic fingerprinting.
All reagents, PCR primers, and methods for DGGE were as previously described (7), with two exceptions: (i) gels were run for 17 h at 60 V and (ii) individual DGGE bands were purified for sequencing by touching them with a sterile disposable plastic 10-μl pipette tip and immersing the tip in a PCR mixture. PCR amplifications (Escherichia coli positions 1070 to 1392) were performed using a Taq polymerase kit and a deoxynucleoside triphosphate mixture (Promega Corp., Fitchburg, Wis.) following instructions provided by the manufacturer. The PCR conditions were 94°C for 5 min, followed by 30 cycles of 92°C for 1 min, 55°C for 1 min, and 72°C for 1 min (for 8 min on a final cycle). The PCR products were visualized on ethidium bromide-stained agarose gels containing a 100-bp DNA ladder (Promega). The presence of a single PCR product was confirmed by reanalysis on DGGE. PCRs showing one major band on DGGE were purified for direct sequencing using a PCR product purification kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
Clone libraries.
PCR was used to amplify a segment of cyanobacterial DNA that included ∼750 bp of the 3′ end of the 16S rRNA gene along with the adjoining internal transcribed spacer (ITS) region. An equal mixture of two forward PCR primers was used: 781f (A) (5′-AATGGGATTAGATACCCCAGTAGTC-3′) and 781f (B) (5′-AAAGGGATTAGATACCCCTGTAGTC-3′). These were designed to specifically amplify cyanobacterial 16S rRNA genes (21). The reverse primer, 23cyR (5′-TGCCTAGGTATCCACC-3′) (R. T. Papke and D. M. Ward, unpublished data), targeted bacterial 23S rRNA gene sequences. Clone libraries were generated using a TOPO-TA PCR product cloning kit (Invitrogen, Carlsbad, Calif.) following the manufacturer's instructions. Plasmid DNA was isolated using a miniprep kit (Qiagen) following the manufacturer's instructions.
Sequencing and sequence analysis.
Purified DGGE bands and plasmid inserts were sequenced on an ABI Prism 310 genetic analyzer using AmpliTaq DNA polymerase, FS, and Big Dye terminators (Applied Biosystems, Foster City, Calif.). Raw sequence data were aligned and edited using Sequencher version 3.1.1 software (Gene Codes Corp., Ann Arbor, Mich.). Sequences were compared to those in the National Center for Biotechnology Information database using BLAST (1). Phylogenetic analysis was performed using the neighbor-joining (29) algorithm with the Jukes-Cantor distance option on PAUP* software [D. L. Swofford, PAUP* phylogenetic analysis using parsimony (*and other methods), version 4, Sinauer Associates, Sunderland, Mass., 1998]. We have the greatest confidence in replicated sequences, as previous studies have shown that PCR and/or cloning artifacts occur as singletons in clone libraries (24, 32).
Microsensor analysis.
Measurements of O2 distribution and oxygenic photosynthesis in the microbial mat (25, 27) were done in situ on 22 August 1999 with fast t90 (90% response time of <0.3 to 0.4 s) Clark-type O2 microsensors (26) with a low (<1 to 2%) stirring sensitivity. The microsensors were connected to a sensitive picoampere meter (PA2000; Unisense A/S, Aarhus, Denmark), and measuring signals were recorded on a strip chart recorder (Servogor 110). Both instruments were battery driven.
The microsensors were linearly calibrated from sensor readings in the anoxic part of the mat and in the overlying spring water, the O2 content of which was determined by Winkler titration measurements (10) with 50-ml Winkler bottles. Readings were done under a cloudless sky with downwelling scalar irradiance ranging from 1,800 to 2,300 μmol of photons m−2 s−1, as determined with a quantum scalar irradiance meter (Biospherical Instruments QSL-101) placed in the spring next to the sampling site. The microsensors were positioned in vertical steps of 100 to 200 μm by a manually operated micromanipulator (MM33; Märtzhäuser GmbH, Wetzlar, Germany), taking care that no self-shadowing affected the measurements. Light-dark shifts to measure photosynthetic rates were done by temporarily shading the measuring area with a nontransparent box (26).
Measurements of spectral light penetration in the mat were done on freshly collected samples in the laboratory with fiber optic scalar irradiance microprobes coupled to a sensitive charge-coupled device-based spectrometer (PMA-11; Hamamatsu Corp., Hamamatsu City, Japan) (11, 12, 14). Undisturbed mat samples were taken from the field site on 22 August 1999 in small glass coring devices and transported to the laboratory within 1 h for further analysis on the same day. The samples were incubated in spring water and illuminated vertically from above with a fiber-optic halogen lamp (KL-1500; Schott GmbH, Mainz, Germany). The scalar irradiance microprobe (with a spherical tip diameter of ∼80 μm) was inserted into the mat at a zenith angle of 145° in steps of 100- to 200-μm vertical distance by using a manually operated micromanipulator (Märtzhäuser). For each step, the spectral scalar irradiance from 380 to 800 nm was recorded on a computer controlling the spectrometer. Reference spectra of incident downwelling irradiance were recorded with the microprobe tip positioned over a black-light trap at a distance from the light source identical to that of the mat surface.
Nucleotide sequence accession numbers.
The sequences have been submitted to GenBank with accession numbers AF550092 to AF550127.
RESULTS
Microscopy.
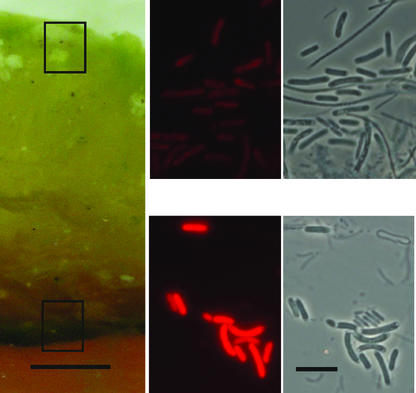
Three distinct layers were visible in cross sections of the mat (Fig. 1): an upper yellow-green ∼2- to 3-mm-thick translucent layer and, below that, a darker-green ∼0.2-mm-thick layer, followed by a multiple-banded orange layer (only the surface of the orange layer is shown in the figure) that extended to the substratum. Synechococcus cells in the surface and subsurface green layers are indistinguishable under phase-contrast microscopy. Each layer contained rod-shaped Synechococcus cells and filamentous bacterial cells. The Synechococcus cells in the subsurface layer were notably more brightly autofluorescent than those in the surface layer. This was similar to previous observations of a 60°C mat sample (25). The surface yellow-green Synechococcus layer at 68°C was notably thicker and appeared more translucent than at 60°C, where the layer was only ∼0.5 mm thick.
FIG. 1.
Cross section of 68°C Mushroom Spring mat showing ∼2- to 3-mm-thick upper translucent yellow-green layer and deeper dark-green layer. The boxes indicate the approximate areas sampled for clone libraries and microscopy; autofluorescence and phase-contrast views are on the right. Bars = 1.0 mm (left panel) and 10.0 μm (photomicrograph panels).
Microsensor analysis.
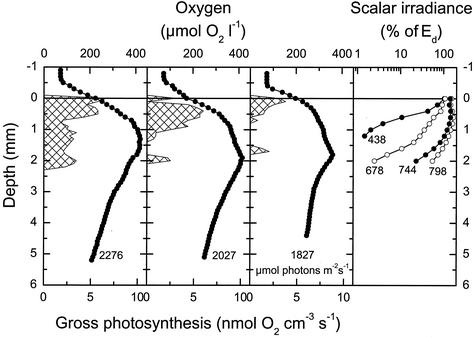
Oxygenic photosynthesis in the upper 2 mm of the mat resulted in supersaturation of the mat with respect to O2 (Fig. 2). Below the photic zone, O2 was only slowly depleted with depth, and the mat was fully oxic under the high-irradiance conditions prevailing during our measurements. Gross photosynthetic activity was confined to a 0.5- to 1-mm-thick surface layer and a less clearly defined subsurface layer ∼1.5 to 2 mm below the mat surface, which corresponded to the depth horizon where cells with high autofluorescence were observed (Fig. 1).
FIG. 2.
Depth profiles of O2 (circles), oxygenic photosynthesis (cross-hatched areas), and scalar irradiance in the 68°C Mushroom Spring microbial mat. O2 and photosynthesis profiles were measured in situ with O2 microsensors in three different positions within the same 1 to 2 cm2 of mat under an ambient downwelling scalar irradiance of 1,827 to 2,276 μmol of photons m−2 s−1. The numbers on light profiles indicate the wavelengths. Scalar irradiance values were normalized to the incident downwelling irradiance, Ed, at the mat surface.
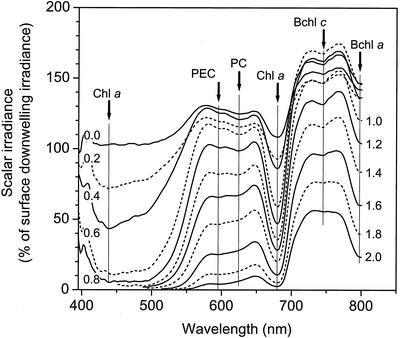
The spectral attenuation of light exhibited different zones of low and high absorbance (Fig. 2 and 3). Blue light (438 nm) was attenuated to 1% of the incident irradiance ∼1 mm below the surface. Light at the absorption maximum of chlorophyll a (678 nm) showed less intense attenuation over the upper part of the photic zone, but attenuation increased significantly ∼1.5 to 2 mm below the mat surface (Fig. 2), encompassing the zone where a dense green band of highly autofluorescent Synechococcus cells was observed (Fig. 1). The scalar irradiance spectra also showed the presence of phycoerythrocyanin (594 nm) and phycocyanin (624 nm), presumably contributed by cyanobacteria, in the upper 1.5 to 2 mm of the mat (Fig. 3).
FIG. 3.
Spectral scalar irradiance measured in the 68°C Mushroom Spring microbial mat. The numbers on the curves indicate the depth below the mat surface, while the arrows and vertical lines indicate the major absorption wavelengths of the predominant photopigments (Chl a, 438 and 678 nm; phycoerythrocyanin (PEC), 594 nm; phycocyanin (PC), 624 nm; Bchl c, 744 nm; Bchl a, 798 nm). The data were normalized to the downwelling spectral irradiance measured at the mat surface.
Scattering of near-infrared (NIR) light in the translucent surface mat resulted in a local maximum of NIR light in the upper 1 to 1.5 mm of the mat, reaching 150 to 170% of the incident irradiance (Fig. 3). Below 1.4 mm, NIR light was attenuated, and we found clear spectral signals of bacteriochlorophyll (Bchl) c (744 nm) and Bchl a (800 nm) absorption in the mat due to the presence of green nonsulfur bacteria and other filamentous phototrophic bacteria (Fig. 2 and 3) (20).
DGGE.
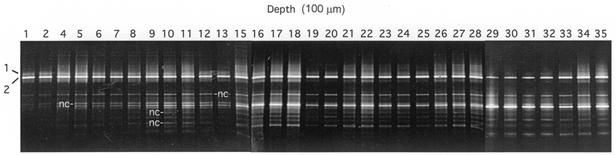
DGGE profiles of a vertical series of 100-μm-thick mat sections were obtained (Fig. 4). Only bands 1 and 2 contained cyanobacterial sequences. The partial (253-bp) sequences obtained from these bands encompass, and are identical to, a short (176-bp) segment that defines an uncultivated cyanobacterial genotype, OS type A′ (GenBank accession number U42374), originally detected in neighboring Octopus Spring (7). The presence of two OS type A′-like sequences in this 68°C region of the Mushroom Spring mat is typical of what we have observed from more extensive DGGE analyses of similar temperature regions of the Octopus Spring mat (7, 9). Although the partial sequences of bands 1 and 2 are identical, they must not be identical over their entire lengths (358 bp), since they migrated to different points in denaturing gradient gels. Band 1 was much less intense than band 2, and bands corresponding in position were present essentially throughout the vertical profile. No pattern in their intensities was observed that might reasonably be interpreted as fluctuations in in situ Synechococcus populations adapted to surface or subsurface conditions.
FIG. 4.
DGGE patterns of PCR-amplified 16S rDNA segments. Each lane represents a 100-μm vertical interval through the microbial mat. Bands 1 and 2 are cyanobacterial sequences; the bands labeled nc are noncyanobacterial sequences; the sequences of the unlabeled bands were not obtained.
Clone libraries.
Nucleic acids were isolated from cells obtained from the translucent yellow-green surface Synechococcus layer and from the dark-green subsurface Synechococcus layer. The approximate locations from which mat material was collected are indicated in Fig. 1. PCR-generated clone libraries were prepared from each of these samples, using primers that amplified a segment of the 16S rRNA gene and the adjacent ITS region. The sequenced 16S rRNA region included a 417-bp segment proximal to position 1085 (E. coli reference), which encompassed the region obtained in DGGE analysis. The sequenced ITS region included a 396-bp segment proximal to the 16S rRNA gene.
Twenty clones from each library were sequenced through the 16S rRNA segment. Thirty-six of these carried cyanobacterial sequences, 19 of which were from the surface library. Less than 1.0% 16S rRNA sequence diversity separated all cyanobacterial clones. Nearly half the clones in the surface and subsurface libraries were identical and matched the OS type A′ sequence and the DGGE bands mentioned above. The second most abundant 16S sequence, represented by five clones, differed from the most abundant sequence by 1 nucleotide and was detected two or more times in each library. Repeated detection of these two sequences in surface and subsurface libraries suggested that neither represented a surface- or subsurface-adapted population. All of the remaining clones differed from each other, and from the most abundant cloned genotype, by at least 1 nucleotide. A phylogram relating the genetic distances between 16S rRNA sequences to their surface and subsurface origins revealed no pattern that suggested genetically distinct Synechococcus populations adapted to surface and subsurface environments (data not shown).
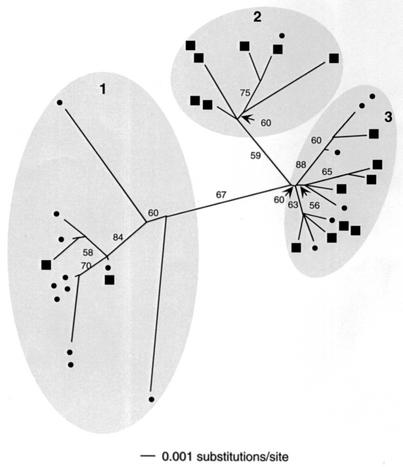
Sequences through the ITS regions of 35 of the above-mentioned clones were obtained. Sequence diversity among ITS segments was higher (up to 4.0%) than that among 16S rRNA segments. A phylogram relating the genetic distance between ITS sequences to their surface or subsurface origin is given in Fig. 5. Three major sequence clusters are indicated. Within cluster 1, most sequences (11 of 13) are from the surface library. Within cluster 2, most (seven of eight) are from the subsurface library. Within cluster 3, both libraries are more evenly represented, with six surface and eight subsurface sequences. Nearly all members of clades 1 and 2 have identical 16S rRNA sequences in the region analyzed. The second well-replicated 16S rRNA genotype was exclusively observed in clade 3.
FIG. 5.
Unrooted neighbor-joining tree of thermophilic Synechococcus sequences based on 396 nucleotides of the ITS region adjacent to the 16S rRNA gene. Circles, surface library sequences; squares, subsurface sequences. Identical sequences are positioned together at terminal branches. Bootstrap values over 50% are shown.
DISCUSSION
This study focused on the Synechococcus organisms in a 68°C region of the Mushroom Spring microbial mat. Microscopic examination revealed Synechococcus layers analogous to those previously reported for a 60°C site in this mat (25). However, the upper Synechococcus layer of the 68°C mat was thicker and more translucent than that in the 60°C mat. The thicker surface layer was underlain by a dark green Synechococcus layer that was ∼2 to 3 mm below the surface as opposed to ∼0.5 mm at the 60°C site. We reasoned that while the expected genetic diversity of Synechococcus 16S rRNA sequences at 68°C is lower than at 60°C (9), the increased distance between the mat's surface and the dark-green layer might enhance our ability to resolve any genetically and/or functionally distinct Synechococcus vertical population structure in this higher-temperature region.
Many physical and physiological measurements, such as highly autofluorescent Synechococcus cells forming a distinct subsurface layer (Fig. 1), subsurface inflections in O2 concentration profiles, and peaks in photosynthetic rates (Fig. 2), provided evidence for the presence of physiologically distinct groups of Synechococcus cells in different depth horizons within the mat at 68°C. Also, notably different light environments measured within the mat's surface and subsurface layers (Fig. 3) clearly identified environmental differences that could have affected the adaptation of Synechococcus populations. However, neither DGGE nor cloning analysis of PCR-amplified 16S ribosomal DNA (rDNA) fragments from the mat's surface and subsurface environments revealed a pattern consistent with vertical stratification of cyanobacterial genotypes.
The lack of a detectable trend in cyanobacterial 16S rRNA DGGE band intensities with depth in the 68°C Mushroom Spring mat contrasts with earlier findings using the same analysis at a 60°C site in the same spring (25). The lack of a trend at the 68°C site could have been an artifact of methodology because of PCR primer competition. The PCR primers used in our DGGE analysis were general bacterial primers that amplified noncyanobacterial as well as cyanobacterial 16S rRNA sequences (7). Template competition from noncyanobacterial sequences, and their concomitant variation in concentration with depth, could have masked any trend in variation of cyanobacterial DGGE band intensities. To eliminate this possibility, we incorporated a cloning approach in our vertical population study that allowed cyanobacterial sequences to be scored independently of noncyanobacterial sequences. Both methods indicated that the mat was dominated by cyanobacterial OS type-A′-like sequences in surface and subsurface layers.
Since different PCR primer sets were used in the DGGE and cloning analyses of the 68°C mat, the observation of OS type A′-like cyanobacterial sequences in surface and subsurface layers provides a measure of assurance that dominant cyanobacterial populations were accounted for and that other dominant non-OS type A′-like populations were not missed due to PCR primer bias. The results also suggest that the abundance of OS type A′-like populations over the vertical interval was a stable characteristic of the late-summer 68°C mat, since samples for DGGE and cloning were collected in July and August of different years. The lack of an apparent light-induced shift in 16S rRNA-defined thermal cyanobacterial-mat populations is not unprecedented. A similar lack of variation in 16S rDNA DGGE band patterns was reported for cyanobacteria in Octopus Spring mats in which population responses to reductions in light intensity and UV light exposure were examined (19).
One interpretation of the lack of a pattern in cyanobacterial 16S rRNA variation with depth is that the physiologically distinct Synechococcus cells occurring at different depths represent populations acclimated to light or other parameters that vary with depth. At low light, cells would be expected to produce more pigment and should therefore exhibit brighter autofluorescence. We explored an alternative hypothesis: that Synechococcus populations evolved by adaptation to fill discrete light-defined niches but the genetic variation of the 16S rRNA sequence is insufficient to reflect the difference. The highly variable ITS region of bacterial rRNA gene operons has been used to study strains whose 16S rRNA gene sequences are nearly identical (3, 4, 13, 15). A recent study of ITS sequences of cultivated open-ocean marine cyanobacterial isolates revealed that subtle genetic divergence patterns within established 16S rRNA clusters are indicative of putative ecotypes whose unique niches have yet to be identified (28). We conducted a phylogenetic analysis using the ITS sequences of Synechococcus populations detected in situ and compared their relatedness to their surface or subsurface origin. Variation among ITS sequences was high (up to 4.0%) relative to that of 16S rRNA gene segments (<1.0%). A pattern emerged in which groups of related sequences shared a common surface or subsurface origin (Fig. 5, shaded regions 1 and 2, respectively). The physical, physiological, and genetic evidence supports the inference that these two ITS clades correspond to distinct ecological populations. Differences in autofluorescence intensities suggest that individual Synechococcus populations are most likely adapted to different light regimes (17), though they are possibly also adapted to other, unmeasured parameters that make the niches at the two vertical positions different. Measurements of physical and chemical parameter and ITS genotype distributions were made in different years; however, we feel the comparison is justified by observations that both types of parameters are stable over a large number of years in cyanobacterial mats in Yellowstone alkaline siliceous springs (33; M. Kühl, unpublished data).
Although the evolution of ecological novelty (i.e., adaptation) is often important for initiating bacterial speciation (5, 6) it takes time for mutations in neutral markers, such as 16S rRNA, to accumulate. Detection of ecotypical populations that are more recently evolved may require more highly variable genetic markers (22). Our results suggest that analysis of ITS sequences is useful in revealing subtle genetic divergence patterns in thermophilic Synechococcus ecotypes that might not be revealed by analysis of 16S rRNA sequences. The existence of a third ITS clade with equal representation from both surface and subsurface samples may suggest that another ecotype exists in the 68°C mat, perhaps exhibiting broader spatial distribution. It is also possible that subcluster branching (Fig. 5) reflects ecotypically different populations that exhibit even less genetic difference at the ITS locus. It is important to detect ecologically distinct populations, no matter how genetically different they are, as these are the populations that will distribute uniquely into different niches and respond uniquely in dynamic environments.
Acknowledgments
This study was funded by awards from the National Science Foundation (D.M.W.; BSR-9708136), the National Aeronautics and Space Administration (D.M.W.; NAG5-8824), and the Danish Natural Science Research Council (M.K.; contract no. 9700549).
We thank the following people for excellent technical assistance: Anni Glud for construction of microsensors and field assistance, Mary Bateson for DNA sequencing, and David Patterson (supported by the Montana State University Thermal Biology Institute) for digital photomicrographs. Unisense A/S is thanked for providing the picoampere meter used in this study. We also thank R. Wiegert for providing access to the University of Georgia research trailer in West Yellowstone.
Footnotes
Journal series no. 2003-02, Montana Agricultural Experiment Station, Montana State University—Bozeman.
REFERENCES
- 1.Altschul, S., T. Madden, A. Schaeffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R., and M. Kühl. 1998. In situ methods for assessment of microorganisms and their activities. Curr. Opin. Microbiol. 1:352-358. [DOI] [PubMed] [Google Scholar]
- 3.Barry, T., G. Colleran, M. Glennon, L. K. Dunican, and F. Gannon. 1991. The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1:51-56. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, H., K. Jørgensen, and J. E. Olsen. 1999. Differentiation of Campylobacter coli and C. jejuni by length and DNA sequence of the 16S-23S rRNA internal spacer region. Microbiology 145:99-105. [DOI] [PubMed] [Google Scholar]
- 5.Cohan, F. M. 2001. Bacterial species and speciation. Syst. Biol. 50:513-524. [DOI] [PubMed] [Google Scholar]
- 6.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Environ. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 7.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferris, M. J., S. C. Nold, N. P. Revsbech, and D. M. Ward. 1997. Population structure and physiological changes within a hot spring microbial mat community following disturbance. Appl. Environ. Microbiol. 63:1367-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferris, M. J., and D. M. Ward. 1997. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 63:1375-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grasshoff, K., M. Ehrhardt, and K. Kernling. 1983. Methods of seawater analysis. Verlag Chemie, Weinheim, Germany.
- 11.Kühl, M., and T. Fenchel. 2000. Bio-optical characteristics and the vertical distribution of photosynthetic pigments and photosynthesis in an artificial cyanobacterial mat. Microb. Ecol. 40:94-103. [DOI] [PubMed]
- 12.Kühl, M., C. Lassen, and B. B. Jørgensen. 1994. Optical properties of microbial mats: light measurements with fiber-optic microprobes, p. 149-165. In Microbial mats: structure, development and environmental significance. L. J. Stal, and P. Caumette, (ed.), Springer, Berlin, Germany.
- 13.Laloui, W., K. A. Palinska, R. Rippka, F. Partensky, N. Tandeau de Marsac, M. Herdman, and I. Iteman. 2002. Genotyping of axenic and non-axenic isolates of the genus Prochlorococcus and the OMF-‘Synechococcus’ clade by size, sequence analysis or RFLP of the internal transcribed spacer of the ribosomal operon. Microbiology 148:453-465. [DOI] [PubMed] [Google Scholar]
- 14.Lassen, C., H. Plough, and B. B. Jørgensen. 1992. A fiber-optic scalar irradiance microsensor: application for spectral light measurements in sediments. FEMS Microbiol. Ecol. 86:247-254. [Google Scholar]
- 15.Leblond-Bourget, N., H. Philippe, I. Mangin, and B. Decaris. 1996. 16S rRNA and 16S to 23S internal transcribed spacer sequence analyses reveal inter- and intraspecific Bifidobacterium phylogeny. Int. J. Syst. Bacteriol. 46:102-111. [DOI] [PubMed] [Google Scholar]
- 16.Miller, S. R., and R. W. Castenholz. 2000. Evolution of thermotolerance in hot spring cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 66:4222-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 18.Moré, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen. 1994. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norris, T. B., T. R. McDermott, and R. W. Castenholz. 2002. The long-term effects of UV exclusion on the microbial composition and photosynthetic competence of bacteria in hot-spring microbial mats. FEMS Microbiol. Ecol. 39:193-209. [DOI] [PubMed] [Google Scholar]
- 20.Nübel, U., M. M. Bateson, V. Vandieken, A. Wieland, M. Kühl, and D. M. Ward. 2002. Microscopic examination of distribution and phenotypic properties of phylogenetically diverse Chloroflexaceae-related bacteria in hot spring microbial mats. Appl. Environ. Microbiol. 68:4593-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palys, T., L. K. Nakamura, and F. M. Cohan. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145-1156. [DOI] [PubMed] [Google Scholar]
- 23.Peary, J. A., and R. W. Castenholz. 1974. Temperature strains of a thermophilic blue-green alga. Nature 202:720-721. [Google Scholar]
- 24.Qiu, X., L. Wu, H. Huang, P. E. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramsing, N. B., M. J. Ferris, and D. M. Ward. 2000. Highly ordered vertical structure of Synechococcus populations within the one-millimeter-thick photic zone of a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 66:1038-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Revsbech, N. P., P. B. Christensen, and L. P. Nielson. 1989. Microelectrode analysis of photosynthetic and respiratory processes in microbial mats, p. 153-162. In Y. Cohen, and E. Rosenberg (ed.), Microbial mats: physiological ecology of benthic microbial communities. American Society for Microbiology, Washington, D.C.
- 27.Revsbech, N. P., and D. M. Ward. 1984. Microelectrode studies of interstitial water chemistry and photosynthetic activity in a hot spring microbial mat. Appl. Environ. Microbiol. 48:270-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocap, G., D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68:1180-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Sheridan, R. P. 1979. Seasonal variation in sun-shade ecotypes of Plectonema notatum (Cyanophyta). J. Phycol. 15:223-226. [Google Scholar]
- 31.Sheridan, R. P., and T. Ulik. 1976. Adaptive photosynthesis responses to temperature extremes by the thermophilic cyanophyte Synechococcus lividus. J. Phycol. 12:255-261. [Google Scholar]
- 32.Speksnijder, A. G. C. L., G. A. Kowalchuk, S. D. Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward, D. M., M. J. Ferris, S. C. Nold, and M. M. Bateson. 1998. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62:1353-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]