Abstract
Insertional mutagenesis was applied for the first time to a fungal biocontrol agent, Pseudozyma flocculosa, in an attempt to obtain mutants with altered antagonistic properties. Transformants were obtained via DNA-mediated transformation. Molecular analyses of the transformants revealed that multiple copies of the plasmid were integrated in tandem at one to many chromosomal loci. The transformants were screened for their biocontrol properties using standard bioassays, and the 160 tested transformants were classified into four groups: group I mutants (22 transformants) showed a stronger antagonistic effect than the wild type (WT) while those of group II (107 transformants) had a comparable antagonistic effect; group III mutants (17 transformants) had a decreased antagonistic effect relative to WT and group IV mutants (14 transformants) had lost their biocontrol properties. Culture extracts of the mutants (group IV) and WT were analyzed and compared for the presence of active metabolites which were then separated by solid-phase extraction and purified using conventional methods. Nuclear magnetic resonance experiments and analytical studies on a metabolite specifically produced by the WT revealed the presence of 2-(2′,4′-diacetoxy-5′-carboxy-pentanoyl) octadecyl cellobioside (flocculosin), a novel glycolipid with strong antifungal properties; the production of this compound would account for the biocontrol activity of P. flocculosa.
Biocontrol agents of plant diseases are a particularly interesting and underinvestigated source of biologically active compounds, because antibiosis is a frequent mechanism by which they exert their antagonistic activities. For example, Pseudozyma flocculosa (Traquair, L. A. Shaw et Jarvis) Boekhout et Traquair (syn. Sporothrix flocculosa Traquair, Shaw et Jarvis) is a basidiomycete fungus related to anamorphs of Ustilaginales (5) which displays strong antagonistic activity against powdery mildew fungi (4). As a promising biocontrol agent, P. flocculosa has been studied extensively with regards to its mode of action. A number of microscopical and chemical studies have suggested that the principal mode of action of P. flocculosa is antibiosis and that production of fatty acids by the fungus plays an important role in its biocontrol potential (2, 4, 14). For this role to be confirmed, the creation of mutants with diminished or no antagonistic properties would be invaluable; however, the molecular biology and genetics inherent to the activity of P. flocculosa are poorly understood. Recently, our laboratory has established transformation-mediated genetic manipulation methods for this fungus (8); this creates opportunities to exploit and characterize the genome structure of this fungus and the molecular and chemical mechanisms involved in biocontrol activity.
Insertional mutagenesis by DNA-mediated transformation has been well developed in recent years. Fungal transformation with DNA that does not exhibit homology with the host's genome results in heterologous integration into the genome; this makes it possible to use the transforming DNA as an insertional mutagen to disrupt genes. Insertional mutagenesis has been used as a genetic tool to mutagenize and tag genes in several fungal species, including Saccharomyces cerevisiae (27), Dictyostelium discoideum (17), Cochliobolus heterostrophus (20), Neurospora crassa (11), Ustilago maydis (6), Alternaria alternata (1), Coprinus cinereus (13), and Magnaporthe grisea (3, 28, 30). However, it has never been applied to fungal biocontrol agents, where it could be a promising approach to unveil genes or gene products involved in the antagonistic activity of the fungus.
In this work, we report the first production of insertional mutants of P. flocculosa generated by genetic transformation. Through this successful transformation, we were able (i) to create several mutants, some of which had completely lost their biocontrol activity, and (ii) to discover an unusual and rare cellobiose lipid with antifungal activity absent in deficient mutants of P. flocculosa.
MATERIALS AND METHODS
Fungal material.
P. flocculosa wild-type (WT) strain PF-1 (ATCC 64874) was used in this study. Stock cultures were maintained at 4°C on YMPD (yeast extract, 3 g/liter; malt extract, 3 g/liter; peptone, 5 g/liter; dextrose, 10 g/liter) agar (Difco, Detroit, Mich.), and biomass was grown in the same medium at 25°C in a broth under constant agitation (150 rpm) in indented flasks. Potato dextrose agar (PDA) (Difco, Sparks, Md.) medium and PDB (Difco, Detroit, Mich.) broth were used for bioassays. Phomopsis sp. was used as a preliminary screening method to evaluate the antagonistic activities of transformants. Sphaerotheca fuliginea (Schlechtend.:Fr.) Pollacci, the fungal pathogen responsible for cucumber powdery mildew, was maintained on Cucumis sativus L. cv. Corona. Pythium aphanidermatum (Edson) Fitzp., Botrytis cinerea Pers.:Fr., Phytophthora infestans (Mont.) de Bary, Fusarium oxysporum f. sp. radicis-lycopersici Jarvis et Shoemaker, and Idriella bolleyi (Sprague) Arx were maintained on PDA medium. Candida albicans (Robin) Berkhout (ATCC 18804 and 66027) cultures were maintained on YMG (yeast extract, 4 g/liter; malt, 10 g/liter; glucose, 4 g/liter) agar medium. These fungi were used in bioassays to investigate the antifungal spectrum of any compound(s) discovered.
Fungal transformation.
Protoplasts of P. flocculosa WT were prepared according to the method Cheng and Bélanger (7). The plasmid pSceI-Hyg, which contains the promoter and terminator sequences of the hsp70 gene from U. maydis and an hph gene from E. coli conferring resistance to hygromycin B (Boehringer Mannheim, Germany), was linearized using the restriction endonuclease XhoI and then transferred into protoplasts by adding polyethylene glycol-CaCl2 (BDH, Poole, England), as described by Cheng et al. (8). Transformants were selected on YMPDA medium amended with 0.8 M sucrose as osmotic stabilizer and 50 μg/ml of hygromycin B as screening agent.
Preliminary screening of transformant antagonistic activity with Phomopsis sp.
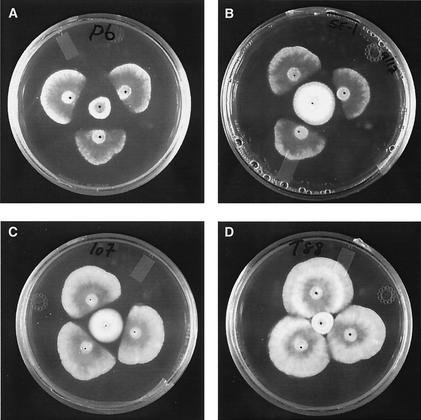
P. flocculosa WT and the transformants were grown on petri plates containing 15 ml of PDA for one week at room temperature. A 5-mm agar plug from actively growing WT or transformant colonies was inoculated in the center of sterile agar plates and incubated at 22°C for 24 h. Three inoculation plugs from an actively growing colony of Phomopsis sp. were then placed around the P. flocculosa colony at a distance of 1.5 cm (Fig. 1). After 72 h of incubation, the distance between the margins of the colonies of transformants and Phomopsis sp. was recorded. The growth of Phomopsis sp. was recorded as the distance from the inoculum to the margin side of the colony facing the transformants of P. flocculosa (Fig. 1). For each transformant, three replicate plates were produced (Table 1).
FIG. 1.
Bioassay of P. flocculosa and transformants against Phomopsis sp. The strains in the center of the petri dishes are PF6 (group I) (A), PF-1 (group II) (B), PF107 (group III) (C), and PF88 (group IV) (D). Three agar plugs of Phomopsis sp. were inoculated 1.5 cm from the center. Results were recorded following incubation for 3 days at 22°C.
TABLE 1.
Growth inhibition of Phomopsis sp. in the presence of P. flocculosa WT and transformant strains
| Group | No. of transformants | Growth of Phomopsis sp. (mm)a |
|---|---|---|
| I | 22 | 0.1 ± 0.2 A (2.6) |
| II | 107 | 1.8 ± 0.7 B (1.6) |
| III | 17 | 3.7 ± 0.8 C (0.3) |
| IV | 14 | 11.4 ± 1.6 D (0.0) |
Data shown are means ± standard deviations based on three replicates and nine observations for each transformant. The WT strain (PF-1) is in group II. The number in parentheses indicates the average distance between the colonies of both fungi at the time of measurement. Within a column, mean values followed by a different letter are significantly different according to Fisher's protected least-significant-difference test (P = 0.05).
Secondary screening of transformant antagonistic activity with S. fuliginea on infected cucumber leaves.
Mycelial disks (5 mm) from P. flocculosa transformants and WT were incubated in PDB broth for 3 days. Foliar disks (5 cm) from cucumber leaves infected with S. fuliginea (disks were cut from leaf portions covered with ca. 95% S. fuliginea colonies) were transferred to petri dishes containing 0.2% N-P-K (20:20:20, percent weight) fertilizer and 0.8% Bacto Agar. Leaf disks were then sprayed with a suspension of transformants and WT containing ca. 104 conidia/ml and incubated at room temperature for 48 h. Sterile PDB broth served as a control. Conidial chains of S. fuliginea were observed using a dissecting microscope (Olympus SZ-PT, Fukuoka, Japan), and rated based on their level of collapse as described by Jarvis et al. (15) (0 = no collapse, 1 = 1 to 25% collapse of conidial chains, 2 = 26 to 50%, 3 = 51 to 75%, 4 = 76 to 100% collapse). This experiment was repeated three times as described in Table 2.
TABLE 2.
Bioassay of P. flocculosa transformants with Sphaerotheca fuliginea
| Strain | No. of strains tested | Mean extent of conidial chain collapse of S. fuliginea ± SDa |
|---|---|---|
| Control (n = 9) | 1 | 0 ± 0 |
| PF-1 (WT strain) (n = 9) | 1 | 4 ± 0 |
| Group I (n = 45) | 5 | 4 ± 0 |
| Group IV (n = 54) | 6 | 0 ± 0 |
Data indicate mean of conidial collapse, where 0 = no effect on S. fuliginea conidial chains, 1 = 1 to 25% collapse 2 = 26 to 50% collapse, 3 = 51 to 75% collapse, and 4 = 76 to 100% collapse. Observations were made after 24 h of treatment. Transformants were cultured in PDB broth for 3 days, and PDB broth without inoculum served as control.
Genetic analyses.
Genomic DNA was extracted from freeze-dried transformants which were maintained on hygromycin B liquid media according to the protocol of Möller et al. (21). Southern blot analysis was performed using standard methods (25). Chromosomal DNA plugs were prepared using previously described methods (8), and contour-clamped homogeneous electric field (CHEF) electrophoresis was carried out in 1% SeaKem GTG (FMC Bioproducts, Rockland, Maine) agarose in a 0.5× Tris-borate-EDTA running buffer at 14°C, using a CHEF-DRIII apparatus (Bio-Rad, Hercules, Calif.). The switching interval used was 90 to 180 s, with total run times of 33 h, at 4.4 V/cm, and the included angle between the electric fields was 120°. The gel was transferred to a GeneScreen plus membrane (NES Research Products, Boston, Mass.) in 0.4 N NaOH as recommended by the manufacturer. Plasmid DNA probes were prepared with 32P using an oligolabeling kit (Pharmacia, Piscataway, N.J.). Hybridization was performed at 65°C overnight, prior to washing the membrane twice with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 1% sodium dodecyl sulfate for 20 min at 65°C.
Comparison and bioassay of WT and transformant extracts to characterize antifungal compounds.
WT and transformant cultures including six stable group IV mutants and three group I mutants were grown at 25°C in petri dishes containing 20 ml of Czapek Dox broth (Difco, Sparks, Md.) supplemented with 0.4% Phytagel (Sigma, Steinheim, Germany) and hygromycin B (50 μg/ml; for transformants only) for 8 days. Cultures as well as medium were collected, freeze-dried, weighed, ground to a fine powder, and subjected to extraction in 80% methanol (MeOH) (ca. 10 g per 100 ml of MeOH). The extracts were then filtered using filter paper (Whatman no. 1), and MeOH was evaporated using a rotary evaporator (model R-114; Büchi, Flawil, Switzerland) until only water remained. Sep-Pak C18 cartridges (Waters, Milford, Mass.) were used to fractionate the remaining aqueous extracts. Extracts were injected individually into preconditioned cartridges, and the overflow (the first fraction) was collected. Cartridges were then rinsed in sequence with five solutions of differing H2O/MeOH ratios: 100% H2O, 80% H2O:20% MeOH, 50% H2O:50% MeOH, 20% H2O:80% MeOH, and 100% MeOH. Each fraction was collected individually, dried, dissolved in MeOH, and bioassayed.
A 10-mm well was made in the center of petri dishes containing 20 ml of PDA medium. A 200-μl aliquot of each fraction was placed in the well of each plate prior to being inoculated with Phomopsis sp. (a 5-mm inoculation plug was placed 2 cm from the well). The growth of Phomopsis sp. was recorded after incubating at room temperature for 3 days. The culture medium (with or without hygromycin B) was used as control. The fraction(s) with the strongest antifungal activities was dried, dissolved in MeOH, and further purified using preparative thin-layer chromatography (PTLC) on silica gel plates (60 F 254, 0.1 mm thick, 5 ×20 cm; Merck, Darmstadt, Germany). The chromatograms were developed in ethyl acetate-formic acid-acetic acid-H2O (100:11:11:27, vol/vol). In order to visualize non-UV absorbing compounds (compounds with no chromatophore), PTLC plates were visualized with iodine vapor and with an anisaldehyde-sulfuric acid spray reagent (12). Chromatophoric bands containing fungal metabolites appeared as dark green spots and their retention values (Rf) were calculated. In particular, one compound (later named flocculosin), present within WT and group I mutant extracts and absent in group IV mutant extracts, was purified using PTLC and Sep Pak C18 cartridges and bioassayed with Phomopsis sp. to evaluate its fungitoxicity.
Structural elucidation of flocculosin. (i) NMR spectra.
1H nuclear magnetic resonance (NMR), correlation spectroscopy (COSY), heteronuclear multiple quantum coherence (HMQC), totally correlated spectroscopy (TOCSY), 13C NMR, distortionless enhancement by polarization transfer (using a 90 and 135° decoupler pulse), and 31P NMR of flocculosin were recorded using a Bruker DMX-600 NMR spectrometer in MeOH-d4 as solvent using a 40-mg sample of pure freeze-dried compound isolated from the WT strain. All chemical shifts are given in parts per million relative to solvent residual peaks, and couplings, where possible, are given in Hz. The mass of flocculosin was determined by fast atom bombardment mass spectroscopy (FABMS) and liquid chromatography mass spectroscopy-electrospray ionization (LCMS-ESI) analysis. FABMS spectra were recorded using a VG Autospec-Q instrument with LS1MS source at 20 kV and 2 μA of current. LCMS-ESI spectra were recorded using a Jaguar ChromaTOF (LECO, St. Joseph, Mich.) instrument with an Agilent (Palo Alto, Calif.) 1100 series high-performance liquid chromatograph (HPLC). LCMS-ESI spectra were processed and analyzed using the Jaguar version 1.00 software. For LCMS-ESI analysis, pure samples were diluted in 50% H2O:50% MeOH to facilitate ionization and analyzed in negative mode with ESI at 75 eV. The infrared spectra of pure freeze-dried flocculosin were recorded using an MB series ABB Bomem Inc. (St. Laurent, Canada) instrument in KBr. Furthermore, the products of acid and base-catalyzed hydrolysis degradation experiments were analyzed using LCMS-ESI and HPLC, Agilent 1100 series equipped with a differential refractometer (model 2142; LKB Bromma, Golden, Colo.) to confirm the identity of sugar moieties and the overall structure of flocculosin derived from the spectroscopic analysis performed.
(ii) Spectrum of antifungal activity for flocculosin.
Bioassays against different fungal species were carried out to determine the antifungal activity of flocculosin. A 5-mm agar plug of P. aphanidermatum, P. infestans, F. oxysporum, or I. bolleyi was inoculated in petri dishes containing 20 ml of PDA. Solutions containing different concentrations (0 to 1,000 μg) of pure flocculosin (isolated from the WT) diluted in 100 μl of MeOH were deposited into wells (5-mm diameter) bored 2 cm from the infected agar plugs in each plate. The plates were incubated at 25°C for 3 to 8 days until inhibition of fungal growth appeared.
For bioassays with C. albicans, a sterile aqueous solution containing yeast cells was combined with YMGA medium, and 20 ml was then poured into a petri plate. A well (5-mm diameter) was bored in the solidified medium, and flocculosin (0 to 1,000 μg concentration) diluted in 100 μl of MeOH was deposited into the well. The plates were incubated at 28°C until inhibition halos were observed. All bioassays were repeated three times for each strain tested, and pure MeOH (100%) served as the control.
RESULTS
Fungal transformation.
Approximately 20 P. flocculosa transformants were obtained per 10 μg of linearized plasmid DNA per 108 protoplasts as described before (8). Such transformation experiments were repeated several times and altogether produced some 600 transformants; 160 of these were selected randomly and subjected to further analyses as described below.
Antagonistic activity of transformants.
As shown in Fig. 1, Phomopsis sp. was inhibited to various degrees when confronted with P. flocculosa WT and the transformants. All transformants tested were classified into four statistically distinct groups based on their antagonistic activity with Phomopsis sp. (Table 1). Most transformants were classified into group II and were indistinguishable from the WT. Some transformants displayed a stronger antagonistic effect than the WT (group I), while others had a lower (relative to the WT) antagonistic effect (group III). Of particular interest, 14 transformants exhibited no activity against Phomopsis sp. (group IV).
To evaluate the stability and reproducibility of the transformants' phenotypes, bioassays were repeated the following year using continuously subcultured mutants. In the absence of selection, some transformants lost their resistance to hygromycin B, and only those mutants retaining antibiotic resistance were reexamined. Out of the 14 mutants in group IV, six retained hygromycin B resistance and showed the same phenotype as observed during the first bioassay. Similarly, of the 12 tested strains in group I which originally had a stronger antagonistic activity compared to the WT, eight had reverted to the same antagonistic level against Phomopsis sp. as the WT. This loss of antagonistic potency also coincided with an inability to grow on selective medium.
Stable transformants were then bioassayed with S. fuliginea, one of the powdery mildew pathogens controlled by P. flocculosa. As shown in Table 2, treatment of powdery mildew colonies with group I transformants caused their collapse within 24 h (90 to 95%), and conidial chains were completely colonized by P. flocculosa. The activity of group I transformants equaled that of the WT in their ability to colonize the conidial chains of the obligate parasite. In contrast, no collapse of powdery mildew chains was observed for mildew colonies treated with group IV transformants or for the control which consisted of infected cucumber leaves treated with PDB broth alone. Taken together, these bioassay results with S. fuliginea paralleled those obtained with Phomopsis sp.
Genetic analysis of mutants.
Southern blot analysis was carried out with 20 transformants chosen randomly from all four groups. Genomic transformant DNA was digested with SacI, an enzyme having one known corresponding restriction site within pSceI-Hyg, and hybridized with the plasmid which served as a probe. In all cases, a strong band at 5.9 kb corresponding to the size of the plasmid was detected, suggesting that the plasmid was inserted in tandem copies within the genome (data not shown).
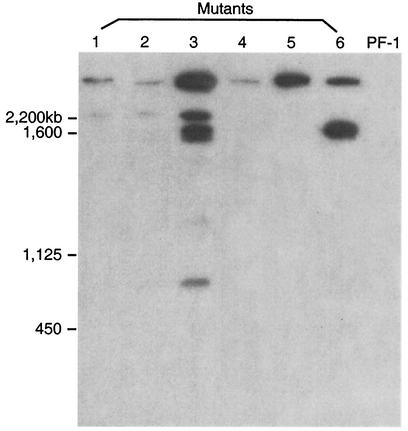
Contour-clamped homogeneous electric field (CHEF) electrophoresis and hybridization were performed for the six group IV mutants that had lost their biocontrol activity. Figure 2 shows the results of hybridization between the electrophoretic karyotypes of P. flocculosa and the plasmid probe. Depending on the transformant analyzed, the number of chromosomes in which the plasmid DNA was integrated varied. For instance, four out of the six mutants demonstrated integration within two or more chromosomes, while the remaining two mutants (PF88 and PF96-1) displayed hybridization within a single chromosome. Interestingly, all mutants harbored at least one insertion on the same chromosome (largest chromosome in Fig. 2). In addition, an ethidium bromide-staining gel showed that mutants PF79, PF96-1, and PF100-1 contained the same amount of genomic DNA (data not shown), and the various intensities of the hybridization bands from these mutants (Fig. 2) strongly suggest the presence of multiple copies of the plasmid inserted at a single chromosomal site and/or multiple integration sites within a single chromosome.
FIG. 2.
Southern hybridization analysis of P. flocculosa (WT) PF-1 chromosomes and deficient mutants with 32P-labeled pSceI-Hyg, separated by CHEF gel electrophoresis. Lanes 1 to 6 correspond to mutants PF78, PF79, PF84, PF88, PF96-1, and PF100-1, respectively.
Extraction and isolation of antifungal metabolites.
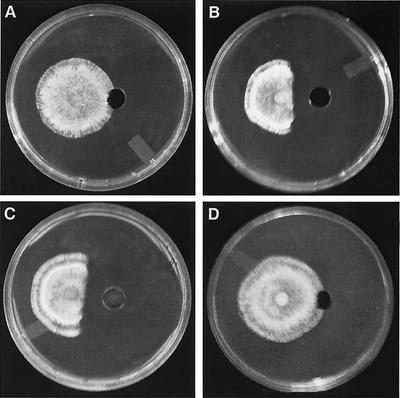
Extracts prepared from group I, group IV, and WT cultures were separated into six fractions; the strongest antifungal activity was found in the 80% MeOH:20% H2O fraction. The same fraction prepared from group I and group IV transformants and the control (medium with hygromycin B) was then compared to this antifungal WT fraction. As shown in Fig. 3, the extracts prepared from group I transformants displayed a stronger level of inhibition compared to the WT, while the same fraction from group IV or from the control showed no sign of fungitoxicity. The 100% H2O fraction eluted from all transformant cultures tested was slightly fungitoxic and was found to contain the hygromycin B added as a selection marker, thereby eliminating the antibiotic as the source of antifungal activity within the 80% MeOH:20% H2O fractions. Using TLC with an anisaldehyde spray reagent, a major chromatophoric band (Rf, 0.74; dark green in color) was observed within WT and group I transformant extracts which was absent in group IV transformant extracts (data not shown). This molecule was then purified and demonstrated potent antifungal activity against Phomopsis sp. at a concentration of 0.2 μg/ml.
FIG. 3.
Bioassay of 80% MeOH:20% H2O fraction prepared from P. flocculosa (WT) PF-1 and transformants with Phomopsis sp. A 200-μl volume of each of the following fractions were bioassayed: control (Czapek Dox medium with hygromycin B) (A), WT (B), transformant PF6 from group I (C), and transformant PF96-1 from group IV (D). Agar plugs inoculated with Phomopsis sp. were placed 2 cm from the center of each well, and results were recorded following incubation for 3 days at 22°C.
Structure elucidation of the purified antifungal molecule.
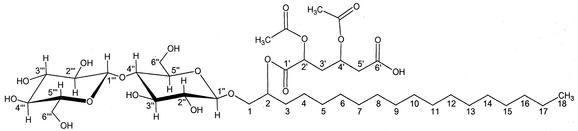
Based on the spectroscopic data and the analysis of acid/base degradation experiments, the chemical structure of the purified molecule was derived (Fig. 4). From the 13C NMR, 1H NMR, COSY, and TOCSY spectra, it was evident that the molecule contained two glucopyranose moieties in a β 1-4 association, as well as an 18 carbon aliphatic chain linked to the anomeric carbon of the first glucose moiety. Since no signals suggesting the presence of double bonds were detected in either the 13C NMR or 1H NMR spectra, and given this molecule's failure to react with iodine vapor when spotted on TLC, we concluded that the aliphatic chain was saturated. The identity and site of attachment of the smaller acetylated chain (C-1′-C-6′) was derived from 13C NMR, HMQC, and TOCSY spectra. No signals were detected for the 31P NMR spectra, confirming the absence of atomic phosphorus from the structure, thus excluding the molecule as a phospholipid. Complete 13C NMR and 1H NMR data are presented in Tables 3 and 4. FABMS analysis gave one signal at 877.5 Da corresponding to a molecular mass of 854.6 Da plus 22.9 Da for sodium. LCMS-ESI analysis of the glycolipid gave a clear molecular ion peak as well as several daughter fragments: (the relative abundance of each peak is expressed as a percentage in parentheses): 853.5 (M−, 18), 836 (5), 759 (5), 753 (19), 711 (100), 669 (21), 651 (15), 605 (14), 573 (28), 517 (11), 507 (28), 350 (8), 143 (9). Since this analysis was performed in negative mode (M−), the actual LCMS-ESI mass obtained, 854.5 Da (853.5 + 1), corroborates both the mass determined using FABMS and the calculated theoretical mass of 854.9 Da. The infrared analysis showed absorptions at 3,422 cm−1, 2,926 cm−1, 2,854 cm−1, 1,744 cm−1, 1,246 cm−1, 1,073 cm−1, 1,044 cm−1. The absorption at 1,744 cm−1 was very strong, suggesting the presence of multiple carbonyl functions (originating from the molecule's acid function and multiple ester bonds). In addition, the signal detected at 3,422 cm−1 was quite wide, indicating multiple hydroxyl groups originating from both glucopyranose moieties. No absorptions were detected in the 1,695 to 1,515 cm−1 range or at 1,425 cm−1 excluding the existence of amide or amine functions. HPLC and LCMS-ESI analysis of the products of acid- and base-catalyzed degradation reactions confirmed the identity of the saturated C-18 aliphatic chain and β-d-glucopyranose as the sole sugar moiety present. Attempts to saponify or hydrolyze and derive the aliphatic C-18 chain by conventional techniques used to analyze fatty acids were unsuccessful, indicating that the aliphatic chain lacked an acid function. The final proposed structure of the molecule is 2-(2′,4′-diacetoxy-5′-carboxy-pentanoyl) octadecyl cellobioside and was subsequently named flocculosin.
FIG. 4.
Proposed structure of flocculosin, 2-(2′,4′-diacetoxy-5′-carboxy-pentanoyl) octadecyl cellobioside (C40 H70 O19), an antifungal cellobiose lipid produced by WT and group I mutants of P. flocculosa.
TABLE 3.
13C NMR spectral data of flocculosin
| Structurea | δ (ppm) |
|---|---|
| C-1 | 75 |
| C-2 | 67 |
| C-3 | 36 |
| C-4 | 34 |
| C-5 | 26 |
| C-6 | 29 |
| C-7 | 29 |
| C-8 | 29 |
| C-9 | 29 |
| C-10 | 29 |
| C-11 | 29 |
| C-12 | 29 |
| C-13 | 29 |
| C-14 | 29 |
| C-15 | 29 |
| C-16 | 32 |
| C-17 | 22 |
| C-18 | 14 |
| C-1′ | 171 |
| C-2′ | 68 |
| C-3′ | 42 |
| C-4′ | 69 |
| C-5′ | 43 |
| C-6′ | 176 |
| C-1′′ | 104 |
| C-2′′ | 73 |
| C-3′′ | 77 |
| C-4′′ | 80 |
| C-5′′ | 73 |
| C-6′′ | 61 |
| C-1′′′ | 101 |
| C-2′′′ | 76 |
| C-3′′′ | 74 |
| C-4′′′ | 70 |
| C-5′′′ | 75 |
| C-6′′′ | 62 |
| -OCOCH3 | 20 |
| -OCOCH3 | 21 |
| -OCOCH3 | 171 |
| -OCOCH3 | 171 |
Underlining indicates atom for which shift is given.
TABLE 4.
1H NMR spectral data of flocculosin
| Protona | δ (ppm) |
|---|---|
| 1-H | 3.8 (d, 2 Hs, J = 10 Hz) |
| 2-H | 3.9 (m, 1 H) |
| 3-H-17-H | 1.3-1.5 (broad doublet, 30 Hs) |
| 18-H | 1.0 (t, 3 Hs, J = 7 Hz) |
| 2′-H | 3.9 (t, 1 H, J = 17 Hz) |
| 3′-H | 2.3 (m, 2 Hs) |
| 4′-H | 4.0 (m, 1 H) |
| 5′-H | 2.6 (d, 2 Hs, J = 4 Hz) |
| 1′′-H | 4.3 (d, 1 H, J = 8 Hz) |
| 2′′-H | 3.5 (dd, 1 H, J = 10 Hz) |
| 3′′-H | 3.6 (dd, 1 H, J = 10 Hz) |
| 4′′-H | 3.5 (dd, 1 H, J = 8 Hz) |
| 5′′-H | 3.6 (m, 1 H) |
| 6′′-H | 3.6, 3.9 (dd, dd, J = 6 Hz, J = 11 Hz, 2 Hs) |
| 1′′′-H | 4.6 (d, 1 H, J = 8 Hz) |
| 2′′′-H | 3.6 (dd, 1 H, J = 10 Hz) |
| 3′′′-H | 3.4 (dd, 1 H, J = 9 Hz) |
| 4′′′-H | 3.4 (dd, 1 H, J = 8 Hz) |
| 5′′′-H | 3.5 (m, 1 H) |
| 6′′′-H | 4.0, 4.3 (dd, dd, J = 6 Hz, J = 13 Hz, 2 Hs) |
| -OCOCH3 | 2.1 (s, 3 Hs) |
| -OCOCH3 | 2.2 (s, 3 Hs) |
Underlining indicates atom for which shift is given.
Spectrum of antifungal activity for flocculosin.
In addition to Phomopsis sp., the antifungal activity of flocculosin was tested against five filamentous fungi and two yeasts. Flocculosin was most effective against P. aphanidermatum, showing potent fungitoxicity at 20 μg (in 100 μl of MeOH). It was equally effective in inhibiting the growth of B. cinerea and P. infestans at 100 μg as well as that of C. albicans. No activity was detected against F. oxysporum and I. bolleyi.
DISCUSSION
In this work, we report the first insertional mutagenesis of a fungal biocontrol agent and the discovery of a rare cellobiose lipid with antifungal activity accounting for, at least part of, the antagonistic properties of P. flocculosa. By using transformation-mediated integration as a mutational tool, we were able to obtain six stable mutants which had lost their biocontrol properties (group IV), as well as 39 other mutants with altered antagonistic properties compared to the WT strain (groups I and III) among 160 tested transformants. This represents an extremely high percentage (28%) of mutants, compared to previous studies with fungal pathogens reporting 2.0% for Colletotrichum magna (23), 1.4% for U. maydis (6), 0.27% for D. discoideum (17), and 0.4% for Colletotrichum gloeosporioides (33). However, if a phenotype is controlled by multiple genes, high mutation frequencies can be expected. In the case of P. flocculosa, multiple genes are likely involved in its biocontrol activity. Alternatively, from the hybridization results of CHEF electrophoresis, we have noticed that all deficient mutants has at least one insertion in the same chromosome (Fig. 2). It is therefore conceivable that there is an integration hot spot in this chromosome, possibly within a gene required for the antagonistic activity, thus leading to multiple hits in the same target.
Although a number of gene disruption techniques have been used to reveal the role of certain enzymes in biocontrol and pathogenic fungal agents (6, 24, 28, 32), insertional mutagenesis has never been used to our knowledge for the study or identification of biocontrol genes or gene products in antagonistic fungi. This may be explained in part by the limited number of fungal biocontrol agents that have been studied in detail and also by the difficulty inherent to achieving a reproducible transformation system for these fungi. However, this report highlights the scientific opportunities that can arise as a result of developing such a system for a fungus displaying biocontrol properties. More specifically, the production of mutants with altered properties provides unique insight into the genetics, the mode of action, the ecology, and the fitness of these beneficial fungi. Incidentally, this opportunity has been exploited in the past for simpler bacterial systems where mutants of antagonistic bacteria were used as tools to discover bioactive toxins and siderophores etc. (16, 19, 31). In our system, group IV mutants have provided direct evidence that the biological control potential of P. flocculosa can be completely inhibited by a rather simple genetic manipulation; alternatively, group I mutants seem to indicate that it is possible to enhance this biocontrol potential. As a result, both groups represent invaluable and unique biological tools to study the genetics and regulation of genes involved in biological control activity.
The deficient group IV mutants of P. flocculosa generated by genetic transformation have conferred a direct way to detect metabolites involved in the antagonistic capacity of the fungus. Based on bioassays that confirmed, both using a test fungus and the target fungus S. fuliginea, that mutants had lost their biocontrol properties, comparative extraction of metabolites from WT and mutant specimens led to the identification of flocculosin, a novel glycolipid. This discovery bears many important implications. First, it describes a simple yet elegant technique to screen for bioactive metabolites from fungi with known biological activity; by blocking the expression of genes responsible for the activity, it becomes much easier to isolate and identify the molecule(s) responsible for such activity as demonstrated in this study. With the current quest for bioactive molecules from natural sources, this approach could be implemented in other fungal systems of interest.
Secondly, it provides biological material with unique genetic keys to decipher the mode of action of biocontrol agents. At this time, there are still very few biopesticides on the market in spite of the increasing demand for biocontrol methods (9). A better understanding of the properties of beneficial agents and more specifically of the genes conferring a given fungus its biocontrol potential would prove useful in both facilitating requirements for registration of such agents and improving their salient properties. This is particularly true in the case of fungi lacking a sexual stage, such as most biocontrol fungi identified to date (e.g., Trichoderma spp.), where it becomes difficult to identify the genes of interest. Indeed, when using DNA-mediated transformation as a mutagen in fungal systems, the ultimate objective is to tag the genes of interest for their subsequent identification and manipulation. However, in similar previous studies, substantial portions (20 to 100%) of generated mutants were not tagged by the transforming DNA (3, 10, 18, 20, 26). It has been proposed that the transformation process itself can cause deletions and chromosomal translocations which may lead to the mutation. Therefore, it is important to confirm that mutations obtained from insertional mutagenesis are associated with an integrated DNA. This can be done by genetic crosses in which the mutant phenotype is shown to cosegregate with the selectable marker on the inserted plasmid. However, for P. flocculosa, a fungus lacking a known sexual stage, the method of choice for verifying that a given mutant has been tagged with the inserted plasmid is the cloning of its DNA flanking the insertion site(s) and subsequent functional complementation by transformation.
Thirdly, the isolation and identification of a rare cellobiose lipid specifically absent in deficient mutant cultures provides many interpretations. Since the molecule is toxic to fungi within the spectrum of activity of P. flocculosa (2), it seems quite evident that it plays a key role in its biocontrol activity. P. flocculosa has long been described as an agent acting by antibiosis against its targets and to date three fatty acids with antifungal activity have been isolated from the culture filtrates of the fungus. However, these fatty acids could only be isolated in detectable quantities following long periods of culture. In light of the proposed structure for the glycolipid identified here, it is possible that the fatty acids previously identified are either products of degradation, products of catalysis, or are additional bioactive molecules produced by P. flocculosa. Another intriguing aspect of this cellobiose lipid is its unusual and rare structure for which there is very little precedent of analogy in the literature. Such glycolipids have been reported recently in Cryptococcus humicola with mycocidal activity against a broad spectrum of both basidiomycetous and ascomycetous yeasts (22). These authors suggest that the glycolipids interfere with membrane integrity which supports the model proposed by Avis and Bélanger (2) for the activity of P. flocculosa. In addition, Spoeckner et al. (29) have described similar compounds produced by U. maydis and some of its anamorphs with no mention of antifungal activity.
In conclusion, this successful insertional mutagenesis of a fungal biocontrol agent, P. flocculosa, has led to the creation of mutants with deficient and enhanced biocontrol properties. This material was used for screening bioactive molecules which allowed the identification of a rare and unusual cellobiose lipid (named flocculosin) that not only explains part of the biocontrol potential of P. flocculosa but confirms phylogenetic link with U. maydis and other Pseudozyma spp. This approach and the biological material thus obtained can find a variety of applications in the discovery and manipulation of gene and gene products from fungal biocontrol agents.
Acknowledgments
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC), the Canada Research Chairs Program (R.R.B.), and Plant Products Co. Ltd.
We thank L. Bernier, D. Auclair, and M. Tremblay for their technical assistance and T. Avis for critical review of the manuscript.
REFERENCES
- 1.Akamatsu, H., Y. Itoh, M. Kodama, H. Otani, and K. Kohmoto. 1997. AAL-toxin deficient mutants of Alternaria alternata tomato phenotype by restriction enzyme-mediated integration. Phytopathology 87:967-972. [DOI] [PubMed] [Google Scholar]
- 2.Avis, T. J., and R. R. Bélanger. 2001. Specificity and mode of action of the antifungal fatty acid cis-9-heptadecenoic acid produced by Pseudozyma flocculosa. Appl. Environ. Microbiol. 67:956-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balhadère, P. V., A. J. Foster, and N. J. Talbot. 1999. Identification of pathogenicity mutants of the rice blast fungus Magnaporthe grisea by insertional mutagenesis. Mol. Plant-Microbe Interact. 12:129-142. [Google Scholar]
- 4.Bélanger, R. R., A. J. Dik, and J. G. Menzies. 1998. Powdery mildews: recent advances toward integrated control, p. 89-109. In G. J. Boland, and L. D. Kuykendall (ed.), Plant-microbe interactions and biological control, Marcel Dekker Inc., New York, N.Y.
- 5.Boekhout, T. 1995. Pseudozyma flocculosa emend. Boekhout, a genus for yeast-like anamorphs of Ustilaginales. J. Gen. Appl. Microbiol. 41:359-366. [Google Scholar]
- 6.Bölker, M., H. U. Bohnert, K. H. Braun, J. Gorl, and R. Kahmann. 1995. Tagging pathogenicity genes in Ustilago maydis by restriction enzyme-mediated integration (REMI). Mol. Gen. Genet. 248:547-552. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, Y. L., and R. R. Bélanger. 2000. Protoplast preparation and regeneration from spores of the biocontrol fungus Pseudozyma flocculosa. FEMS Microbiol. Lett. 190:287-291. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, Y. L., F. Belzile, P. Tanguay, L. Bernier, and R. R. Bélanger. 2001. Establishment of a gene transfer system for Pseudozyma flocculosa, an antagonistic fungus of powdery mildew fungi. Mol. Genet. Genomics 266:96-102. [DOI] [PubMed] [Google Scholar]
- 9.Cook, R. J. 1993. Making greater use of intoduced microorganisms for biological control of plant pathogens. Annu. Rev. Phytopathol. 31:53-80. [DOI] [PubMed] [Google Scholar]
- 10.Epstein, L., K. Lusnak, and S. Kaur. 1998. Transformation-mediated developmental mutants of Glomerella graminicola (Collectotrichum graminicola). Fungal Genet. Biol. 23:189-203. [DOI] [PubMed] [Google Scholar]
- 11.Garnand, K., and M. A. Nelson. 1995. The effect of DNA structure and restriction enzymes on transformation efficiencies in Neurospora crassa. Fungal Genet. Newsl. 42:29-31. [Google Scholar]
- 12.Gibbons, S., and A. I. Gray. 1998. Isolation by planar chromatography, p. 209-245 In R. J. P. Cannell (ed.), Methods in biotechnology, vol. 4. Humana Press Inc., Ottawa, N.J.
- 13.Granado, J. D., K. Kertesz-Chaloupková, M. Aebi, and U. Kües. 1997. Restriction enzyme-mediated DNA integration in Coprinus cinereus. Mol. Gen. Genet. 256:28-36. [DOI] [PubMed] [Google Scholar]
- 14.Hajlaoui, M. R., N. Benhamou, and R. R. Bélanger. 1992. Cytochemical study of the antagonistic activity of Sporothrix flocculosa on rose powdery mildew. Sphaerotheca pannosa var. rosae. Phytopathology 82:583-589. [Google Scholar]
- 15.Jarvis, W. R., J. A. Shaw, and J. A. Traquair. 1989. Factors affecting antagonism of cucumber powdery mildew by Stephanoascus flocculosa and S. rugulosus. Mycol. Res. 92:162-165. [Google Scholar]
- 16.Kloepper, J. W., J. Leong, M. Teintze, and M. N. Schroth. 1980. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286:885-886. [Google Scholar]
- 17.Kuspa, A., and W. F. Loomis. 1992. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc. Natl. Acad. Sci. USA 89:8803-8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linnemannstöns, P., T. Vob, P. Hedden, P. Gaskin, and B. Tudzynski. 1999. Deletions in the gibberellin biosynthesis gene cluster of Gibberella fujikuroi by restriction enzyme-mediated integration and conventional transformation-mediated mutagenesis. Appl. Environ. Microbiol. 65:2558-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loper, J. E. 1988. Role of fluorescent siderophore production in biological control of Pythium ultimum by a Pseudomonas fluorescens strain. Phytopathology 78:166-172. [Google Scholar]
- 20.Lu, S., L. Lyngholm, G. Yan, C. Bronson, O. C. Yoder, and B. G. Turgeon. 1994. Tagged mutation at the Tox1 locus of Cochliobolus heterostrophus by restriction enzyme-mediated integration. Proc. Natl. Acad. Sci. USA 91:12649-12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Möller, E. M., G. Bahnweg, H. Sandermann, and H. H. Geiger. 1992. A simple and efficient protocol for isolation of the high molecular weight DNA from filamentous fungi, fruit bodies and infected plant tissues. Nucleic Acids Res. 20:6115-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puchkov, E. O., U. Zähringer, B. Lindner, T. V. Kulakovskaya, U. Seydel, and A. Wiese. 2002. The mycocidal, membrane-active complex of Cryptococcus humicola is a new type of cellobiose lipid with detergent features. Biochim. Biophys. Acta 1558:161-170. [DOI] [PubMed] [Google Scholar]
- 23.Redman, R. S., J. C. Ranson, and R. J. Rodriguez. 1999. Conversion of the pathogenic fungus Colletotrichum magna to non-pathogenic endophytic mutualist by gene disruption. Mol. Plant-Microbe Interact. 12:969-975. [Google Scholar]
- 24.Rey, M., J. J. Delgado, and T. Benitez. 2001. Improved antifongique activity of a mutant of Trichoderma harangueuse CECT 2413 which produces more extra cellular proteins. Appl. Microbiol. Biotechnol. 55:604-608. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 26.Sanchez, O., R. E. Navarro, and J. Aguirre. 1998. Increased transformation frequency and tagging of developmental genes in Aspergillus nidulans by restriction enzyme-mediated integration (REMI). Mol. Gen. Genet. 258:89-94. [DOI] [PubMed] [Google Scholar]
- 27.Schiestl, R. H., and T. D. Petes. 1991. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88:7585-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi, Z., D. Christian, and H. Leung. 1995. Enhanced transformation in Magnaporthe grisea by restriction enzyme mediated integration of plasmid DNA. Phytopathology 85:329-333. [Google Scholar]
- 29.Spoeckner, S., V. Wray, M. Nimtz, and S. Lang. 1999. Glycolipids of the smut fungus Ustilago maydis from cultivation on renewable resources. Appl. Microbiol. Biotechnol. 51:33-39. [Google Scholar]
- 30.Sweigard, J. A., A. M. Carroll, L. Farrall, F. G. Chumley, and B. Valent. 1998. Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol. Plant-Microbe Interact. 11:404-412. [DOI] [PubMed] [Google Scholar]
- 31.Thomashow, L. S., and D. M. Weller. 1988. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170:3499-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woo, S. L., B. Donzelli, F. Scala, R. Mach, G. E. Harman, C. P. Kubicek, G. Del sorbo, and M. Lorito. 1999. Disruption of the ech42 (endochitinase-encoding) gene effects biocontrol activity in Trichoderma harzianum P1. Mol. Plant-Microbe Interact. 12:419-429. [Google Scholar]
- 33.Yakoby, N., R. Zhou, I. Kobiler, A. Dinoor, and D. Prusky. 2001. Development of Colletotrichum gloeosporioides restriction enzyme-mediated integration mutants as biocontrol agents against anthracnose disease in avocado fruits. Phytopathology 91:143-148. [DOI] [PubMed] [Google Scholar]