Abstract
Cryptosporidium parvum oocyst DNA samples (n = 184) from humans with cryptosporidiosis contracted during foreign travel or during outbreaks in the United Kingdom were characterized genetically and categorized by single-strand conformation polymorphism (SSCP)-based analysis of the small-subunit gene (pSSU) (∼300 bp) and second internal transcribed spacer (pITS-2) (∼230 bp) of nuclear ribosomal DNA. The two recognized genotypes (types 1 and 2) of C. parvum could be readily differentiated by a distinct electrophoretic shift in the pSSU SSCP profile, associated with a nucleotide difference of ∼1.3 to 1.7%. Of the 102 samples from cases contracted during foreign travel, 88 (86.3%) were identified as C. parvum type 1 and 14 (13.7%) were identified as type 2. For outbreak samples, unequivocal differentiation between type 1 (n = 20; one child nursery outbreak) and type 2 (n = 62; two waterborne outbreaks) was also achieved. Nucleotide variation in pITS-2 (both within and among samples representing each genotype) was substantially greater (10 to 13 different profiles for each genotype, relating to sequence differences of ∼1 to 42%) than that in pSSU. SSCP analysis of pITS-2 for all samples revealed that some profiles had a broad geographical distribution whereas others were restricted to particular locations, suggesting a link between some subgenotypes and the geographical origin or source. Comparative denaturing polyacrylamide gel electrophoretic analysis revealed the same genotypic identification and a similar subgenotypic classification of samples as SSCP analysis. The findings of this study, particularly the detection of intragenotypic variation by SSCP, should have significant diagnostic implications for investigating transmission patterns and the monitoring of outbreaks.
Species of Cryptosporidium are oocyst-forming apicomplexan parasites which infect a wide range of vertebrate hosts. Intestinal disease caused by these parasites is called cryptosporidiosis and is transmitted via the fecal-oral route (e.g., in child day care centers) and can be associated with waterborne outbreaks (27). The clinical signs of cryptosporidiosis in humans are mainly diarrhea, dehydration, malabsorption, weight loss, and/or wasting (27). In many cases, infection is self-limiting and resolves spontaneously, although chronic infections may develop in high-risk patient groups, particularly neonates and immunocompromised individuals (those with congenital or acquired immunodeficiencies or those undergoing immunosuppressive chemotherapy). The accurate identification of Cryptosporidium to the species or “strain” (i.e., genotypic) level is important for studying transmission patterns and genetic structures and is central to the control of cryptosporidiosis (9, 26, 34). Currently, 11 species of Cryptosporidium are recognized (9, 10), of which C. parvum is of major significance in humans. However, these species cannot be distinguished from one another simply on the basis of host occurrence and/or parasite morphology (9, 27). Consequently, a range of different molecular methods have been developed for the genetic characterization and classification of members of the genus Cryptosporidium (9, 29, 34).
PCR techniques have found broad applicability because their sensitivity permits the analysis of DNA from minute amounts of parasite material (34). A range of different genetic loci have been used specifically for the molecular identification or differentiation of Cryptosporidium samples. These include regions of nuclear ribosomal DNA (rDNA) and the genes for the Cryptosporidium oocyst wall protein (cowp), heat shock protein 70 (hsp70), and thrombospondin-related adhesive protein (trap) (9). In particular, studies have demonstrated that a portion of the small-subunit (SSU) ribosomal gene provides useful genetic markers for the specific identification of Cryptosporidium because of a relatively low level of intraspecific variation in sequence and significant differences among genotypes (9, 26). The internal transcribed spacers (ITS) of rDNA also provide useful genetic markers for genotypic identification, because they have been shown to be considerably more variable in sequence within a species (4, 25, 28).
Among other methods, DNA sequencing and PCR-coupled restriction fragment length polymorphism (PCR-RFLP) have been widely employed for the genotypic identification of Cryptosporidium isolates (34). While useful, such approaches do not necessarily allow sequence and length variation within an amplicon to be analyzed accurately (12). For example, direct sequencing of the ITS regions is difficult or impossible because of sequence variability (polymorphism) within an oocyst isolate (references 4 and 22 and unpublished results), and PCR-RFLP does not accurately resolve the variation present. In spite of such impediments, sequence variation within the ITS indicates that this ribosomal spacer region should be particularly well suited to investigate population genetic structures and to characterize genetic variability within and among isolates. To overcome limitations in the analysis of sequence variability both within and among amplicons, we have recently employed the mutation scanning approach of single-strand conformation polymorphism (SSCP) for the display of genetic variation in nuclear gene regions (SSU and hsp70) of Cryptosporidium with a small number of well-defined oocyst DNA samples (18). However, to date, mutation scanning has not been exploited to investigate population variation within C. parvum by employing genetic loci with higher levels of intraspecific sequence variability, such as the ITS, and larger sample sizes. While some earlier publications (reviewed in reference 15) indicated that genetic variation within both of the currently recognized genotypes of C. parvum (H or type 1, infecting humans and nonhuman primates, and C or type 2, found in both humans and animal hosts) was limited, significant intragenotypic variability has been detected by using microsatellite analyses (2, 11, 21). In the present study, we employed SSCP and denaturing polyacrylamide gel electrophoresis (DPGE) to investigate the magnitude of nucleotide variation in portions of the SSU and second ITS (ITS-2) of rDNA within and among a relatively large number of C. parvum isolates from humans, and we discuss the implications of the findings and the approach.
MATERIALS AND METHODS
Isolates, DNA isolation, and enzymatic amplification.
One hundred and eighty-four Cryptosporidium isolates were obtained from the national collection of oocysts in the United Kingdom (6). These isolates originated from humans with clinical cryptosporidiosis reported to be contracted during recent international travel (n = 102) or during three different outbreaks (n = 82) in England (Tables 1 and 2). One of the outbreaks was in a day care nursery in Middlesex, the second was a drinking water outbreak in Clitheroe, and the third was a swimming pool-associated outbreak in Cleethorpes. Previously, genomic DNA samples were purified from individual oocyst isolates by employing a standard approach (5), and their genotypes were determined by PCR-RFLP of a 519-bp region of the cowp gene (32).
TABLE 1.
Results from SSCP analyses of amplicons produced from 102 C. parvum DNA samples purified from oocyst isolates from humans returning to the United Kingdom following recent travel to different countriesa
| Profile | Travel destination (no. of samples) | No. of samples | |
|---|---|---|---|
| pSSU | pITS-2 | ||
| 1 | 1A | Spain (18), Greece (2), Cyprus (3), Egypt (1), Pakistan (4), United States (1), Jamaica (1), Cuba (1), Peru (1), Mexico (1) | 33 |
| 1B | Denmark (1), France (2), Spain (18), Cyprus (2), Greece (2), Turkey (3), Pakistan (3), India (1), Mexico (2), Carribean Islands (1) | 35 | |
| 1C | Kenya (1), Gambia (1) | 2 | |
| 1D | Kenya (1) | 1 | |
| 1E | Pakistan (1) | 1 | |
| 1F | Pakistan (2), United States (1) | 3 | |
| 1G | Nigeria (1), Pakistan (2), India (1), Caicos Islands (1), Equador (1) | 6 | |
| 1H | Spain (2) | 2 | |
| 1I | Jamaica (1) | 1 | |
| 1J | Spain (2), Greece (1) | 3 | |
| 1K | Menorca (1) | 1 | |
| 2 | 2A | Spain (3) | 3 |
| 2B | Spain (1) | 1 | |
| 2C | Portugal (2) | 2 | |
| 2D | Portugal (1), Uganda (1), Saudi Arabia (1) | 3 | |
| 2E | Africa (1) | 1 | |
| 2F | Cyprus (1)b | 1 | |
| 2G | Turkey (1) | 1 | |
| 2H | Pakistan (1) | 1 | |
| 2I | Mexico (1) | 1 | |
Samples were categorized to the genotypic level (types 1 and 2) based on pSSU profiles and to the subgenotypic level (1A to 1K and 2A to 2I) based on pITS-2 profiles.
This sample (c35) from Cyprus differed slightly in profile from all other type 2 samples examined; this difference related to one A-to-T transversion in the pSSU sequence.
TABLE 2.
Results from SSCP analyses of amplicons produced from 82 C. parvum (type 1 or type 2) oocyst DNA samples purified from oocyst isolates from humans associated with three different cryptosporidiosis outbreaks in the United Kingdoma
| SSCP profile | Outbreak type | No. of samples | |
|---|---|---|---|
| pSSU | pITS-2 | ||
| 1 | 1A | Child day care nursery, Middlesex | 13 |
| 1B | Child day care nursery, Middlesex | 1 | |
| 1L | Child day care nursery, Middlesex | 5 | |
| 1M | Child day care nursery, Middlesex | 1 | |
| 2 | 2J | Drinking water, Clitheroe | 39 |
| Swimming pool, Cleethorpes | 23 | ||
Samples were categorized to the genotypic level (types 1 and 2) based on pSSU profiles and to the subgenotypic level (1A, 1B, 1L, 1M, and 2J) based on pITS-2 profiles.
From each genomic DNA sample, two regions of the nuclear genome were PCR amplified separately by using oligonucleotide primers 18SiF (forward, 5′-AGTGACAAGAAATAACAATACAGG-3′) and 18SiR (reverse, 5′-CCTGCTTTAAGCACTCTAATTTTC-3′) (∼300-bp region of the SSU rRNA gene, designated pSSU) (24) and primers YA56F (forward, 5′-GGCGCTACTTCATATAATATAATGTTTTTT-3′) and YA54R (reverse, 5′-GGCGCTAATTTTAACTTAAATTGGTTAAGAAA-3′) (∼230-bp region of ITS-2, designated pITS-2). A nontarget, GC-rich sequence (first 5 bases 5′) was included in the latter two primers to achieve an increase in the predicted melting temperature (from ∼43 to 46°C to ∼53 to 55°C) and improved PCR stringency and amplification efficiency. Primers were end labeled with [33P]ATP (NEN, DuPont) by using T4 kinase (Promega, Madison, Wis.).
PCR amplification was performed in 50-μl volumes with 25 pmol of each primer, a 250 μM concentration of each deoxynucleoside triphosphate, 3 mM MgCl2, and 1 U of Taq polymerase (Promega) under the following conditions: after an initial denaturation at 94°C for 5 min, reaction mixtures were subjected to 30 cycles of 94°C for 30 s (denaturation), 55°C (pSSU) or 50°C (pITS-2) for 30 s (annealing), and 72°C for 30 s (extension), followed by a final extension at 72°C for 5 min in a 480 thermocycler (Perkin-Elmer Cetus, Norwalk, Conn.). The amount of genomic DNA template added to the PCR mixture was usually ∼1 pg to 20 ng. Control samples without DNA were included in each PCR run. To test the specificity of the PCR, 19 control samples, including DNA from feces or blood from a human without any evidence of parasitic infection and DNAs from the bacteria Escherichia coli, Lactobacillus acidophilus, and Lactobacillus gasseri; the yeast Saccharomyces cerevisiae; the protozoan parasites Giardia duodenalis and Entamoeba histolytica; and the helminth parasites Schistosoma japonicum, Schistosoma mansoni, Hymenolepis nana, Taenia saginata, Taenia solium (Platyhelminthes), Ascaris sp., Ancylostoma caninum, Necator americanus, Strongyloides stercoralis, Oesophagostomum bifurcum, and Trichuris trichiura (Nematoda), were subjected to the same amplification procedure (with both primer sets).
After thermocycling, individual amplicons were mixed with an equal volume of loading buffer (10 mM NaOH, 95% formamide, and 0.05% of both bromophenol blue and xylene cyanole), and the intensities of selected samples were verified on ethidium bromide-stained 2.5% agarose gels with TBE (65 mM Tris-HCl, 27 mM boric acid, 1 mM EDTA [pH 9]) (Bio-Rad, Richmond, Calif.) as the buffer and φX174-HaeIII (Promega) as a size marker.
Electrophoresis.
Radiolabeled amplicons were denatured at 95°C for 5 min and snap cooled on a freeze block (−20°C) for 5 min prior to SSCP analysis or DPGE in a conventional sequencing rig (S2; Life Technologies). SSCP analysis was performed in 0.4-mm-thick, 0.6× mutation detection enhancement gels (MDE; FMC BioProducts, Rockland, Maine), cast according to the manufacturer's protocol. Gels were poured (with insertion of a 60-tooth well-comb; well width of 3 mm), allowed to polymerize at 24°C for 70 min, and then subjected to electrophoresis in an air conditioned room (18°C constant temperature) with TBE as the buffer. Gels were prerun at 30 W (constant) for 20 min (until the gel had reached a temperature of ∼24°C), and samples (1.7 μl) were loaded into wells and subjected to electrophoresis under the same conditions for 5 h. DPGE was carried out in 0.4-mm-thick, 5% polyacrylamide gels containing 42% urea. Samples (3 μl each) were loaded into shark-tooth comb wells (4 mm wide) and subjected to electrophoresis at 50 W for 3 h at 24°C (achieving a glass plate temperature of ∼39 to 40°C), with TBE. For both electrophoretic procedures, gels were dried onto 3MM filter paper (Whatman) and usually subjected to autoradiography for 24 h with Curix-Blue film (Agfa). Electrophoretic profiles obtained by using each of the two approaches were demonstrated to be reproducible on different days with amplicons produced on different days (results not shown). While Taq polymerase may introduce artifactual nucleotide misincorporations during amplification to a rate of ∼10−4 per base, such errors occur randomly within the amplicons and are thus not detected as discrete bands on electrophoretic gels (12).
Since bands at different positions in an SSCP gel represent distinct types or conformations of a molecule (36), the banding profiles provide a high-resolution fingerprint of sequence variation within individual samples. Hence, each band within an SSCP profile can be considered a genetic marker. SSCP profiles were scored separately, such that each band at a defined migration position in the gel was a character and thus coded 1 for presence and 0 for absence. The assumption is that each band is a dependent character (i.e., represents a single-stranded, molecular variant or conformer). Pairwise comparisons of the differences (percent) in SSCP banding profiles between samples were calculated by using the formula (I/P) × 100, where P is the total number of bands representing the entire population (or sample size) under study and I is the number of bands differing between two samples (17). Pairwise comparisons of the level of sequence differences were made by using the GAP program available at the Australian National Genomic Information Service (http://mel1.angis.org.au/WebANGIS/WebFM [institutional access required]). For both profile data and sequence data, phenograms were constructed by using the unweighted pair group method using arithmetic averages (31).
DNA sequencing.
Selected pSSU amplicons were purified over minicolumns (Wizard PCR Prep; Promega), eluted in 30 μl of H2O, and then subjected to automated sequencing (BigDye chemistry; Applied Biosystems) in both directions, using the same primers as for PCR. SSCP bands representing selected pITS-2 amplicons were PCR enriched as described previously (14) and then sequenced via cloning. In brief, after autoradiographic exposure of SSCP gels, selected single-strand bands were excised with a scalpel, suspended in 50 μl of water, and incubated at 4°C for 2 h. Then, 2 μl of this suspension was added to 98 μl of water, and 1 to 2 μl thereof was subjected to PCR (employing the same primer set and conditions as used for primary amplification). An aliquot of column-purified amplicon (∼100 ng of DNA) was then cloned into the pGEM-T Easy plasmid vector (Promega). Clones were isolated, and inserts were amplified and subsequently sequenced (in both directions) by employing the vector primers SP6 and T7 (Promega). Nucleotide sequences were subjected to BLASTP and BLASTX similarity searches of the nonredundant GenBank database (NCBI Basic BLAST; http://www.ncbi.nlm.nih.gov/BLAST/).
Nucleotide sequence accession numbers.
Individual band sequences for the bands referred to in Fig. 3 and 4 are available from the EMBL, GenBank, and DDJB databases under accession numbers AJ539183 to AJ539222.
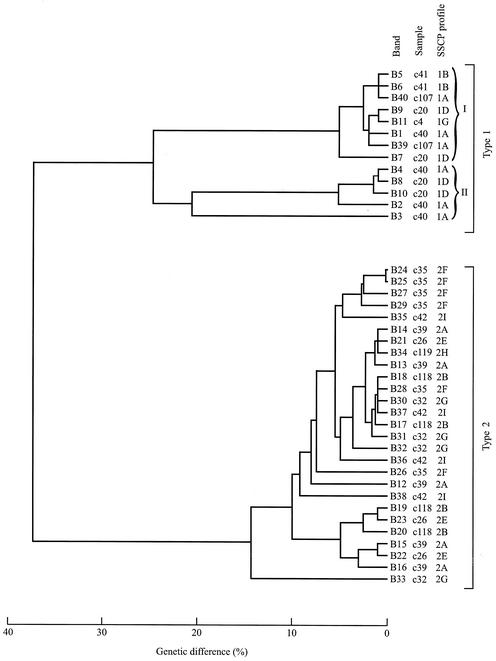
FIG. 3.
Phenograms depicting the genetic difference (in percent, upon pairwise comparison) among pITS-2 sequences representing 40 bands individually excised from an SSCP gel (Fig. 1). Thirteen and 27 band sequences represent C. parvum type 1 and type 2, respectively. SSCP profiles 1A, 1B, 1D, and 1G relate to samples c107 (United Kingdom), c41 (Spain), c20 (Kenya), and c4 (Caicos Islands), respectively, and profiles 2A, 2B, 2E, 2F, 2G, 2H, and 2I relate to samples c39 (Spain), c118 (Spain), c26 (Africa), c35 (Cyprus), c32 (Turkey), c119 (Pakistan), and c42 (Mexico), respectively. Type 1 sequences separate into two distinct clusters (I and II) (using a threshold of ∼22% difference).
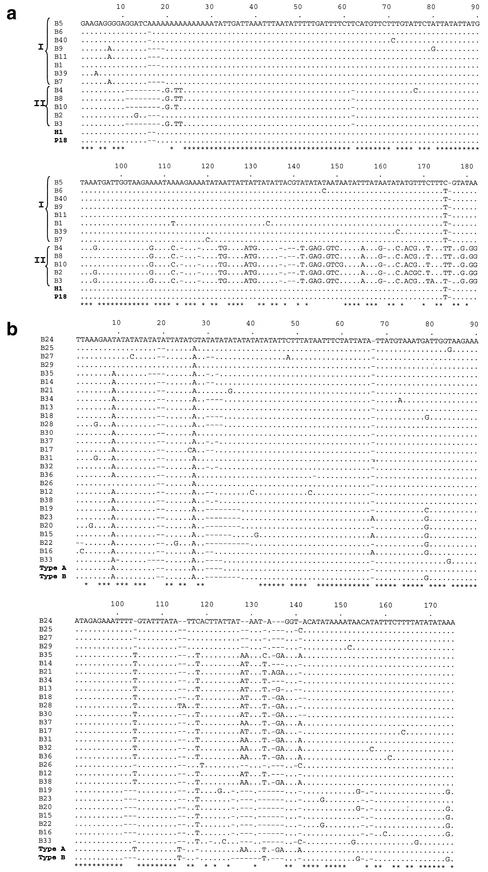
FIG. 4.
Alignment of pITS-2 band sequences representing C. parvum type 1 over 182 positions (a) and of pITS-2 sequences representing C. parvum type 2 over 175 positions (b), determined following PCR-based enrichment from SSCP bands. The sequences of the bands are presented in the same order (top to bottom) as in Fig. 3. A dot indicates a base identical to that in the top sequence, a dash indicates an insertion-deletion event, and an asterisk indicates sequence identity among all sequences of a particular genotype. Sequences representing C. parvum type 1 (codes H1 and P18 [25]) and C. parvum type 2 (codes type A and type B [22]) have been included for comparative purposes.
RESULTS
Specificity of the PCR and sensitivity of amplification.
The specificity of the primer sets 18SiF-18SiR (pSSU) and YA56F-YA54R (pITS-2) and the PCR conditions for the amplification of C. parvum pSSU and pITS-2 were assessed with a range of control DNA samples, followed by electrophoretic analysis and autoradiography. Two positive control samples (codes c107 and c232) representing C. parvum type 1 and type 2, respectively, were selected and employed. DNA samples prepared from human blood or feces (from a person with no evidence of any parasitic infection) and 17 additional samples from a range of prokaryotes (bacteria) and eukaryotes (protozoa, trematodes, cestodes, and nematodes), developmental stages of which can occur in the intestinal tracts of humans, were also subjected to PCR. With ∼0.5 to 5 ng of genomic DNA template per PCR, products of the expected size were amplified from samples c107 and c232 (positive controls) and detected by electrophoresis, whereas no bands were detectable for any of the other 19 negative control samples tested (not shown).
The sensitivity of the PCR was determined by titration of genomic DNAs representing samples c107 and c232, followed by SSCP analysis (not shown). The smallest amount of C. parvum genomic DNA detectable by amplification with primer set 18SiF-18SiR (pSSU) or YA56F-YA54R (pITS-2) and subsequent detection on agarose gels was estimated at 1 to 2 pg, which is comparable to that in a previous study of Eimeria species (Protozoa: family Eimeriidae) (35).
Having demonstrated the specificity and sensitivity of the PCR and the conditions, 184 C. parvum DNA samples from humans representing sporadic cases of cryptosporidiosis following recent overseas travel (n = 102) or cases linked to cryptosporidiosis outbreaks in the United Kingdom (n = 82) were subjected to PCR. As expected, amplicons were ∼300 bp (pSSU) or ∼230 bp (pITS-2) in size on agarose gels, and no unequivocal size variation was detectable among any of the amplicons representing each of the rDNA loci (not shown). Direct sequencing of selected pSSU and pITS-2 amplicons and comparison of sequence data with those in current databases verified their identity. All 184 samples were then subjected to high-resolution electrophoretic analyses.
SSCP-based analyses of pSSU and pITS-2 amplicons.
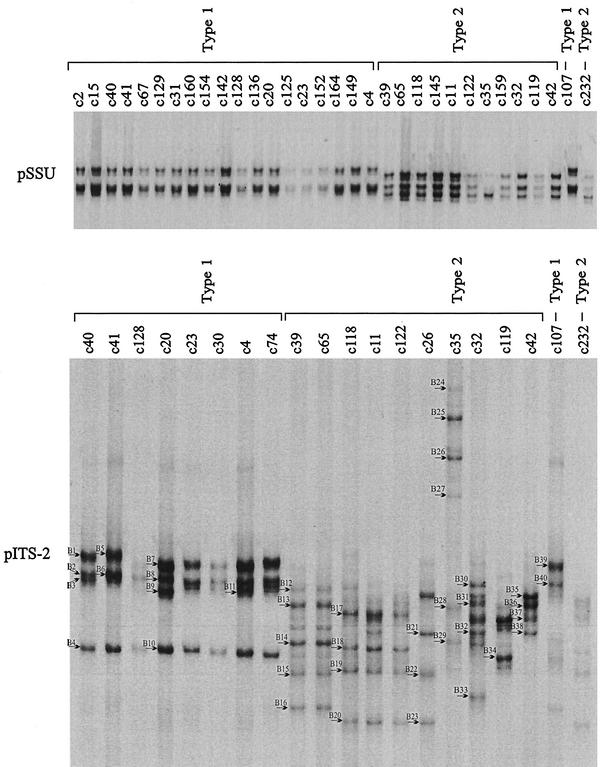
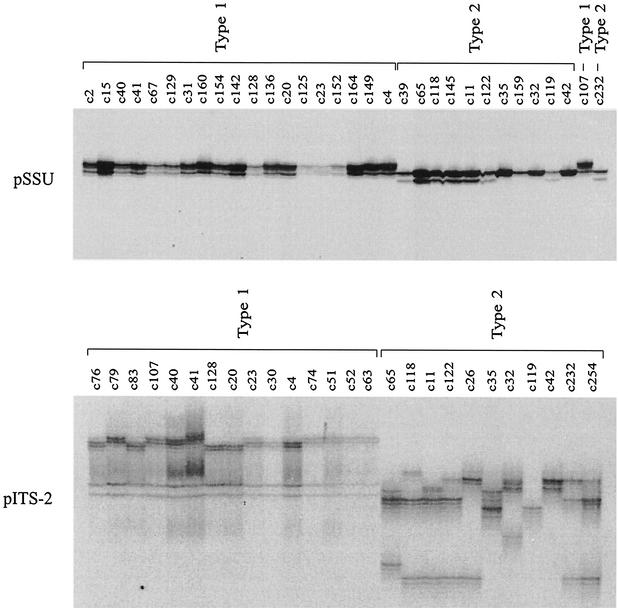
Results of the SSCP analyses of the pSSU and pITS-2 amplicons are shown in Tables 1 and 2, and representative electrophoretic gels are shown in Fig. 1. In the SSCP analysis of pSSU, 88 (86.3%) of the 102 samples from cases contracted during foreign travel could be identified as type 1, whereas 14 samples (13.7%) were identified as type 2. No variation in SSCP profiles was detectable among any of the samples representing type 1, and only one type 2 sample (c35) from Cyprus differed slightly in profile from all other samples of that genotype from various other geographical regions (Fig. 1). Sequencing of representative samples (codes c11, c32, c35, c39, c42, c65, c118, c119, c122, c145, c159, and c232) (Fig. 1) revealed one nucleotide difference (A-to-T transversion) at alignment position 230 between sample c35 and all other type 2 samples sequenced (the alignment available from the authors). BLAST analyses revealed a perfect match of the c35 sequence (over alignment positions 142 to 269) with that under accession number AF133842 (N. J. Pieniazek et al., unpublished data), which is distinct from the majority of other type 2 sequences reported to date. In contrast, no nucleotide difference in pSSU was detected among any of the type 1 samples sequenced (codes c2, c4, c15, c20, c23, c31, c40, c41, c67, c125, c128, c129, c136, c142, c149, c152, c154, c160, and c164), the sequences of which were in accordance with a range of sequences representing type 1 contained in current databases (e.g., accession number AF093491). Irrespective of the mutation at alignment position 230 detected within type 2 by SSCP analysis of the pSSU locus, a distinct difference in the position of bands (relating to a 4- to 5-bp difference over 269 bp, excluding primers) enabled the unequivocal identification of and delineation between C. parvum types 1 and 2 by SSCP analysis (Fig. 1). Also for outbreak samples (Table 2), unequivocal differentiation of type 1 (n = 20; child day care nursery outbreak in Middlesex, United Kingdom) from type 2 (n = 62; waterborne outbreaks in Clitheroe and Cleethorpes, United Kingdom) was achieved. For all samples examined in this study, the genotypic classification based on the SSCP analysis of pSSU was the same as that achieved by PCR-RFLP analysis of the cowp gene (32).
FIG. 1.
SSCP analysis of sequence variation in the two loci pSSU (top) and pITS-2 (bottom) within and among C. parvum samples originating from humans. One hundred eighty-four samples were initially scanned by SSCP, and sample subsets are included on the gel images. Samples c107 and c232 from humans from the United Kingdom were included as type 1 and type 2 reference controls, respectively. For pSSU, the first 19 samples (c2 to c4) were identified as type 1, and the following 11 samples (c39 to c42) were identified as type 2. For pITS-2, the first 8 samples (c40 to c74) were identified as type 1, and the following 10 samples were identified as type 2. Significant pITS-2 profile variation was detected within both types 1 and 2. Forty different bands (B1 to B40) (arrows) were excised from the pITS-2 SSCP gel and then subjected to sequencing.
While no or a limited variation in banding profile was detectable within each of the two genotypes in the SSCP analysis of pSSU (Fig. 1), totals of 13 and 10 different pITS-2 profiles (∼4 to 12 bands per profile) were displayed for all 184 samples representing C. parvum types 1 and 2, respectively (Tables 1 and 2). A representative SSCP gel displaying some of these pITS-2 profiles and the genetic relationship of the samples (based on 75 dependent characters recorded for all SSCP profiles obtained) are shown in Fig. 1. Usually, a lower degree of profile variation was displayed among type 1 than among type 2 samples, and the former genotype formed three main groups (represented by profiles 1A and 1B; 1C, 1D, 1E, 1F, and 1 M; and 1G, 1H, 1K, 1I, 1J, and 1L), to the exclusion of the latter, which could be divided into six groups (2A, 2H, and 2I; 2B, 2C, and 2D; 2J; 2G; 2F; and 2E).
For samples from cases contracted during foreign travel (Table 1), pITS-2 SSCP profiles 1A and 1B were represented by 33 (37.5%) and 35 (39.8%) of the 88 C. parvum type 1 samples analyzed, respectively, whereas the other nine profiles (1C to 1K) were represented by 1 to 6 (1.1 to 6.7%) of those samples (Table 1). For type 1 samples, profiles 1A and 1B displayed the widest geographical distribution, followed by profiles 1G, 1C, 1F, and 1J, whereas profiles 1D, 1E, 1H, 1I, and 1K were each unique to a particular geographical region. Although the sample size representing type 2 was ∼16% of that representing type 1, the SSCP analysis of pITS-2 amplicons revealed nine distinct profiles (2A to 2I) (Table 1). For these samples, profile D had the widest geographical distribution and profiles 2A to 2C occurred on the Iberian Peninsula, whereas profiles 2E to 2I were each unique to a particular location. Profile 2F (representing sample c35) was the most distinct with respect to all other type 2 samples, in accordance with the SSCP and sequence data for the pSSU locus for this sample. Overall, the SSCP results for the samples from foreign travelers would suggest an association between some pITS-2 profiles and geographical origin, in spite of a bias in the numbers of samples from particular countries or geographical regions (Table 1). However, given the complexity of some pITS-2 SSCP profiles (particularly within type 2) (Fig. 1), it cannot necessarily be ruled out that more than one genetic variant (i.e., subgenotype) may occur within a particular sample.
For outbreak samples (Table 2), no variation in pITS-2 SSCP profile (2J, representing type 2) was detected among the 62 samples representing the waterborne outbreaks in Clitheroe and Cleethorpes, whereas four different profiles (1A, 1B, 1L, and 1M, representing type 1) were detected among the 20 samples representing the Middlesex nursery outbreak. Thus, the same subgenotype was associated with both of the waterborne outbreaks, whereas the latter findings would suggest that at least four different sources of C. parvum had contributed to the nursery outbreak, which is the subject of further epidemiological investigation. In spite of intragenotypic profile variability detected within both the foreign travel and outbreak sample groups, the two genotypes of C. parvum could be distinguished based on the pITS-2 SSCP profiles (Fig. 1 and 2). Importantly, the genotypic categorization of samples as type 1 or type 2 based on the SSCP analysis of pITS-2 was in accordance with the results achieved by using the pSSU locus.
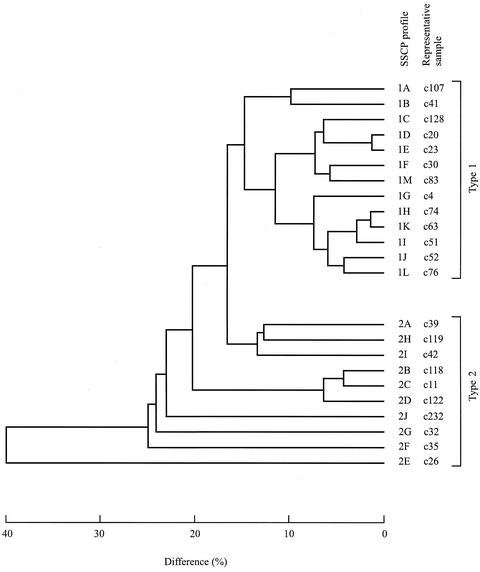
FIG. 2.
Phenogram depicting the differences (in percent) among the 23 different pITS-2 SSCP profiles detected for the 184 C. parvum samples examined. Profiles 1A to 1M are represented by type 1 samples c107 (United Kingdom), c41 (Spain), c128 (Gambia), c20 (Kenya), c23 (Pakistan), c30 (Pakistan), c4 (Caicos Islands), c74 (Spain), c51 (Jamaica), c52 (Menorca), c63 (Menorca), c76 (United Kingdom), and c83 (United Kingdom), respectively. Profiles 2A to 2J are represented by type 2 samples c39 (Spain), c118 (Spain), c11 (Portugal), c122 (Portugal), c26 (Africa), c35 (Cyprus), c32 (Turkey), c119 (Pakistan), c42 (Mexico), and c232 (United Kingdom), respectively.
Characterization of nucleotide variation in pITS-2 within and among representative C. parvum samples subjected to SSCP analysis.
In order to verify that the bands within individual SSCP profiles did indeed represent ITS-2 of C. parvum, 40 different bands (indicated in Fig. 1) representing a significant spectrum of variability were individually excised and then subjected to PCR enrichment and sequencing. Each of the bands was unique in sequence, and no polymorphic sites were detected in the sequences. The sequences derived from individual bands (bands B1 to B11, B39, and B40 representing type 1, and bands B12 to B38 representing type 2) were then subjected to database searches, with the highest identity (84 to 99%) being found with ITS sequences of C. parvum, including those with accession numbers AF015774, AF093008, AF093012, and/or AF040725. Sequence identities of ∼76 to 99% for type 1 and ∼75 to 99% for type 2 were recorded, thus providing clear support that individual bands represented the ITS-2 region. While the alignment between type 1 and 2 sequences was not entirely informative due to the high level of nucleotide difference (∼30 to 56%) between them, alignment within a genotype was readily achieved, which facilitated pairwise comparisons of sequences and the characterization of the extent and nature of nucleotide variation. Pairwise comparisons among the band sequences representing each of the genotypes (accession numbers AJ539183 to AJ539222) revealed differences varying from ∼1 to 38% (type 1) and ∼1 to 56% (type 2). A phenogram displaying the genetic differences among all of the excised bands is shown in Fig. 3. Given the complexity of SSCP profiles (Fig. 1 and 2) and the presence of multiple pITS-2 sequence types within a sample (Fig. 3 and 4), the topology of the phenogram based on SSCP profiles was not concordant with that based on sequence data (Fig. 2 and 3).
For type 1 samples (n = 5), the phenogram based on sequence data (Fig. 3) revealed two clusters (I and II) of sequence, whereas a greater number of clusters was evident for the type 2 samples (n = 7), for which more bands were sequenced (because of a higher degree of profile variation detected by SSCP [Fig. 1]). For instance, sequences belonging to both type 1 clusters I and II (differing by ∼20 to 35%) were detected within each C. parvum sample c40 and c20 (representing profiles 1A and 1D, respectively) for which the majority of SSCP bands was excised and sequenced (Fig. 3). Within cluster I, length variation in pITS-2 related mainly to the number of A's between alignment positions 15 and 20 (Fig. 4a). Ten substitutions were detected, of which A-to-G transitions occurred at positions 4, 7, and 80; C-to-T transitions occurred at positions 71, 120, 134, 147, 164, and 175; and an A-to-T transversion occurred at position 112 (Fig. 4a). Among sequences representing cluster II (alignment length of 172 bp), length variation was predominantly attributable to the presence of a unique tract, 11GGGTC15, in the sequence of band B2 which was absent from all other band sequences (Fig. 4a). A-to-G transitions were detected at positions 20, 94, and 151; C-to-T transitions were detected at positions 76 and 169; and A-to-T transversions were detected at positions 22, 23, and 172. Comparison between the consensus sequences representing clusters I and II revealed unequivocal nucleotide differences at alignment positions 107, 124, 144, 146, 148, 160, 179, 181, and 182 (A-to-G); 123, 129, 130, and 166 (A-to-T); 171 (C-to-T); 167 (G-to-C); and 131 and 168 (G-to-T), as well as insertion-deletion events at seven positions (Fig. 4). Comparison among all 13 type 1 band sequences revealed nucleotide variation at 57 of 182 alignment positions (Fig. 4a).
The pattern of nucleotide variation among type 2 sequences differed from that among the type 1 sequences in that there was no clear separation into two clusters (Fig. 4b). The length and nucleotide variation detected within type 2 was associated predominantly with insertion-deletion events, microsatellite variation [predominantly AA, AT, TA, T, or poly(T)], and point mutations. The microsatellite variability occurred chiefly in two highly AT-rich regions between alignment positions 1 and 40 and between positions 101 and 138, whereas point mutations were mainly between alignment positions 1 and 27, 35 and 53, 73 and 84, 118 and 124, and 146 and 174 (Fig. 4b). A selected analysis of nucleotide variation within each of four type 2 samples (c39, c118, c35, and c42) for which the majority of dominant SSCP bands were sequenced (alignments not shown) revealed four to six distinct sequence variants per sample, differing by 3 to 13% (c39), 3 to 10% (c118), 2 to 6% (c35), or 4 to 9% (c42) upon pairwise comparison. Comparison among all of the 29 type 2 band sequences in the alignment (Fig. 4b) revealed nucleotide variation at 65 of 175 positions. Based on these findings, the spectrum of nucleotide variation among bands within some samples examined was greater, for example, than that reported previously by Le Blancq et al. (22), where two ITS sequence types (A and B) were detected within a bovine isolate (KSU-1) of C. parvum by using a molecular cloning approach (Fig. 4b).
Adaptation to DPGE analysis.
After having conducted the SSCP analyses and characterized nucleotide and/or length variation in both the pSSU and pITS-2 loci between and/or within genotypes (Fig. 1 to 4), we aimed at decreasing the electrophoretic running time required for analysis of amplicons. We therefore evaluated whether reliable differentiation between type 1 and type 2 of C. parvum could also be achieved by DPGE using either or both of the ribosomal loci. We also assessed whether the intragenotypic variability detectable in the pITS-2 locus by SSCP analysis (Fig. 1) could be displayed reliably. DPGE analysis of the samples representing the entire spectrum of variability (for the 184 samples and the two ribosomal loci) revealed the same genotypic identification and a similar subgenotypic classification as did the SSCP analysis (Fig. 5). While the subgenotypic classification of the type 2 samples by DPGE was the same as that for SSCP analysis, ∼30% less profile variation (9 instead 13 profiles) was detectable among the type 1 samples, which is attributable to a lack of length (rather than sequence) variation among some of the samples. These results reinforced the superior ability of SSCP to detect subtle nucleotide variation among molecules of the same length within an amplicon (13, 35) and reflected the finding of increased sequence and length variability in pITS-2 within and among type 2 samples compared with type 1.
FIG. 5.
Representative DPGE analysis of sequence variation in the two loci, pSSU (top) and pITS-2 (bottom), within and among C. parvum samples from humans. Subsets from the 184 samples scanned at both loci by SSCP, and representing the entire spectrum of sequence variation detected have been included. For pSSU, the first 19 samples (c2 to c4) were identified as type 1, and the following 11 samples (c39 to c42) were identified as type 2 (Fig. 1). Samples c107 and c232 from humans from the United Kingdom were included as type 1 and type 2 reference controls, respectively. For pITS-2, the first 4 samples (c76 to c107) from a nursery outbreak in Middlesex (United Kingdom) and the next 11 samples (c40 to c63) from humans following recent foreign travel were identified as type 1. Another nine samples (c65 to c42) from patients following overseas travel and two samples (c232 and c254) from a waterborne outbreak at Clitheroe (United Kingdom) were identified as type 2. Significant pITS-2 profile variation was detected within both types 1 and 2. The level of sequence variation displayed within and among samples representing type 2 was greater than that for type 1, in accordance with SSCP analysis (Fig. 1).
DISCUSSION
In this investigation, significant population variation (i.e., sequence and/or length variation) was detected within each of the two C. parvum genotypes, which contrasts with some of the original proposals of a limited intragenotypic variability (9, 15) and supports a number of recent studies (1-3, 11, 21, 28) revealing considerable levels of genetic variation within the species. For example, a recent investigation (2) using microsatellite analysis of 94 C. parvum samples from humans and animals revealed two subgenotypes within type 1 and four subgenotypes within type 2. Some of the subgenotypes had a broad geographical distribution, whereas others were limited to specific locations. In another study, two distinct subgenotypes were identified within each type by employing nine microsatellite loci (1). Interestingly, some of the sequence heterogeneity in pITS-2 detected in the present study within and among type 2 samples of C. parvum was also associated with microsatellite variability [predominantly AT, TA, T, or poly(T)], which is consistent with the AT richness of the C. parvum genome (2). In addition to its significance for investigating population genetic structures and possibly transmission patterns of C. parvum, the accurate display of sequence heterogeneity in the ITS may be of fundamental relevance for studying molecular evolutionary processes and inheritance in C. parvum. In a detailed study of C. parvum (using a molecular cloning approach), Le Blancq et al. (22) demonstrated that there are at least five copies of the rDNA unit per haploid genome, which are not organized in a conventional head-to-tail tandem array. There was evidence that the rDNA units are dispersed (as single copies) to at least three different chromosomes. Two structurally distinct types of rDNA unit were detected, namely, type A (four copies) and type B (one copy), and the most significant sequence variability between the two types related to the ITS. While the magnitude of sequence divergence in the ITS region of C. parvum appears to be similar to that in the related apicomplexan parasite Plasmodium berghei (7), the functional significance of the two types of rDNA in C. parvum is presently unknown, although in the malarial parasite, the different sets of rRNA genes give rise to structurally distinct, stage-specific ribosomes (20, 23). It is also unclear whether such heterogeneity relates to sequence variation within or among individual oocysts in an isolate. A micromanipulation, PCR-based SSCP approach could be employed to address this question through the analysis of single Cryptosporidium oocysts (33).
The nature of nucleotide variation in pITS-2 within C. parvum detected here was distinct from that found in previous studies (22, 25) in that novel sequences were detected (Fig. 4a). The most likely reason for this is that the design of the primers allowed the specific amplification of more sequence types from the C. parvum samples. Also, the magnitude of nucleotide variation detected here within both type 1 and type 2 of C. parvum was greater than that described in previous reports (22, 25), which most likely relates to the use of PCR-based enrichment of SSCP bands prior to sequencing rather than direct sequencing from cloned amplicons. In the present investigation, none of the SSCP band sequences had 100% identity with other bands within a particular sample (for which multiple bands were sequenced) (e.g., bands B1 to B4 in sample c40 [Fig. 3]), which was not surprising because no attempt was made to unequivocally define sense and antisense bands on SSCP gels (16, 36).
The relatively high degree of sequence heterogeneity in the ITS-2 region, which is predicted to be present at a low copy number and as single copies on different chromosomes (22), indicates that sequence homogenization (“concerted evolution” [8]) is less effective in C. parvum (Fig. 1) than in some other organisms with a high copy numbers (on a single locus) organized in tandem arrays, where sequence homogeneity is maintained by intrachromosomal exchange processes (16, 19, 30). The sequence heterogeneity in the ITS of some apicomplexan parasites appears to be constrained by selection to maintain distinct sets of rRNA genes (23, 35). While intraisolate (or intraorganism) variation can represent a practical problem for genetic analyses of oocyst DNA samples by using some PCR approaches (12), such as PCR-RFLP and direct sequencing, it presents a distinct advantage for the detection of population variation within Cryptosporidium by mutation scanning, as demonstrated clearly here. Indeed, the PCR-based enrichment-cloning approach employed prior to the sequencing of SSCP bands indicates that sequence heterogeneity is greater (in that there are more than two sequence types per genotype) than predicted previously from direct cloning (22, 25). Although the biological and epidemiological significance of the subgenotypic variability in pITS-2 is currently unknown, some subgenotypes showed a relatively broad geographical distribution, whereas others were restricted to particular regions (Table 1). Nonetheless, further study is needed to establish whether there is a definite link to the geographical origin of an isolate. If this is indeed the case, this would have significant implications for the tracking of original sources of infection.
In conclusion, the results of the present study demonstrate clearly the applicability of both SSCP and DPGE to screen C. parvum DNA samples for genetic variation and to categorize them to both the genotypic and subgenotypic levels. These electrophoretic methods have significant advantages over some other molecular diagnostic approaches applied to Cryptosporidium. In contrast to some PCR-based approaches which rely on the genotypic identification of Cryptosporidium based solely on the detection of bands of particular sizes on agarose gels (providing a phenotypic rather than a genotypic readout), SSCP enables a detailed, qualitative analysis of amplicons for both length and sequence variability. This also has the advantage that any eventual contaminating or erroneous amplicons of the same size as those of C. parvum could be differentiated from specific ones based on the migratory characteristics of bands in the profiles. The present SSCP was able to detect a transversion event (A to T) in pSSU between type 2 samples, demonstrating its high mutation detection rate. This information, together with preliminary findings for C. felis, C. serpentis, and C. baileyi (18), reinforces the view that SSCP analysis of pSSU should enable the specific identification of all currently recognized species of Cryptosporidium (9, 10), although this proposal still requires testing. The concordance of the pSSU and pITS-2 SSCP results in the genotypic categorization of the C. parvum samples and the ability to directly fingerprint intragenotypic nucleotide variability in the ITS-2 region clearly reinforce the usefulness of mutation scanning for detecting population variation within Cryptosporidium species. By using SSCP or DPGE, large numbers of samples can be scanned directly for genetic variation without the need for DNA sequence analysis first, which reduces time, labor, and cost. The approaches are also considerably less expensive than, for example, real-time PCR, because they use conventional equipment readily available in most molecular biology laboratories. Therefore, the present tools should be useful for studying the epidemiology and the genetic makeup of Cryptosporidium populations as well as for the diagnosis and monitoring of cryptosporidiosis outbreaks. Future work will be aimed at the automation of electrophoretic analysis, which would increase throughput capability and should simplify the recording and storage of electrophoretic profile data. Similar approaches should be applicable to a range of other pathogens of socioeconomic importance.
Acknowledgments
Kristin Elwin and Anne Thomas (PHLS Cryptosporidium Reference Unit) are gratefully acknowledged for maintaining the collection of oocyst isolates and the initial genotypic classification thereof. Thanks are extended to local microbiology laboratories for providing fecal specimens, to Shuja Shafi (Central Middlesex PHL) for assisting in the identification of human cases associated with the nursery outbreak, to Xingquan Zhu for preliminary work, to Neil Chilton for constructive criticisms on the final manuscript, and to other colleagues for previously providing some parasites used for preparing control DNA samples in the present study.
Project support through GeneType Pty Ltd, the University of Melbourne (Collaborative Research Program scheme), and DEFRA is gratefully acknowledged.
REFERENCES
- 1.Aiello, A. E., L. Xiao, J. R. Limor, C. Liu, M. S. Abrahamsen, and A. A. Lal. 1999. Microsatellite analysis of the human and bovine genotypes of Cryptosporidium parvum. J. Eukaryot. Microbiol. 46:46S-47S. [PubMed]
- 2.Caccio, S., W. Homan, R. Camilli, G. Traldi, T. Kortbeek, and E. Pozio. 2000. A microsatellite marker reveals population heterogeneity within human and animal genotypes of Cryptosporidium parvum. Parasitology 120:237-244. [DOI] [PubMed] [Google Scholar]
- 3.Caccio, S., F. Spano, and E. Pozio. 2001. Large sequence variation at two microsatellite loci among zoonotic (genotype C) isolates of Cryptosporidium parvum. Int. J. Parasitol. 31:1082-1086. [DOI] [PubMed] [Google Scholar]
- 4.Carraway, M., S. Tzipori, and G. Widmer. 1996. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl. Environ. Microbiol. 62:712-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalmers, R. M., K. Elwin, W. J. Reilly, H. Irvine, A. L. Thomas, and P. R. Hunter. 2002. Cryptosporidium in farmed animals: the detection of a novel isolate in sheep. Int. J. Parasitol. 32:21-26. [DOI] [PubMed] [Google Scholar]
- 6.Chalmers, R. M., K. Elwin, A. Thomas, and D. H. M. Joynson. 2002. Infection with unusual types of Cryptosporidium is not restricted to immunocompromised patients. J. Infect. Dis. 185:270-271. [DOI] [PubMed] [Google Scholar]
- 7.Dame, J. B., M. Sullivan, and T. F. McCutchan. 1984. Two major sequence classes of ribosomal RNA genes in Plasmodium berghei. Nucleic Acids Res. 12:5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elder, J. F., and B. J. Turner. 1995. Concerted evolution of repetitive DNA sequences in eukaryotes. Q. Rev. Biol. 70:297-320. [DOI] [PubMed] [Google Scholar]
- 9.Fayer, R., U. Morgan, and S. J. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol. 30:1305-1322. [DOI] [PubMed] [Google Scholar]
- 10.Fayer, R., J. M. Trout, L. Xiao, U. M. Morgan, A. A. Lal, and J. P. Dubey. 2001. Cryptosporidium canis n. sp. from domestic dogs. J. Parasitol. 87:1415-1422. [DOI] [PubMed] [Google Scholar]
- 11.Feng, X., S. M. Rich, D. Akiyoshi, J. K. Tumwine, A. Kekitiinwa, N. Nabukeera, S. Tzipori, and G. Widmer. 2000. Extensive polymorphism in Cryptosporidium parvum identified by multilocus microsatellite analysis. Appl. Environ. Microbiol. 66:3344-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasser, R. B. 1997. Mutation scanning methods for the analysis of parasite genes. Int. J. Parasitol. 27:1449-1463. [DOI] [PubMed] [Google Scholar]
- 13.Gasser, R. B., and N. B. Chilton. 2001. Applications of single-strand conformation polymorphism (SSCP) to taxonomy, diagnosis, population genetics and molecular evolution of parasitic nematodes. Vet. Parasitol. 101:201-213. [DOI] [PubMed] [Google Scholar]
- 14.Gasser, R. B., and J. R. Monti. 1997. Identification of parasitic nematodes by PCR-SSCP of ITS-2 rDNA. Mol. Cell Probes 11:201-209. [DOI] [PubMed] [Google Scholar]
- 15.Gasser, R. B., and P. O'Donoghue. 1999. Isolation, propagation and characterisation of Cryptosporidium. Int. J. Parasitol. 29:1379-1413. [DOI] [PubMed] [Google Scholar]
- 16.Gasser, R. B., J. R. Monti, B.-Z. Qian, A. M. Polderman, P. Nansen, and N. B. Chilton. 1998. A mutation scanning approach for the identification of hookworm species and analysis of population variation. Mol. Biochem. Parasitol. 92:303-312. [DOI] [PubMed] [Google Scholar]
- 17.Gasser, R. B., X. Q. Zhu, I. Beveridge, and N. B. Chilton. 2001. Mutation scanning analysis of sequence heterogeneity in the second internal transcribed spacer (rDNA) within some members of the Hypodontus macropi (Nematoda: Strongyloidea) complex. Electrophoresis 22:1076-1085. [DOI] [PubMed] [Google Scholar]
- 18.Gasser, R. B., X. Q. Zhu, S. Caccio, R. Chalmers, G. Widmer, U. M. Morgan, R. C. A. Thompson, E. Pozio, and G. F. Browning. 2001. Genotyping Cryptosporidium parvum by single-strand conformation polymorphism analysis of ribosomal and heat shock gene regions. Electrophoresis 22:433-437. [DOI] [PubMed] [Google Scholar]
- 19.Gasser, R. B., X. Q. Zhu, N. B. Chilton, L. A. Newton, T. Nedergaard, and P. Guldberg. 1998. Analysis of sequence homogenisation in rDNA arrays of Haemonchus contortus by denaturing gradient gel electrophoresis. Electrophoresis 19:2391-2395. [DOI] [PubMed] [Google Scholar]
- 20.Gunderson, J. H., M. L. Sogin, G. Wollett, M. Hollingdale, V. F. De La Cruz, A. P. Waters, and T. F. McCutchan. 1987. Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science 238:933-937. [DOI] [PubMed] [Google Scholar]
- 21.Guyot, K., A. Follet-Dumoulin, E. LeLievre, C. Sarfati, M. Rabodonirina, G. Nevez, J. C. Cailliez, D. Camus, and E. Dei-Cas. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39:3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Blancq, S. M., N. V. Khramstov, F. Zamani, S. J. Upton, and T. W. Woo. 1997. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol. Biochem. Parasitol. 90:463-478. [DOI] [PubMed] [Google Scholar]
- 23.McCutchan, T. F., J. Li, G. A. McConkey, M. J. Rogers, and A. P. Waters. 1995. The cytoplasmic ribosomal RNAs of Plasmodium spp. Parasitol. Today 11:134-138. [DOI] [PubMed] [Google Scholar]
- 24.Morgan, U. M., C. C. Constantine, D. A. Forbes, and R. C. A. Thompson. 1997. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J. Parasitol. 83:825-830. [PubMed] [Google Scholar]
- 25.Morgan, U. M., P. Deplazes, D. A. Forbes, F. Spano, H. Hertzberg, K. D. Sargent, A. Elliot, and R. C. A. Thompson. 1999. Sequence and PCR-RFLP analysis of the internal transcribed spacers of the rDNA repeat unit in isolates of Cryptosporidium from different hosts. Parasitology 118:49-58. [DOI] [PubMed] [Google Scholar]
- 26.Morgan, U. M., L. Xiao, R. Fayer, A. A. Lal, and R. C. A. Thompson. 1999. Variation in Cryptosporidium: towards a taxonomic revision of the genus. Int. J. Parasitol. 29:1733-1751. [DOI] [PubMed] [Google Scholar]
- 27.O'Donoghue, P. J. 1995. Cryptosporidium and cryptosporidiosis in man and animals. Int. J. Parasitol. 25:139-195. [DOI] [PubMed] [Google Scholar]
- 28.Ong, C. S., D. L. Eisler, A. Alikhani, V. W. Fung, J. Tomblin, W. R. Bowie, and J. L. Isaac-Renton. 2002. Novel cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a cervine genotype. Emerg. Infect. Dis. 8:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quintero-Betancourt, W., E. R. Peele, and J. B. Rose. 2002. Cryptosporidium parvum and Cyclospora cayetanensis: a review of laboratory methods for detection of these waterborne parasites. J. Microbiol. Methods 49:209-224. [DOI] [PubMed] [Google Scholar]
- 30.Schlötterer, C., and D. Tautz. 1994. Chromosomal homogeneity of Drosophila ribosomal DNA arrays suggests intrachromosomal exchanges drive concerted evolution. Curr. Biol. 4:777-783. [DOI] [PubMed] [Google Scholar]
- 31.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy: the principles and practice of numerical classification. W. H. Freeman, San Francisco, Calif.
- 32.Spano, F., L. Putignani, J. McLauchlin, D. P. Casemore, and A. Crisanti. 1997. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 150:207-217. [DOI] [PubMed] [Google Scholar]
- 33.Sturbaum, G. D., C. Reed, P. J. Hoover, B. H. Jost, M. M. Marshall, and C. R. Sterling. 2001. Species-specific, nested PCR-restriction fragment length polymorphism detection of single Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 67:2665-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Widmer, G. 1998. Genetic heterogeneity and PCR detection of Cryptosporidium parvum. Adv. Parasitol. 40:223-239. [DOI] [PubMed] [Google Scholar]
- 35.Woods, W. G., D. G. Richards, K. G. Whithear, G. R. Anderson, W. K. Jorgensen, and R. B. Gasser. 2000. High-resolution electrophoretic procedures for the identification of five Eimeria species from chickens, and detection of population variation. Electrophoresis 21:3558-3563. [DOI] [PubMed] [Google Scholar]
- 36.Zhu, X. Q., and R. B. Gasser. 1998. Single-strand conformation polymorphism (SSCP)-based mutation scanning approaches to fingerprint sequence variation in ribosomal DNA of ascaridoid nematodes. Electrophoresis 19:1366-1373. [DOI] [PubMed] [Google Scholar]