Abstract
Traditional microscope-based estimates of species richness of aquatic hyphomycetes depend upon the ability of the species in the community to sporulate. Molecular techniques which detect DNA from all stages of the life cycle could potentially circumvent the problems associated with traditional methods. Leaf disks from red maple, alder, linden, beech, and oak as well as birch wood sticks were submerged in a stream in southeastern Canada for 7, 14, and 28 days. Fungal biomass, estimated by the amount of ergosterol present, increased with time on all substrates. Alder, linden, and maple leaves were colonized earlier and accumulated the highest fungal biomass. Counts and identifications of released conidia suggested that fungal species richness increased, while community evenness decreased, with time (up to 11 species on day 28). Conidia of Articulospora tetracladia dominated. Modifications of two molecular methods—denaturing gradient gel electrophoresis (DGGE) and terminal restriction fragment length polymorphism (T-RFLP) analysis—suggested that both species richness and community evenness decreased with time. The dominant ribotype matched that of A. tetracladia. Species richness estimates based on DGGE were consistently higher than those based on T-RFLP analysis and exceeded those based on spore identification on days 7 and 14. Since traditional and molecular techniques assess different aspects of the fungal organism, both are essential for a balanced view of fungal succession on leaves decaying in streams.
Aquatic hyphomycetes belong to a phylogenetically heterogeneous group of true fungi that dominate the breakdown of leaves and other allochthonous detritus in streams and rivers (5, 15). Their activity increases the palatability of the substrate to detritus feeders. Macroinvertebrates discriminate between, and show preferences for, particular fungal species (2, 6, 26). To fully characterize the fungal contribution to leaf decay and invertebrate nutrition, the aquatic hyphomycete community must be subdivided into its constituent species. The same principle applies when investigating relationships between biodiversity and ecological functions—a topic that has attracted increasing attention in the last decade (18, 33).
Most of the fungal biomass on decaying leaves consists of vegetative (nonreproducing) hyphae that cannot be identified through conventional microscopy. To characterize community structure, leaf surfaces often are examined for sporulating structures (e.g., see references 7 and 30), or conidia released from the leaf are collected, counted, and identified (e.g., see reference 3). The obvious shortcoming to these protocols is that absence of conidia might be due to the absence of species or to the presence of nonsporulating mycelium. In the initial phase of fungal colonization, between the landing of conidia and their growth into a sporulating colony, newly arrived species will escape detection by traditional microscope-based techniques.
Molecular approaches characterize nucleic acids that are present in all stages of the fungal life cycle, and could circumvent the problems associated with the microscopy-based techniques. In bacterial ecology, two methods have been particularly useful: terminal restriction fragment length polymorphism (T-RFLP) analysis (20) and denaturing gradient gel electrophoresis (DGGE) (23). In T-RFLP analysis, PCR of the extracted community DNA is performed with one primer labeled fluorescently at the 5′ end. The PCR products are digested with a restriction enzyme, and the labeled terminal restriction fragments are separated and detected with a DNA sequencer. The number of different-size fragments provides an estimate of the minimum number of strains present in the community (20). In DGGE, the mixture of PCR products of the microbial community is separated based on differences in the ease of denaturation arising from sequence variability (23). Even though T-RFLP analysis and DGGE were developed to study bacterial populations, they also can be used in studies of fungal ecology (19). They have been used, for example, to characterize fungi associated with corn silage (22), with roots of Scots pine seedlings (17), and with Spartina alterniflora decaying in salt marshes (10).
Here, we report an investigation into the early stages of colonization of five leaf species and birch wood by aquatic hyphomycetes. Results obtained with T-RFLP analysis and DGGE were compared with results from traditional microscope-based techniques. If, as generally accepted, aquatic hyphomycetes dominate leaf decay, we expect the discrepancy between molecular and traditional estimates of fungal diversity to be highest on slowly decaying substrates. Limited fungal growth is likely to delay the onset of reproduction. Molecular data will also provide an alternative estimate of fungal diversity on leaves and other substrates and potentially alert us to the presence of fungi other than aquatic hyphomycetes.
MATERIALS AND METHODS
Study site.
The field experiment was conducted in May 2002 in Boss Brook, a small stream in Fenwick, Nova Scotia, Canada (45°43.000′N, 064°09.56′W) (4). This first-order stream runs through a mixed forest dominated by white birch (Betula papyrifera Marsh), several maple species (Acer rubrum L., Acer saccharum Marsh., and Acer spicatum Lam.), and white spruce [Picea glauca (Moench) Voss]. The stream bed consists of stones and gravel. At the sampling site, the stream is 2 to 3 m wide and 20 to 50 cm deep.
Substrates.
Senescent leaves from red maple (A. rubrum), linden (Tilia cordata Mill.), alder [Alnus glutinosa (L.) Gaertn.], beech (Fagus sylvatica L.), and oak (Quercus rubra L.) were collected from single trees in the fall of 2001. They were leached in tap water for 2 h, cut into 18-mm-diameter disks with a corkborer, and dried at room temperature. Wooden Popsicle sticks (B. papyrifera) were cut into sections 1 by 6 cm. Sets of 20 disks or eight sticks were placed in nylon mesh bags (20 by 15 cm; mesh size, 2 mm), which were attached to bricks in the stream. Three arbitrarily chosen bags of each substrate were recovered after 1, 2, or 4 weeks of submergence. On each sampling date, air and water temperature and water pH were measured. Ten leaf disks or four wood sections from each bag were used to induce sporulation; the other 10 were freeze-dried for ergosterol analysis and DNA extraction.
Spore production.
Ten leaf disks from each bag were rinsed with running tap water to remove adherent material and incubated with shaking in 200 ml of autoclaved distilled water for 2 days at 16°C to induce spore formation (21). The water was filtered through an 8-μm-pore-size membrane filter (Millipore Corporation, Bedford, Mass.) that was stained with cotton blue in lactophenol (50 mg liter−1). One quarter of the filter was fixed on a slide with 3 drops of lactic acid, and the section was observed through a light microscope (magnification, 400×). All spores on the section were identified and counted. The leaf or wood material used for spore induction was freeze-dried and weighed, and the results are expressed as number of conidia day−1 milligram (dry mass)−1.
Ergosterol analysis.
The method for microwave-assisted ergosterol extraction was modified from that of Young (36). Freeze-dried leaf disks (50 to 100 mg) were ground in liquid nitrogen, suspended in a solution containing 2.0 ml of methanol and 0.5 ml of 2 M NaOH, and sealed in 15-ml glass tubes. Six extraction tubes were placed in a 250-ml capped plastic bottle. The samples were microwaved at 50% power for 95 s in a microwave oven (Sears Kenmore model 85055; Sears Canada, Inc., Toronto, Ontario, Canada) and removed from the plastic bottle after cooling to room temperature. The solution was neutralized with 1 ml of 1 M HCl, and ergosterol was extracted with three consecutive hexane washes. The combined hexane fractions were evaporated to dryness, and the residue was reextracted in 1 ml of methanol. The solution was injected into a high-performance liquid chromatography C18 column (Varian, Palo Alto, Calif.) and eluted with methanol at 1.5 ml min−1 at 20°C (elution time, 5.3 min). Ergosterol content was estimated by comparison of peak areas with those of external standards.
DNA sources.
Cultures of Anguillospora crassa Ingold (strain F-15283), Anguillospora filiformis Greathead (F-20687), Anguillospora furtiva Descals et Marvanová (F-20483) Anguillospora longissima (Sacc. et Syd.) Ingold (F-00980), Anguillospora rosea Descals et Marvanová (F-08983), Anguillospora rubescens Gulis et Marvanová (F-11298), Clavariopsis aquatica De Wild. (F-10791), Colispora elongata Marvanová (F-08382), Tetracladium marchalianum De Wild. (F-11391), Tetracladium setigerum (Grove) Ingold (F-20987), and Tumularia aquatica (Ingold) Descals et Marvanová (F-02081) were provided by L. Marvanová from the Czech Collection of Microorganisms. A culture of Heliscus lugdunensis Sacc. et Thérry (CS-950) was provided by G. Krauss from UFZ-Centre for Environmental Research, Halle, Germany. Aspergillus niger (85 W 4100), Escherichia coli (85 W 0400), Chlamydomonas reinhardtii (86 W 0102), and Rhizopus stolonifer (85 W 4900) were obtained from Ward's Supply Company (St. Catharines, Ontario, Canada), and Pythium irregulare (BA-15-6214) was from Carolina Biological Supplies (Burlington, N.C.). The cultures were grown in 1% malt broth for 3 weeks, and the mycelia were filtered, freeze-dried, and used for DNA extraction. Thalassiosira pseudonana CCMP 1007 was obtained from the Center for Culture of Marine Phytoplankton (West Woothbay Harbor, Maine). Agaricus bisporus was obtained from a local grocery store. Leaves of A. rubrum were collected from a single tree on the university campus, while specimens of Corophium volutator were collected from mudflats in the Bay of Fundy, near Dorchester, New Brunswick, Canada.
DNA extraction and amplification.
DNA was isolated from 25 mg of freeze-dried leaf material with a soil DNA extraction kit (MoBio Laboratories, Solana Beach, Calif.) or from 30 mg of freeze-dried pure fungal cultures following phenol-chloroform extraction (24). The 5′ end of the 18S ribosomal DNA (rDNA) was amplified with the primer pair NS1 (35) and the fungus-specific primer GCfung (22). The 3′ end of the 18S rDNA of pure aquatic hyphomycete cultures was amplified with NS5 (35) and with primer D (12) labeled with the fluorescent dye Cy5 (12). The 3′ end of 18S rDNA from environmental samples was amplified with the fungus-specific primer F1300 (5′ GATAACGAACGAGACCTTAAC 3′) and Cy5-labeled primer D. PCR was carried out with Ready-To-Go PCR beads (Amersham Biosciences, Piscataway, N.J.) with a 0.2 μM concentration of each primer and 50 ng of DNA. Initial denaturation for 2 min at 95°C was followed by 35 cycles of denaturation for 30 s at 95°C, primer annealing for 30 s at 55°C, and extension for 1 min at 72°C. A final extension for 5 min at 72°C completed any partial polymerizations.
T-RFLP analysis.
The 680-bp product of the amplification with NS5 and D was purified with the GFX DNA purification kit (Amersham Biosciences). Restriction digests were carried out in a 15-μl volume containing 10 ng of purified PCR product and 10 U of a restriction enzyme. The following enzymes were used: CfoI, DdeI, HaeIII, HinfI, MspI, and RsaI (Roche Diagnostics, Indianapolis, Ind.). Digests were performed at 37°C for 2 h. Formamide loading dye (3 μl) was added to 5 μl of digested DNA. Terminal restriction fragment lengths were determined on a Long Read Tower DNA sequencer (Visible Genetics, Suwanee, Ga.) in fragment analysis mode.
T-RFLP analysis of environmental samples.
The 450-bp product of the amplification with F1300-D was purified, digested, and analyzed as described above. Restriction digests were carried out with DdeI (Roche Diagnostics).
DGGE.
The 370-bp product from the amplification with NS1 and GCfung was analyzed with the DCode mutation detection system (Bio-Rad, Hercules, Calif.). Samples (3 μl standard or 10 μl environmental sample) were loaded on 8% (wt/vol) polyacrylamide gels in 1× Tris-acetate-EDTA with denaturing gradient from 20 to 55% (100% denaturant corresponds to 40% formamide and 7 M urea). The gels were run in 1× TAE at 70 V and 56°C for 12 h and stained with 1× GelStar (BioWhittaker Molecular Applications, Rockland, Maine). The gel images were captured under UV light with a Sony digital video camera and analyzed with the computer program NIH Image (National Institutes of Health, Bethesda, Md.).
To ensure that the efficiency of amplification was consistent for all templates, we carried out experiments where mycelia from A. longissima and C. aquatica were mixed in ratios of decreasing concentration of A. longissima (such that it was 50, 33, 25, 20, 10, 7, 5, 3, 2, and 1% of the total mycelia). The DNA from the mycelia was extracted, amplified, and separated on DGGE. The gel images were analyzed with NIH Image software (National Institutes of Health) to determine if the ratios of band intensities reflected the initial template ratios. The extraction and amplification efficiencies were consistent. When A. longissima was present as 1 and 2% of the total mycelial mixture, it was not detected on DGGE. The lowest detectable level was 3% of the total PCR product.
RESULTS
Ergosterol accumulation and spore production.
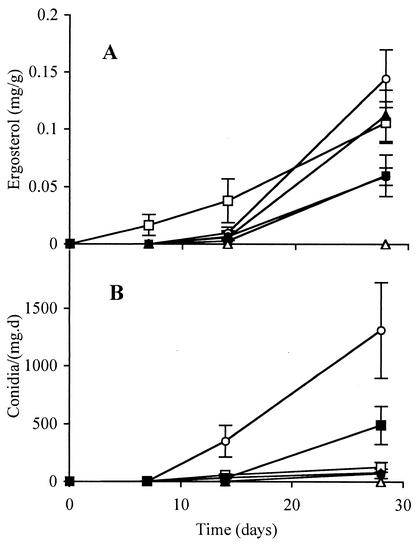
Ergosterol content and spore production both increased with time of submergence in the stream (Fig. 1). Alder, beech, and linden accumulated the highest ergosterol levels. Alder and maple released the greatest number of spores after 4 weeks. The more recalcitrant substrates such as oak and wood accumulated fungal biomass more slowly. The ergosterol concentration extracted from wood was low but detectable. During the decomposition period 11 fungal species were identified (Table 1). After 1 week the spore counts were very low, and on leaves (except beech) they were fairly evenly distributed among a few species. By contrast, the community on birch wood was dominated by H. lugdunensis. After 4 weeks the species richness was higher, but evenness was lower (except on beech). The spore counts from all substrates were dominated by Articulospora tetracladia.
FIG. 1.
Ergosterol content (A) (in milligrams gram of dry mass−1) and conidium production (B) (conidia day−1 milligram of dry mass−1) in linden (□), maple (▪), alder (○), oak (•), and beech (▴) leaves and birch wood sticks (▵) decaying in Boss Brook between 30 April and 28 May 2002. Results are averages of three replicates ± standard errors of the means (error bars).
TABLE 1.
Percentage of contributions of aquatic hyphomycete species to spore production from leaves and birch wood exposed in Boss Brook for 7, 14, or 28 days
| Substrate | % Contribution to spore production after exposure (days)
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Linden
|
Maple
|
Birch wood
|
Alder
|
Beech
|
Oak
|
|||||||||||||
| 7 | 14 | 28 | 7 | 14 | 28 | 7 | 14 | 28 | 7 | 14 | 28 | 7 | 14 | 28 | 7 | 14 | 28 | |
| Alatospora acuminata | 8 | 2 | 3 | 5 | 4 | 6 | 1 | 2 | 16 | 6 | 13 | 4 | ||||||
| Anguillospora longissima | 22 | 11 | 10 | 15 | 17 | 7 | 25 | 10 | 16 | 6 | 6 | 3 | 5 | 12 | 18 | 4 | 11 | |
| Anguillospora filiformis | 33 | 2 | 3 | 10 | 5 | 1 | 23 | 1 | 5 | 1 | 1 | 10 | 4 | 4 | 17 | 2 | ||
| Articulospora tetracladia | 22 | 43 | 78 | 18 | 42 | 74 | 17 | 23 | 61 | 42 | 61 | 71 | 13 | 16 | 58 | 36 | 29 | 62 |
| Clavariopsis aquatica | 3 | 2 | 1 | 3 | 1 | 5 | 1 | 1 | ||||||||||
| Flagellospora curvula | 13 | 2 | 3 | 3 | 8 | 8 | 9 | 8 | 2 | 9 | 2 | 6 | 18 | 17 | 11 | |||
| Heliscus lugdunensis | 22 | 2 | 2 | 43 | 9 | 1 | 50 | 31 | 9 | 21 | 20 | 2 | 67 | 58 | 2 | 27 | 21 | 2 |
| Tetrachaetum elegans | 3 | 3 | 1 | 5 | 1 | 1 | 1 | 3 | ||||||||||
| Tetracladium marchalianum | 3 | 1 | 2 | 1 | 3 | 1 | 1 | 2 | ||||||||||
| Tricladium sp. | 15 | 0 | 8 | 3 | 8 | 2 | 2 | 2 | 3 | 7 | 2 | |||||||
| Varicosporium elodeae | 3 | 13 | 6 | 2 | 8 | 1 | 5 | 2 | 1 | 1 | ||||||||
| Total no. of species | 4 | 9 | 8 | 6 | 10 | 11 | 4 | 6 | 9 | 7 | 11 | 10 | 7 | 6 | 11 | 4 | 6 | 11 |
Fungus-specific primer design.
The 18S rDNA sequences from 40 true fungi were aligned with sequences from groups we wanted to exclude from the amplification, e.g., plant substrate, algae, diatoms, bacteria and possibly microinvertebrates. The designed primer F1300 binds approximately 450 bases upstream from the well-described primer NS8 (35) whose sequence partially coincides with the primer D used in this study. We performed PCRs with template DNA from all pure aquatic hyphomycete controls, plants and representative species from other taxa. The primer pair amplified DNA from all pure aquatic hyphomycete controls, as well as ascomycetes (Aspergillus niger) and basidiomycetes (Agaricus bisporus), but excluded zygomycetes (Rhizopus stolonifer), oomycetes (Pythium irregulare), bacteria (Escherichia coli), diatoms (Thalassiosira pseudonana), algae (Chlamydomonas reinhardtii), plants (Acer rubrum), and animals (Corophium volutator). The optimal annealing temperature of the primer pair F1300 and D was empirically found to be 55°C; annealing temperatures below 45°C resulted in unspecific amplification of the plant substrates and algae. The amplification was unsuccessful at annealing temperatures above 60°C. The Ready-To-Go PCR beads (Amersham Biosciences) contained Mg2+ concentration of 1.5 mM which was sufficient for specific amplification of fungal DNA. Other Mg2+ concentrations were not tested.
Terminal-RFLP analysis.
We used the amplification product of F1300 and D for T-RFLP analysis of the aquatic hyphomycete standards. For all six restriction enzymes used, we observed higher variability in restriction fragment lengths at the 3′ end of the amplification product than at the 5′ end. The restriction enzyme DdeI gave the most variation of restriction fragment lengths in the pure aquatic hyphomycete controls, followed by CfoI, HaeIII, and HinfI. MspI and RsaI had conserved restriction sites. With DdeI, we could not differentiate between the two Tetracladium species (T. marchalianum and T. setigerum), or between C. aquatica, A. furtiva, and T. aquatica, or between A. filiformis and A. rosea. Thus, the 12 species of aquatic hyphomycetes tested yielded a total of eight distinct fragments.
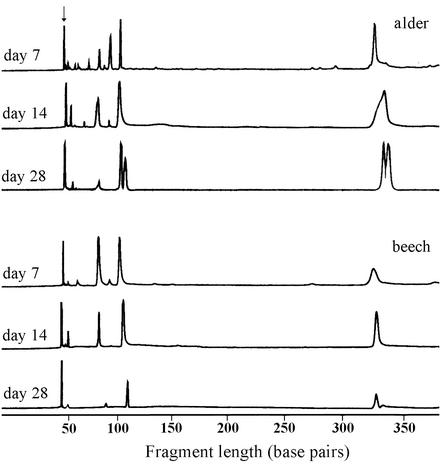
We digested the environmental samples amplified with F1300 with DdeI and detected the length of the 3′ end of the PCR product (e.g., Fig. 2). The highest ribotype richness and community evenness occurred after 1 week for all substrates except wood (Table 2). The ribotype richness decreased with time on all substrates except wood and oak, and the fungal community on all substrates was dominated by one ribotype after four weeks. The length of the dominant ribotype (115 bp) coincides with that of Articulospora tetracladia, whose spores dominated the fungal community (Table 1).
FIG. 2.
T-RFLP chromatograms from the fungal communities associated with alder and beech leaves after 7, 14, and 28 days of exposure in Boss Brook. The 3′ end of the 18S rRNA gene was amplified with a primer pair specific for ascomycetes, basidiomycetes, and chytridiomycetes and digested with DdeI. The peak designated by the arrow is unextended primer and was not considered for the numbers of ribotypes. The peak of 115 bp matches with the one from Articulospora tetracladia.
TABLE 2.
Number of fungal ribotypes identified by T-RFLP or DGGE (in parentheses) on plant material after stream exposure for 7, 14, or 28 days
| Substrate | No. of ribotypes identified by T-RFLP (DGGE) analysis after exposure (days)
|
||
|---|---|---|---|
| 7 | 14 | 28 | |
| Linden | 6 (16) | 5 (12) | 1 (9) |
| Maple | 5 (11) | 4 (11) | 2 (8) |
| Wood | 5 (16) | 8 (14) | 4 (9) |
| Alder | 10 (19) | 6 (11) | 6 (10) |
| Beech | 6 (10) | 4 (11) | 4 (7) |
| Oak | 5 (11) | 6 (11) | 3 (9) |
DGGE analysis.
The denaturing gradient gel electrophoresis of pure aquatic hyphomycete controls with the primer pair NS1 and GCfung separated 10 ribotypes out of the 12 pure fungal standards available (T. aquatica and C. aquatica migrated to the same position as did the two Tetracladium species). When mycelia from A. longissima and C. aquatica were mixed at known ratios, amplified and separated on DGGE, band intensities reflected these ratios (linear regression: y = 0.99, x = − 0.26, R2 = 0.998; P < 0.0001). Therefore, band intensities on DGGE may be useful indicators of biomass distribution among individual fungal species on decaying leaves.
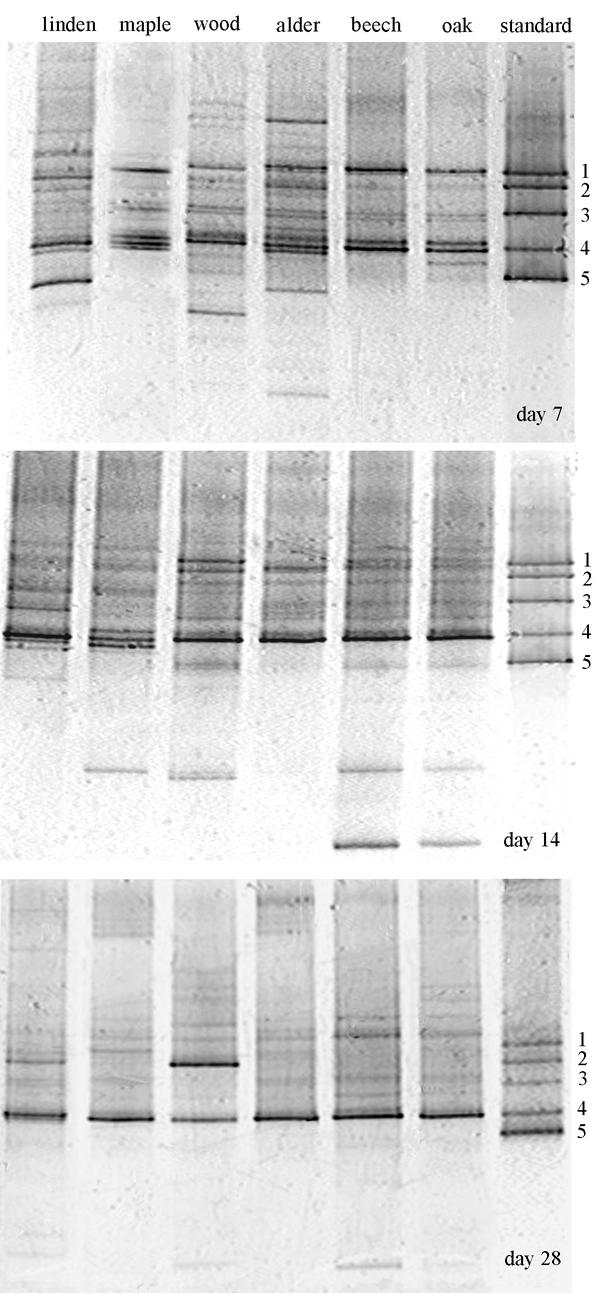
The DGGE analysis of environmental samples showed high species richness and community evenness after one week of submergence in all substrates (Fig. 3). On some substrates we found up to 19 bands representing different fungal ribotypes. Ribotype richness declined with time (a maximum of 10 ribotypes were present after four weeks) as did community evenness. After four weeks, one ribotype dominated on all substrates except wood, where two ribotypes were dominant. The dominant band had the same migration distance through the gel as that of Articulospora tetracladia. The two dominant bands on wood matched Articulospora tetracladia and H. lugdunensis.
FIG. 3.
DGGE gels of the 5′ end of 18S rDNA amplified with fungus-specific primers and run on a 20 to 55% gradient gel. Amplification products from pure cultures of aquatic hyphomycetes were run as standards. Bands: 1, Anguillospora furtiva; 2, H. lugdunensis; 3, T. marchalianum; 4, Articulospora tetracladia; and 5, T. aquatica.
DISCUSSION
The analysis of DNA sequences present in a community can be a powerful ecological tool. The stretches of DNA sequences that are best suited for analysis must have highly conserved regions separated by more variable sequences. This pattern permits efficient amplification by PCR and postamplification separation and detection. Our analysis with T-RFLP initially focused on the region of the 18S rRNA gene amplified by the primers NS5 (35) and D (12). This primer pair, however, amplified DNA from pure cultures of oomycetes and diatoms, and DNA isolated from the plant substrates themselves. To overcome these drawbacks we designed a fungus-specific primer for the T-RFLP analysis of the community. Our goal was to amplify DNA from as many fungi from the targeted groups as possible without amplification of DNA from the plant substrate or other taxa present in the stream. We targeted the 3′ end of the 18S rDNA because it generally exhibits moderate interspecies variability but no variability among strains or isolates from the same species.
Based on information from databases and tests with a variety of organisms, these primers selectively amplify DNA from true fungi. Unfortunately, the resolution among aquatic hyphomycetes was limited: T-RFLP analysis yielded only eight distinct fragments for the 12 species investigated. The two Tetracladium species produced identical fragments, which is not surprising considering the very high similarity among 18S and internal transcribed spacer sequences of five congeneric species of Tetracladium (24). Another fragment of identical size was formed by two species (C. aquatica and T. aquatica) associated with the same teleomorph genus (Massarina, Dothideales, Ascomycota) (34) and a third species (A. furtiva) associated with a different teleomorph (Rutstroemia, Helotiales, Ascomycota) (34). The third common fragment was formed by A. filiformis (unknown affinities) and A. rosea (teleomorph genus: Orbilia, Helotiales, Ascomycota). Resolution was better with DGGE, but was still insufficient to differentiate the two Tetracladium species or T. aquatica and C. aquatica. Both molecular techniques therefore potentially underestimate species diversity.
With the traditional technique based on spore release, all substrates accumulated fungal biomass during the first 4 weeks of immersion (Fig. 1). Ergosterol levels rose more quickly and reached higher levels on fast-decaying, e.g., alder leaves, than on slow-decaying, e.g., birch wood, substrates. A similar pattern was found when comparing the release of conidia as a function of immersion time. After 7 days, a few species (between four and seven), none of them clearly dominant, released a small number of conidia (Table 1). With one exception (beech leaves), the number of species was higher after 14 days and remained at a similar level after 28 days. At this point, however, one species, Articulospora tetracladia, was clearly dominant and contributed 60 to 80% of all conidia. Such increasing dominance at later stages of decomposition has been observed in many other studies; over 90% of released conidia often belong to two to four species (7, 13, 14, 21, 27, 29, 30).
As our T-RFLP procedure did not allow us to completely differentiate our 12 test species, similar overlaps in the fungal communities on the stream-exposed substrates seem likely. This lack of resolution probably resulted in underestimation of the number of different strains or species on the environmental samples. On the other hand, the molecular techniques also may include data from strains that did not sporulate. Interestingly, estimates based on T-RFLP peaks and conidium identification were essentially the same for 7-day samples (average for T-RFLP and conidia, 5.5 ribotypes and 5.3 species, respectively). After 14 and 28 days, however, the number of T-RFLP peaks declined. This decline may be attributed at least in part to the high signal-to-noise ratio of the instrument. Fungal ribotypes that are not relatively common (<3%) will not exhibit a distinct peak but will remain hidden in the baseline (20). Furthermore, the detection setup in T-RFLP analysis could be saturated by the signal from the fragment, such that above a threshold level, an increase in concentration of a fragment was not matched by a proportional increase in the signal. While the falling number of peaks strongly suggests a decline in fungal diversity, it is not clear whether this was due to lower species number or to lower evenness. The dominant peak on all substrates, however, matched Articulospora tetracladia, and in the case of birch wood the dominant peaks matched Articulospora tetracladia and H. lugdunensis, which is consistent with the results from the spore examination and DGGE.
Our DGGE procedure was more sensitive, but was still unable to differentiate among all our test strains. This result again implies that the number of bands underestimates the number of species. Nevertheless, DGGE provided valuable additional information, e.g., that species diversity was highest after only 1 week of stream exposure (maximum number of ribotypes, 19 (on alder); average number of ribotypes for all substrates, 14 ribotypes) and subsequently declined. Even after 28 days, estimates based on DGGE (average = 8.7 ribotypes) were close to those based on conidium identification (average = 10). As with the traditional and T-RFLP techniques, DGGE results suggest an increasing dominance of the community by one or two species. In contrast to conidial counts, however, it indicates that a substantial number of species present after 7 days have dropped out by day 14 and 28. There are two possible explanations. (i) Autumn-shed leaves carry propagules or dormant hyphae from terrestrial fungi (7, 30). During the early stages of immersion in a stream, these propagules germinate, raising their DNA levels above the threshold of detection for DGGE. Terrestrial species, however, are not very competitive in the stream, and, under normal circumstances, are quickly displaced by aquatic hyphomycetes (7, 30). (ii) During an initial phase, propagules of many species of aquatic hyphomycetes land on the leaf and quickly germinate (25), resulting in a rapid increase in species diversity. Many of these newly established minicolonies fail to gain a firm foothold and perish. The inhospitality of the substrate surface, microbivory, or active interference among fungal species during conidial attachment or germination might be responsible for such a decline. At least during later stages, there is little evidence for interference between mycelia of different species of aquatic hyphomycetes, since considerable interspecific intermingling of hyphae occurs at very small spatial scales (11, 28). We are currently examining the earliest phase of fungal colonization to determine if this stage presents a bottleneck for later fungal diversity on decaying leaves. If so, it would be an important step forward in understanding how the aquatic hyphomycete community on a substrate is regulated, and how this regulation affects their ecological functions.
All three methods point to decreased evenness in the fungal community after 14 and 28 days, and all suggest that Articulospora tetracladia becomes one of the dominant species. There are nevertheless substantial differences. For example, DGGE bands from wood after 28 days suggest that H. lugdunensis is at least as common as Articulospora tetracladia (Fig. 3). Conidium production data show a clear dominance by Articulospora tetracladia (61%), with three others each contributing approximately 10% (A. longissima, Flagellospora curvula, and H. lugdunensis [Table 1]). While pure culture controls showed a strong correlation between the initial biomass of a given species and the brightness of its DGGE band, it is premature to conclude that this relationship will be preserved during DNA extraction, PCR and DGGE from environmental samples. Nevertheless, discrepancies between the traditional techniques and the two molecular methods are not surprising. When counting conidia, we capture those parts of the mycelia that are sporulating (potentially important when evaluating the leaf-to-leaf dispersal of a species in a stream). The molecular techniques rely on DNA present throughout the mycelium, and more closely reflect total biomass contributions of the various species (relevant when investigating the effects of fungal colonization on leaf-feeding invertebrates). Discrepancies also were found when conidium production from leaves was compared with biomass estimates based on quantitative enzyme-linked immunosorbent assay (8). For example, A. acuminata was estimated to have the highest biomass but produced few conidia. The opposite was true for T. marchalianum. The preferred method depends on the objectives of the study; generally, a more complete understanding of fungal colonization of leaves will result when several of these approaches are combined.
Molecular techniques based on DNA analysis have most frequently been applied to communities of bacteria, at least in part because many bacteria cannot be cultivated and identified by traditional methods (>85 to 99.999% [1, 16, 31, 32]), but these techniques are equally suitable for investigating fungal ecology. For example, cloning fungal rRNA genes amplified from soil samples revealed a very different community than a culture-based approach (9). On the other hand, there was a general consistency between molecular (internal transcribed spacer-based) and direct-microscopic methods in describing the ascomycete community on decaying Spartina alterniflora blades (10). As in the present study, the molecular approach (10) revealed more distinct strains than the spore-based approach during the early phase (seven and two species, respectively); in a later phase, the difference was less pronounced (eight and five species, respectively). There also were clear differences in the relative frequencies of the various species, as estimated by the two techniques.
To characterize the fungal community on plant material decaying in streams, a combination of traditional and molecular techniques is most useful. Both T-RFLP analysis and DGGE are medium- to high-throughput techniques to analyze fungal community richness and evenness. Although both techniques require expensive equipment, many samples can be processed in a short time, and once the process is optimized the operating costs per sample are low. Both molecular techniques allow profiling of the fungal community. One advantage of DGGE over T-RFLP analysis is that it also can be used for analysis of the phylogenetic relationships among the species comprising the community (22). Moreover, bands excised from a denaturing gel can be used to design species-specific molecular probes to detect spatial patterns of fungal taxa on the plant substrate.
In general terms, the biology and ecological roles of aquatic hyphomycetes are well understood (5). Many details concerning the roles and interactions of different species and strains, potential trade-offs between growth and reproduction, and the roles of sexual and asexual reproduction, however, remain elusive. Molecular methods promise data that can be used to fill some of these gaps.
Acknowledgments
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microb. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arsuffi, T. L., and K. Suberkropp. 1984. Leaf processing capabilities of aquatic hyphomycetes: interspecific differences and influence on shredder feeding preferences. Oikos 42:144-154. [Google Scholar]
- 3.Bärlocher, F. 1982. Conidium production from leves and needles in four streams. Can. J. Bot. 60:1487-1494. [Google Scholar]
- 4.Bärlocher, F. 1987. Aquatic hyphomycete spora in 10 streams in New Brunswick and Nova Scotia. Can. J. Bot. 65:76-79. [Google Scholar]
- 5.Bärlocher, F. 1992. The ecology of aquatic hyphomycetes. Springer-Verlag, Berlin, Germany.
- 6.Bärlocher, F., and B. Kendrick. 1973. Fungi and food preferences of Gammarus pseudolimnaeus. Arch. Hydrobiol. 72:501-516. [Google Scholar]
- 7.Bärlocher, F., and B. Kendrick. 1974. Dynamics of the fungal population on leaves in a stream. J. Ecol. 62:761-791. [Google Scholar]
- 8.Bermingham, S., L. Maltby, and F. M. Dewey. 1997. Use of immunoassays for the study of natural assemblages of aquatic hyphomycetes. Microb. Ecol. 33:223-229. [DOI] [PubMed] [Google Scholar]
- 9.Borneman, J., and R. J. Hatrin. 2000. PCR primers that amplify fungal rRNA genes from environmental samples. Appl. Environ. Microbiol. 66:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchan, A., S. Y. Newell, J. I. L. Moreta, and M. A. Moran. 2002. Analysis of internal transcribed spacer (ITS) regions of rRNA genes in fungal communities in a southeastern U.S. salt marsh. Microb. Ecol. 43:329-340. [DOI] [PubMed] [Google Scholar]
- 11.Chamier, A.-C., P. A. Dixon, and S. A. Archer. 1984. The spatial distribution of fungi on decomposing alder leaves in a freshwater stream. Oecologia 64:92-103. [DOI] [PubMed] [Google Scholar]
- 12.Elwood, H. J., G. J. Olsen, and M. L. Sogin. 1985. The small-subunit ribosomal RNA gene sequences from the hypotrichous ciliates Oxytricha nova and Stylonychia pustulata. Mol. Biol. Evol. 2:399-410. [DOI] [PubMed] [Google Scholar]
- 13.Garnett, H., F. Bärlocher, and D. Giberson. 2000. Aquatic hyphomycetes in Catamaran Brook: colonization dynamics, seasonal patterns, and logging effects. Mycologia 92:29-41. [Google Scholar]
- 14.Gessner, M. O., M. Thomas, A.-M. Jean-Louis, and E. Chauvet. 1993. Stable successional patterns of aquatic hyphomycetes on leaves decaying in a summer cool stream. Mycol. Res. 97:163-172. [Google Scholar]
- 15.Gessner, M. O., F. Bärlocher, and E. Chauvet. Qualitative and quantitative analyses of aquatic hyphomycetes in streams. In C. K. M. Tsui, K. D. Hyde, and W. H. Ho (ed.), Freshwater mycology: a practical approach, in press. Fungal Diversity Press, University of Hong Kong Press, Hong Kong, Hong Kong.
- 16.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature (London) 345:60-63. [DOI] [PubMed] [Google Scholar]
- 17.Heinonsalo, J., K. S. Jørgensen, and R. Sen. 2001. Microcosm-based analyses of Scots pine seedling growth, ectomycorrhizal fungal community structure and bacterial carbon utilization profiles in boreal forest humus and underlying alluvial mineral horizons. FEMS Microb. Ecol. 36:73-84. [DOI] [PubMed] [Google Scholar]
- 18.Kinzig, A. P., S. W. Pacala, and D. Tilman. 2001. The functional consequences of biodiversity. Princeton University Press, Princeton, N.J.
- 19.Kowalchuk, G. A. 1999. New perspectives towards analyzing fungal communities in terrestrial environments. Curr. Opin. Biotechnol. 10:247-251. [DOI] [PubMed] [Google Scholar]
- 20.Liu, W.-T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maharning, A. R., and F. Bärlocher. 1996. Growth and reproduction in aquatic hyphomycetes. Mycologia 88:80-88. [Google Scholar]
- 22.May, L. A., B. Smiley, and M. G. Schmidt. 2001. Comparative denaturing gradient gel electrophoresis of fungal communities associated with whole plant corn silage. Can. J. Microbiol. 47:829-841. [DOI] [PubMed] [Google Scholar]
- 23.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikolcheva, L. G., and F. Bärlocher. 2002. Phylogeny of Tetracladium based on 18S rDNA. Czech Mycol. 53:285-295. [Google Scholar]
- 25.Read, S. J., S. T. Moss, and E. B. G. Jones. 1992. Attachment and germination of conidia. In: F. Bärlocher (ed.), The ecology of aquatic hyphomycetes. Springer-Verlag, Berlin, Germany, pp. 135-151.
- 26.Rong, Q., K. R. Sridhar, and F. Bärlocher. 1995. Food selection of three leaf-shredding invertebrates. Hydrobiologia 316:173-181. [Google Scholar]
- 27.Shearer, C. A. 1992. The role of woody debris, p. 77-98. In F. Bärlocher (ed.), The ecology of aquatic hyphomycetes. Springer-Verlag, Berlin, Germany.
- 28.Shearer, C. A., and L. Lane. 1983. Comparison of three techniques for the study of aquatic hyphomycete communities. Mycologia 75:498-508. [Google Scholar]
- 29.Shearer, C. A., and J. Webster. 1985. Aquatic hyphomycete communities in the River Teign. I. Longitudinal distribution patterns. Trans. Br. Mycol. Soc. 84:489-501. [Google Scholar]
- 30.Suberkropp, K., and M. J. Klug. 1976. Fungi and bacteria associated with leaves during processing in a woodland stream. Ecology 57:707-719. [Google Scholar]
- 31.Torsvik, V., J. Gokosoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature (London) 345:63-65. [DOI] [PubMed] [Google Scholar]
- 33.Wardle, D. A. 2002. Communities and ecosystems. Princeton University Press, Princeton, N.J.
- 34.Webster, J. 1992. Anamorph-teleomorph relationships, p. 99-117. In F. Bärlocher (ed.), The ecology of aquatic hyphomycetes. Springer-Verlag, Berlin, Germany.
- 35.White, T. J., T. Bruns, S. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, N.Y.
- 36.Young, C. J. 1995. Microwave-assisted extraction of the fungal metabolite ergosterol and total fatty acids. J. Agric. Food Chem. 43:2904-2910. [Google Scholar]