Abstract
The gene encoding a poly(dl-lactic acid) (PLA) depolymerase from Paenibacillus amylolyticus strain TB-13 was cloned and overexpressed in Escherichia coli. The purified recombinant PLA depolymerase, PlaA, exhibited degradation activities toward various biodegradable polyesters, such as poly(butylene succinate), poly(butylene succinate-co-adipate), poly(ethylene succinate), and poly(ε-caprolactone), as well as PLA. The monomeric lactic acid was detected as the degradation product of PLA. The substrate specificity toward triglycerides and p-nitrophenyl esters indicated that PlaA is a type of lipase. The gene encoded 201 amino acid residues, including the conserved pentapeptide Ala-His-Ser-Met-Gly, present in the lipases of mesophilic Bacillus species. The identity of the amino acid sequence of PlaA with Bacillus lipases was no more than 45 to 50%, and some of its properties were different from those of these lipases.
Recently, some types of aliphatic polyesters were developed as biodegradable plastics. Poly(lactic acid) (PLA), poly(butylene succinate) (PBS), and poly(butylene succinate-co-adipate) (PBSA) are the most promising materials of the commercially available synthetic polyesters. PLA has been used mainly as a biomedical material, such as for drug delivery microcapsules (14). Because of its superior physical properties, PBSA is expected to become widely used as an alternative to ordinary plastics.
It has been confirmed that PLA and PBSA are naturally degraded in soil or compost (5). While various microorganisms have been isolated as PBS degraders (12, 23, 30, 31), it is known that PLA is less susceptible to degradation than other aliphatic biodegradable plastics in the natural environment (24). Although Suyama et al. tested three kinds of soils for screening of aliphatic polyester-degrading bacteria, they could not isolate any PLA-degrading bacteria, despite the presence of bacteria degrading other polyesters (30). As reasons for the lower susceptibility of PLA to degradation, two possibilities are proposed; one is that the physical properties of PLA prevent biodegradation, and the other is that PLA degradation is made difficult by the absence of naturally existing enzymes.
Enzymatic degradation of aliphatic polyesters, such as PBS and poly(ɛ-caprolactone) (PCL), has been reported with the use of commercially available esterases and lipases; however, PLA is considered to be degraded by proteinase-like enzymes (15, 36). In addition, it was reported that the lipase PL, a commercially available lipase, degraded PLA; however, it required the sequential reactions of chemical hydrolysis at higher pH and temperature (7). Some researchers reported a relationship between PLA degradation and silk fibroin degradation in PLA-degrading Amycolatopsis sp. and Tritirachium album (8, 25). Pranamuda et al. purified from an Amycolatopsis sp. a PLA-degrading enzyme which showed protease activity toward silk and synthetic protease substrates (26). A 24-kDa PLA-degrading enzyme which showed degrading activity toward casein and fibrin but not triolein and tributyrin was purified from Amycolatopsis sp. strain K104-1 by Nakamura et al., and a lactic acid monomer was detected as the PLA degradation product (19). The relationship of an esterase with PLA degradation has been reported only for the PLA-degrading enzyme from Bacillus smithii, a thermophilic PLA-degrading bacterium (28). The enzyme purified from this strain was 62.5 kDa and was active against various fatty acid esters at 60°C. To our knowledge, there is no information on the gene encoding the PLA-degrading enzyme.
The PLA-degrading bacterium Paenibacillus amylolyticus strain TB-13 has been isolated and found to have the ability to degrade various aliphatic polyesters, such as PBS, PBSA, PCL, and poly(ethylene succinate) (PES) (32). It was suggested that the esterase and/or protease in the culture broth may be related to the degradation of these polyester-based plastics. It is unclear whether several enzymes are related to the degradation.
In this study, we report the molecular cloning and sequence analysis of the PLA-degrading enzyme (PLA depolymerase) gene in P. amylolyticus strain TB-13 as well as its functional expression in Escherichia coli. Furthermore, the properties of the recombinant enzyme were characterized. This is the first report on the molecular analysis of the PLA depolymerase gene.
MATERIALS AND METHODS
Chemicals.
PLA samples with weight-average molecular weights of 0.5 × 104 (PLA05), 1.0 × 104 (PLA10), 1.5 × 104 (PLA15), and 2.0 × 104 (PLA20) were purchased from Wako Pure Chemical Co. (Tokyo, Japan). PBS with the trade names BIONOLLE 1020 (PBS1020; weight-average molecular weight, 1.4 × 105) and BIONOLLE 1001 (PBS1001; weight-average molecular weight, 2.6 × 105) and PBSA with the trade names BIONOLLE 3020 (PBSA3020; weight-average molecular weight, 1.4 × 105) and BIONOLLE 3001 (PBSA3001; weight-average molecular weight, 2.5 × 105) were supplied by Showa Highpolymer Co., Ltd. (Tokyo, Japan). PES was provided by Nippon Shokubai Co., Ltd. (Tokyo, Japan). Poly(3-hydroxybutyric acid-co-3-hydroxyvaleric acid) [P(HB-co-HV)] was purchased from Sigma-Aldrich Chemical Co. (St. Louis, Mo.). PCL and proteinase K from T. album were purchased from Wako. Plysurf A210G was obtained from Daiichi Kougyou Seiyaku Co. (Tokyo, Japan). All other compounds used were standard commercial preparations.
Bacterial strains, plasmids, and media.
The culture conditions for P. amylolyticus strain TB-13 have been described previously (32). E. coli XL10-Gold (Stratagene, La Jolla, Calif.) was used for the isolation of recombinant derivatives of pUC18 (37). E. coli BL21(DE3) and pET25b(+) (Novagen, Madison, Wis.) were used as the expression host and plasmid, respectively. E. coli strains carrying plasmids were grown at 37°C in Luria-Bertani (LB) medium (17) containing 0.1 mg of ampicillin/ml. For the detection of recombinants with PLA-degrading activity, emulsified PLA solution (0.5% [wt/vol]) containing 1.5% agar was prepared as described previously (32) and overlaid on an LB agar plate.
Construction of the genomic library.
Genomic DNA was extracted from strain TB-13 by a modification of the method of Saito and Miura (27). The DNA was completely digested with HindIII, and fragments of between 2 and 4 kbp were ligated into HindIII-digested and dephosphorylated pUC18. The plasmids were transformed into E. coli XL10-Gold. A transformant which showed a translucent clear zone on the indicator plate was isolated as the positive clone.
DNA sequencing and data analysis.
DNA sequencing was carried out with an ABI Prism 310 DNA sequencer (Perkin-Elmer Applied Biosystems, Foster City, Calif.). The nucleotide and amino acid sequences were analyzed by using the GENETYX-MAC program, version 8 (Software Development Co., Ltd., Tokyo, Japan), and FASTA and BLAST on the DDBJ/GenBank/EMBL nucleotide sequence databases. Selected amino acid sequences were aligned by using the Clustal X program (9).
Plasmid construction for overexpression in E. coli.
The gene encoding PLA depolymerase was subcloned in vector pET25b(+) by PCR amplification. The restriction sites for EcoRI and NheI were incorporated into the forward and the reverse primer sequences, respectively. In order to amplify from Ala1 to Glu170, the following primers were used: forward, 5′-CCGGAATTCGGCAACTGAACGCACGCCA-3′, and reverse, 5′-CCTAGCTAGCTTCGATAAGTGCGGCCTT-3′ (EcoRI and NheI restriction sites, respectively, are underlined). The PCR conditions were as follows: 1 cycle of 2 min at 94°C and 30 cycles each of 15 s at 96°C, 30 s at 60°C, and 1 min at 68°C. The construct was named pLA-EN.
Expression of the PLA depolymerase gene.
E. coli BL21(DE3) transformed with pLA-EN was used to inoculate 100 ml of LB medium (containing 100 μg of ampicillin/ml). After incubation for 3 h at 32°C, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM, and the mixture was incubated for 3 h at 32°C. Cells were harvested by centrifugation at 8,000 × g for 10 min at 4°C, and the periplasm fraction was extracted by the method of Neu and Heppel (20).
The collected periplasm fraction was loaded onto an Ni2+-immobilized chelating Sepharose Fast Flow (Pharmacia Biotech, Upsala, Sweden) column (1.0 by 10 cm) equilibrated with 20 mM potassium phosphate buffer (pH 7.0) containing 0.1 M NaCl. The column was eluted with a linear 0 to 200 mM imidazole gradient for 1 h at a flow rate of 2.0 ml/min. The fractions containing the PLA-degrading activity were pooled and concentrated by ultrafiltration (YM10 membrane; Millipore, Bedford, Mass.).
Enzyme activity assays. (i) PLA- and PBSA-degrading activities.
The degrading activities with various molecular weights for PLA, PBS, and PBSA were determined by measuring the decrease in the turbidity of their emulsions. PLA, PBS, and PBSA (each at 0.5% [wt/vol]) emulsified with Plysurf A210G (0.02% [wt/vol]) in 100 mM sodium phosphate-NaOH buffer (pH 10.0) for PLA depolymerase and 100 mM Tris-HCl buffer (pH 8.0) for proteinase K were used as substrates. The emulsions were diluted to provide approximately 1,000 mg of the total organic carbon (TOC) concentration/liter. The reaction was started by adding 0.05 ml of enzyme solution containing 30 μg of protein to 1.95 ml of emulsion and incubating the mixture at 37°C. When the reaction was finished, the turbidity was measured at a wavelength of 580 nm. The reaction mixture was centrifuged at 15,000 × g for 10 min, and TOC in the supernatant was measured by using a TOC-V analyzer (Shimadzu, Kyoto, Japan). The concentrations of dl-lactic acid and succinic acid were determined by using an F-kit (R-Biopharm, Darmstadt, Germany).
(ii) Esterase activity.
Esterase activity was measured by the method of Kay et al. with p-nitrophenyl acetate as the substrate (10). One unit was defined as the amount of enzyme required to liberate 1 μmol of p-nitrophenol per min.
Activities for various substrates. (i) p-Nitrophenyl esters.
The assay for esterase activity with p-nitrophenyl esters of acetate (C2), butyrate (C4), caprylate (C8), caprate (C10), palmitate (C16), and stearate (C18) was performed by the method of Eggert et al. (4). Alternatively, gum arabic and deoxycholate were omitted from the reaction mixture, and the reaction was performed with 100 mM potassium phosphate buffer (pH 7.0) at 37°C. One unit was defined as the amount of enzyme required to liberate 1 μmol of p-nitrophenol per min.
(ii) Triglycerides.
The activities for triolein and tributyrin were measured by a titration method. The substrate emulsion was prepared by sonication of 25% triglycerides and 1.5% gum arabic for 5 min. Enzyme was added to a 2.0-ml reaction mixture consisting of 1.0 ml of emulsion, 10 mM morpholinepropanesulfonic acid (MOPS) (pH 7.0), and 10 mM CaCl2. The amount of released oleic acid or butyric acid was measured by titration with 0.1 N NaOH on a DL55 Titrator (Mettler-Toledo, Schwerzenbach, Switzerland).
(iii) Other polyesters.
Emulsions of PCL, PES, and P(HB-co-HV) were prepared as described above. The decrease in turbidity was measured at a wavelength of 580 nm.
Protein assay.
The protein concentration was determined as described by Lowry et al. (16). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed with a 12.5% separating gel as described by Laemmli (13).
Nucleotide sequence accession number.
The nucleotide sequence of plaA was submitted to GenBank and received the accession number AB093482.
RESULTS
Cloning of the gene encoding PLA depolymerase.
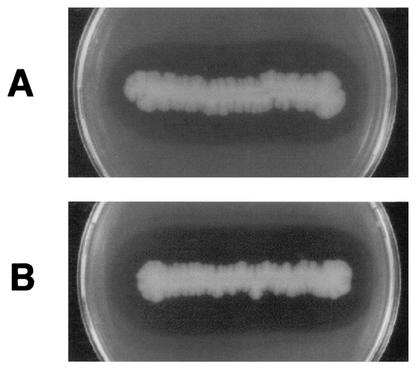
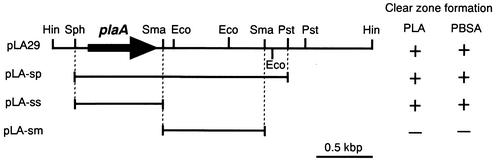
From the genomic library of strain TB-13, one clone forming a clear zone on an emulsified PLA-containing agar plate was obtained (Fig. 1). The recombinant also formed a clear zone on an agar plate containing emulsified PBSA. The recombinant plasmid in the clone, named pLA29, contained an insert of 2.9 kbp. The restriction map of this insert is shown in Fig. 2. In order to locate the region containing the PLA depolymerase gene, the insert was fragmented by restriction enzymes and subcloned into the appropriate site of pUC18. The resultant plasmids were tested for their abilities to form a clear zone on an agar plate containing emulsified PLA or PBSA. The gene responsible for forming a clear zone was located in pLA-ss, containing the insert DNA of 0.9 kbp.
FIG. 1.
Clear-zone formation obtained with recombinant E. coli XL10-Gold on agar plates containing emulsified PLA05 (A) and PBSA3020 (B). The plates were incubated at 37°C for 48 h.
FIG. 2.
Restriction map of the 2.9-kbp HindIII fragment from pLA29, showing the positions of the HindIII (Hin), SphI (Sph), SmaI (Sma), EcoRI (Eco), and PstI (Pst) restriction sites. The location of the plaA gene is indicated by an arrow. E. coli XL10-Gold transformed with each plasmid was grown on an agar plate containing emulsified PLA05 or PBSA3020 at 37°C for 24 h.
Nucleotide sequence and deduced amino acid sequence.
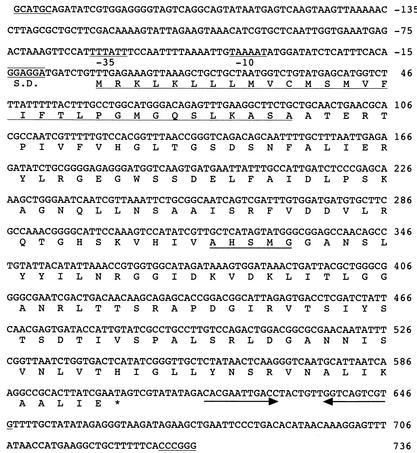
The nucleotide sequence of the 0.9-kbp DNA fragment inserted in pLA-ss was determined for both strands. Portions of the nucleotide sequence of the sense strand and the deduced amino acid sequence are shown in Fig. 3. One open reading frame (ORF) was located between nucleotides 1 and 606, and its molar G+C content was 46.4%. The 606-bp ORF has a putative TTG start codon 8 bp downstream from a potential ribosome binding site (−14GGAGGA−9). Potential −35 and −10 promoter sequences were observed, as was a stem-loop structure downstream from the stop codon at position 604. This ORF was named plaA.
FIG. 3.
Nucleotide sequence of the PLA depolymerase gene. The deduced amino acid sequence is shown below the nucleotide sequence. A putative promoter region (−10 and −35), a Shine-Dalgarno (S.D.) sequence, and a ρ-independent terminator (horizontal arrows) are indicated. A sequence that exhibits the characteristics of a signal peptide is underlined, and the putative active site is doubly underlined. SphI and SmaI restriction sites at the beginning and end of the sequence, respectively, are underlined.
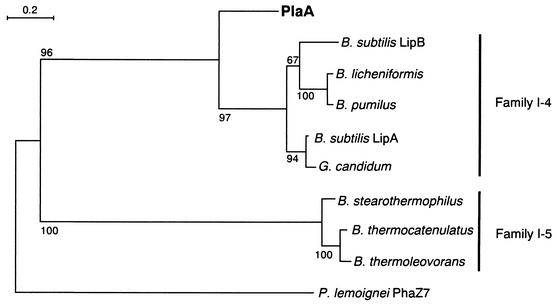
The deduced amino acid sequence of the preprotein is composed of 201 amino acids with a predicted molecular weight of 21,661. The putative cleavage site of the signal peptide was located between Ala31 and Ala32 according to the rules for a signal peptide sequence. The mature protein is composed of 170 amino acids with a predicted molecular weight of 18,232 and an isoelectric point of 9.22. An Ala-X1-Ser-X2-Gly pentapeptide conserved in Bacillus lipases and Pseudomonas lemoignei poly(hydroxyalkanoate) (PHA) depolymerase was observed between positions 107 and 110 (X1; His, X2; Met) (Table 1). The deduced amino acid sequence showed 45 to 50% sequence identity with mesophilic Bacillus family I-4 lipases (3, 18, 21, 34), ∼20% sequence identity with thermophilic Bacillus family I-5 lipases (2, 11, 29), but no identity with PHA depolymerase from P. lemoignei, except for the conserved pentapeptide (6).
TABLE 1.
Conserved sequences in family I-4 and I-5 lipases
| Enzyme | Amino acid sequencea | Identity (%) | Accession no. |
|---|---|---|---|
| PlaA | HIVAHSMGGANS | AB093482 | |
| Family I-4 lipases | |||
| B. pumilus | DIVAHSMGGANT | 50.3 | A34992 |
| B. licheniformis | DIVAHSMGGANT | 51.4 | CAB95850 |
| B. subtilis LipA | DIVAHSMGGANT | 49.0 | M74010 |
| B. subtilis LipB | DIVAHSMGGANT | 44.9 | C6952 |
| Geotrichum candidum | DIVAHSMGGANT | 46.2 | A02813 |
| Consensus sequenceb | dIVAHSMGGANt | ||
| Family I-5 lipases | |||
| B. thermoleovorans | HIIAHSQGGQTA | 20.0 | AF134840 |
| B. thermocatenulatus | HIIAHSQGGQTA | 21.6 | X95309 |
| B. stearothermophilus | HIIAHSQGGQTA | 20.5 | U78785 |
| Consensus sequencec | -I-AHSQGG--- | ||
| P. lemoignei PhaZ7 | DIVAHSMGVSMS | AY026355 |
The consensus pentapeptide is shown in bold type.
Consensus sequence for PlaA and family I-4 lipases. Residues conserved only in family I-4 lipases are indicated by lowercase letters.
Consensus sequence for PlaA, family I-4 lipases, and I-5 lipases.
Functional expression and purification of the recombinant enzyme in E. coli.
Expression vector pLA-EN was constructed by ligating the PCR-amplified EcoRI/NheI fragment to EcoRI/NheI-digested pET25b(+). This construct, which placed the plaA gene under the control of the T7 promoter and which contained an in-frame fusion with the N-terminal PelB leader peptide for periplasmic secretion and with C-terminal six-His residues, was transformed into E. coli BL21(DE3). Esterase activity was detected in the culture broth after induction by IPTG. After 3 h of induction, the periplasm fraction was extracted, and 180 U of enzyme was obtained from 100 ml of culture. Purification of the recombinant enzyme was facilitated by affinity chromatography with the His-tagged protein. The His-tagged enzyme was purified 9.4-fold with a yield of 22% and a specific activity of 16.0 U/mg. The purified PLA depolymerase migrated as a single band with a molecular mass of approximately 22 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown).
Properties of the recombinant enzyme. (i) Substrate specificity.
The substrate specificity of the purified PLA depolymerase is summarized in Table 2. The enzyme degraded various biodegradable polyesters, PBS, PCL, and PES, as well as PLA and PBSA, but not P(HB-co-HV). It was observed that PLA with a lower molecular weight showed a higher degradability. Similar results were observed when PBS and PBSA were used as substrates. For the triglycerides, PLA depolymerase degraded triolein and tributyrin, the typical substrates for lipase and esterase, respectively. The enzyme exhibited esterase activity toward various p-nitrophenyl alkyl esters, showing the highest activity for butyl ester, with a specific activity of 184.6 U/mg.
TABLE 2.
Substrate specificity of purified PLA depolymerase
| Substrate | Relative activity |
|---|---|
| Polyestersa | |
| PLA05 | ++ |
| PLA10 | ++ |
| PLA15 | + |
| PLA20 | + |
| PBSA3001 | ++ |
| PBSA3020 | ++ |
| PBS1001 | + |
| PBS1020 | + |
| PCL | ++ |
| PES | ++ |
| P(HB-co-HV) | − |
| Triglyceridesb | |
| Triolein | + |
| Tributyrin | + |
| p-Nitrophenyl estersc | |
| C2 | 0.60 |
| C4 | 1 |
| C8 | 0.79 |
| C10 | 0.56 |
| C16 | 0.50 |
| C18 | 0.27 |
Decrease in the turbidity of the emulsion of >60% (++) or <60% (+) or no decrease (−) after incubation at 37°C for 1 h.
+, NaOH consumption on titration of between 0.1 and 0.2 ml.
Relative specific activies, with the value of the C4 ester set at 1.
(ii) Time course of PLA and PBSA degradation.
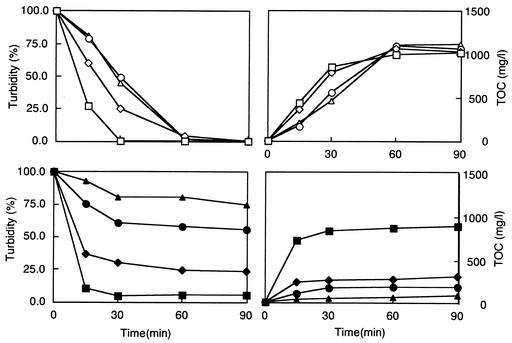
The degradation activities toward PLA and PBSA with various molecular weights were examined. The purified enzyme showed degradation activities toward both PLA and PBSA emulsions (Fig. 4). Chemical hydrolysis was not observed during the reaction period unless the enzyme was added. For PLA, a higher degradability was observed at a lower molecular weight. The emulsion of PLA with a molecular weight of 5,000 was completely degraded in 30 min, and all of the other PLAs were degraded in 90 min. An increase in the TOC concentration in the supernatant of the reaction solution was correlated with a decrease in the turbidity. The degradabilities of PBS and PBSA were affected by their components and molecular weights. PBSA showed a higher degradability than PBS, and a higher degradability was observed at a lower molecular weight. The concentrations of dl-lactic acid at 90 min were 0.89, 1.08, 1.18, and 1.16 g/liter from PLA05, PLA10, PLA15, and PLA20, respectively. It took 6 h for the emulsions of PBSA3020 and PBSA3001 to be completely degraded and 24 h for that of PBS1020; however, 33% of the emulsion of PBS1001 remained even after 24 h of incubation. Proteinase K degraded PLA05 with a 61.5% decrease in turbidity, and 380 mg of TOC/liter was detected at 90 min. It took over 20 h for PLA05 to be completely degraded. Proteinase K could not degrade any PBSA or PBS at all.
FIG. 4.
PLA and PBSA degradation by the purified recombinant PLA depolymerase. Emulsions of PLA05 (open square), PLA10 (open diamond), PLA15 (open circle), PLA20 (open triangle), PBSA3020 (closed square) PBSA3001 (closed diamond), PBS1020 (closed circle), and PBS1001 (closed triangle) were used as substrates. The decrease in the turbidity at a wavelength of 580 nm (left) and the TOC concentrations (right) were measured as described in Materials and Methods.
(iii) Optimal conditions for and stability of the purified enzyme.
The degradability of emulsified PLA was assayed under various pH and temperature conditions. The enzyme preferred a high pH range, exhibiting maximal activity at pH 10.0, and no activity was detected under acidic conditions. The enzyme was active over a wide temperature range, exhibiting maximal activity at 45 to 55°C. The enzyme was remarkably stable at an alkaline pH, showing maximal stability at pH 10.0 for 30 min of incubation. The temperature stability decreased at temperatures over 50°C.
DISCUSSION
Some microorganisms which degrade PLA have been isolated from the natural environment (8, 19, 24, 25, 28, 33); however, the isolates are mostly derived from the actinomycetes, and there are few reports for other bacteria. Furthermore, no information is available concerning the gene encoding the PLA-degrading enzyme. A PLA-degrading bacterium, P. amylolyticus strain TB-13, was isolated and found to degrade various biodegradable polyesters as well as PLA (32). In this study, we cloned the structural gene encoding PLA depolymerase from strain TB-13 and analyzed its sequence. Recombinant E. coli harboring the plaA gene formed a clear zone on an agar plate containing emulsified PLA or PBSA. This result revealed that this gene encodes the PLA depolymerase, and its expression product could also degrade PBSA.
The purified recombinant PLA depolymerase showed a wide substrate specificity for polyester-based plastics, such as PLA, PBS, PBSA, PCL, and PES, and for the triglycerides tributyrin and triolein, the substrates for esterase and lipase, respectively (Table 2). From the substrate specificity, it is estimated that PLA depolymerase is a type of lipase. A higher esterolytic activity was observed with the shorter fatty acid chain lengths of the tested p-nitrophenyl esters. Considering these results, we assume that the PlaA would not be categorized as a true lipase, which prefers a long chain length of p-nitrophenyl ester. There are very few reports on a PLA-degrading esterase or lipase. A PBSA-degrading esterase from Acidovorax delafieldii strain BS-3 could not degrade a PLA emulsion (unpublished data). Sakai et al. reported the purification of a thermophilic PLA depolymerase from B. smithii and found that it exhibited esterase activity; however, the degradability of the other polyesters has remained unknown (28). The physicochemical properties of B. smithii esterase are different from those of PlaA, e.g., molecular weight, optimal conditions, and stability. Unfortunately, genetic information on B. smithii esterase is lacking; however, it is clear that these enzymes are quite different from each other.
During the enzymatic degradation of the PLA emulsions, lactic acid was detected, and its concentration was equivalent to approximately 40% the TOC concentration. This result suggests the existence of a water-soluble oligomer as the degradation product. On the contrary, no succinic acid was detected after PBSA degradation, suggesting that the degradation product is a water-soluble oligomer. It is estimated that the difference in the degradation patterns between PLA and PBSA is caused by their polymer structures: a homopolymer for PLA and a heteropolymer for PBSA. The detailed compositions of the degradation products are still unknown. It is known that PLA is degraded by some commercially available proteases (15, 36). In comparison with the proteases, PLA depolymerase from strain TB-13 showed a rate of degradation superior to that obtained with the same amount of proteinase K.
The amino acid sequence of PlaA showed similarity to those of Bacillus lipases but no more than 45 to 50% identity. However, the pentapeptide around the catalytic Ser was highly conserved. Most mammalian and bacterial lipases conserve a Gly-X-Ser-X-Gly pentapeptide containing the catalytic Ser residue; however, the first Gly residue is replaced with an Ala residue in these Bacillus lipases (3). Arpigny and Jaeger classified bacterial lipases into eight families based on a comparison of their amino acid sequences and biochemical properties (1). Family I was divided into six subfamilies, and the Bacillus lipases were classified into subfamilies I-4 and I-5, including lipases from mesophilic Bacillus and from thermophilic Bacillus or Staphylococcus, respectively. PlaA showed sequence similarity with family I-4 enzymes but less identity than is seen among the other family I-4 lipases, forming another branch separate from the other lipases in the phylogenetic tree (Table 1 and Fig. 5). The PHA depolymerase from P. lemoignei, PhaZ7, is included in the lipases possessing an Ala-His-Ser-Met-Gly pentapeptide. Because of its unique sequence, Handrick et al. suggested that PhaZ7 should be classified into a new family of carboxylesterases (6). The purified recombinant PLA depolymerase could not degrade P(HB-co-HV) (Table 2), and the entire amino acid sequences of PlaA and PhaZ7 were different, except for the catalytic pentapeptide.
FIG. 5.
Dendrogram of the PLA depolymerase from P. amylolyticus strain TB-13 (PlaA) and related lipases. On the basis of the amino acid alignment, the dendrogram was constructed as described in Materials and Methods. The numbers at the nodes indicate the percent recovery in 100 bootstrap resamplings. The accession numbers for the amino acid sequences are indicated in Table 1.
For the mammalian and bacterial lipases, it is known that the active site is composed of Ser, Asp, and His residues forming a catalytic triad (22). Recently, the three-dimensional structure of B. subtilis LipA was determined, and its catalytic triad was identified as Ser77, Asp133, and His156 (35). Among the family I-4 lipases, these amino acids are highly conserved. The catalytic triad of PlaA is predicted to be Ser78, Asp128, and His151. The crystal structure of B. subtilis LipA also revealed the amino acids forming an oxyanion hole to be Ile12 and Met78, which are necessary for the activation of the serine hydrolases. These amino acids are highly conserved in family I-4 lipases, except for PlaA, in which the first Ile is replaced with Leu and some amino acids around it are also changed. Concerning their sequence similarities, it is estimated that the family I-4 lipases share common three-dimensional structures with B. subtilis LipA. However, we assumed that the detailed structure of PlaA is rather different from that of the other lipases.
The purified PLA depolymerase showed maximal activity at pH 10.0; such a high optimal pH is a common characteristic of family I-4 lipases. It is notable that their activities decreased significantly at temperatures above 45°C; however, the activity of PlaA was not affected even when it was incubated for 30 min at 50°C and decreased only by 20% at 55°C. The thermostability of PlaA may be related to its sequence uniqueness in relation to the sequences of family I-4 lipases. From these results, it appears that PlaA from P. amylolyticus strain TB-13 correlates with the family I-4 lipases; however, some of its properties are different from those of typical family I-4 lipases.
In this study, we report for the first time the cloning and sequencing analysis of a PLA-degrading enzyme. The gene, named plaA, encoded a lipase, and the expression product showed a wide range of substrate specificities for polyester-based biodegradable plastics. Thus far, study of the protease has been focused on the enzymatic degradation of PLA (36). Considering that strain TB-13 produces lipase for PLA degradation, it is necessary to consider that microorganisms producing lipase and/or esterase are also distributed as PLA degraders in the natural environment.
Acknowledgments
We thank Showa Highpolymer Co., Ltd., for supplying PBS and PBSA and Nippon Shokubai Co., Ltd., for supplying PES.
REFERENCES
- 1.Arpigny, J. L., and K.-E. Jaeger. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343:177-183. [PMC free article] [PubMed] [Google Scholar]
- 2.Cho. A. R., S. K. Yoo, and E. J. Kim. 2000. Cloning, sequencing and expression in Escherichia coli of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol. Lett. 186:235-238. [DOI] [PubMed] [Google Scholar]
- 3.Dartois, V., A. Baulard, K. Schanck, and C. Colson. 1992. Cloning, nucleotide sequence and expression in Escherichia coli of a lipase gene from Bacillus subtilis 168. Biochim. Biophys. Acta 1131:253-260. [DOI] [PubMed] [Google Scholar]
- 4.Eggert, T., G. Pencreac'h, I. Douchet, R. Verger, and K.-E. Jaeger. 2000. A novel extracellular esterase from Bacillus subtilis and its conversion to a monoacylglycerol hydrolase. Eur. J. Biochem. 267:6459-6469. [DOI] [PubMed] [Google Scholar]
- 5.Ghorpade, V. M., A. Gennadios, and M. A. Hanna. 2001. Laboratory composting of extruded poly(lactic acid) sheets. Bioresour. Technol. 76:57-61. [DOI] [PubMed] [Google Scholar]
- 6.Handrick, R., S. Reinhardt, M. L. Focarete, M. Scandola, G. Adamus, M. Kowalczuki, and D. Jendrossek. 2001. A new type of thermoalkalophilic hydrolase of Paucimonas lemoignei with high specificity for amorphous polyesters of short chain-length hydroxyalkanoic acids. J. Biol. Chem. 276:36215-36224. [DOI] [PubMed] [Google Scholar]
- 7.Hoshino, A., and Y. Isono. 2002. Degradation of aliphatic polyester films by commercially available lipases with special reference to rapid and complete degradation of poly(l-lactide) film by lipase PL derived from Alcaligenes sp. Biodegradation 13:141-147. [DOI] [PubMed] [Google Scholar]
- 8.Jarerat, A., and Y. Tokiwa. 2001. Degradation of poly(l-lactide) by a fungus. Macromol. Biosci. 1:136-140. [Google Scholar]
- 9.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 10.Kay, M. J., R. W. McCabe, and L. H. G. Morton. 1993. Chemical and physical changes occurring in polyester polyurethane during biodegradation. Int. Biodeterior. Biodegrad. 31:209-225. [Google Scholar]
- 11.Kim, H. K., S. Y. Park, J. K. Lee, and T. K. Oh. 1998. Gene cloning and characterization of thermostable lipase from Bacillus stearothermophilus L1. Biosci. Biotechnol. Biochem. 62:66-71. [DOI] [PubMed] [Google Scholar]
- 12.Kleeberg, I., C. Hetz, R. M. Kroppenstedt, R. J. Muller, and W. D. Deckwer. 1998. Biodegradation of aliphatic-aromatic copolyesters by Thermomonospora fusca and other thermophilic compost isolates. Appl. Environ. Microbiol. 64:1731-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Leenslag, J. W., A. J. Pennings, R. R. Boss, F. R. Rozema, and G. Boering. 1987. Resorbable materials of poly(l-lactide). Biomaterials 8:311-314. [DOI] [PubMed] [Google Scholar]
- 15.Li, S. M., I. Molina, M. B. Martinez, and M. Vert. 2002. Hydrolytic and enzymatic degradations of physically crosslinked hydrogels prepared from PLA/PEO/PLA triblock copolymers. J. Mater. Sci. Mater. Med. 13:81-86. [DOI] [PubMed] [Google Scholar]
- 16.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 17.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Möler, B., R. Vetter, D. Wilke, and B. Foullois. October 1991. Alkaline Bacillus lipases, coding DNA sequences and bacilli which produce the lipase. Patent WO9116422.
- 19.Nakamura, K., T. Tomita, N. Abe, and Y. Kamio. 2001. Purification and characterization of an extracellular poly(l-lactic acid) depolymerase from a soil isolate, Amycolatopsis sp. strain K104-1. Appl. Environ. Microbiol. 67:345-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neu, H. C., and L. A. Heppel. 1965. The release of enzyme from Escherichia coli by osmotic shock during the formation of spheroplast. J. Biol. Chem. 240:3685-3692. [PubMed] [Google Scholar]
- 21.Nthangeni, M. B., H. G. Patterton, A. van Tonder, W. P. Vergeer, and D. Litthauer. 2001. Over-expression and properties of a purified recombinant Bacillus licheniformis lipase: a comparative report on Bacillus lipases. Enzyme Microb. Technol. 28:705-712. [DOI] [PubMed] [Google Scholar]
- 22.Ollis, D. L., E. Cheah, M. Cygler, B. Dijkstra, F. Frolow, S. M. Franken, M. Harel, S. J. Remington, I. Silman, J. Schrag, J. L. Sussman, K. H. Verschueren, and A. Goldman. 1992. The α/β hydrolase fold. Protein Eng. 5:197-211. [DOI] [PubMed] [Google Scholar]
- 23.Pranamuda, H., Y. Tokiwa, and H. Tanaka. 1995. Microbial degradation of an aliphatic polyester with a high melting point, poly(tetramethylene succinate). Appl. Environ. Microbiol. 61:1828-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pranamuda, H., Y. Tokiwa, and H. Tanaka. 1997. Polylactide degradation by an Amycolatopsis sp. Appl. Environ. Microbiol. 63:1637-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pranamuda, H., and Y. Tokiwa. 1999. Degradation of poly(l-lactide) by strains belonging to genus Amycolatopsis. Biotechnol. Lett. 21:901-905. [Google Scholar]
- 26.Pranamuda, H., A. Tsuchii, and Y. Tokiwa. 2001. Poly(l-lactide)-degrading enzyme produced by Amycolatopsis sp. Macromol. Biosci. 1:25-29. [Google Scholar]
- 27.Saito, H., and K. Miura. 1963. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72:619-629. [PubMed] [Google Scholar]
- 28.Sakai, K., H. Kawano, A. Iwami, M. Nakamura, and M. Moriguchi. 2001. Isolation of a thermophilic poly-l-lactide degrading bacterium from compost and its enzymatic characterization. J. Biosci. Bioeng. 92:298-300. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt-Dannert, C., M. L. Rua, H. Atomi, and R. D. Schmid. 1996. Thermoalkalophilic lipase of Bacillus thermocatenulatus. I. Molecular cloning, nucleotide sequence, purification and some properties. Biochim. Biophys. Acta 1301:105-114. [DOI] [PubMed] [Google Scholar]
- 30.Suyama, T., Y. Tokiwa, P. Ouichanpagdee, T. Kanagawa, and Y. Kamagata. 1998. Phylogenetic affiliation of soil bacteria that degrade aliphatic polyesters available commercially as biodegradable plastic. Appl. Environ. Microbiol. 64:5008-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tansengeo, M. L., and Y. Tokiwa. 1998. Thermophilic microbial degradation of polyethylene succinate. World J. Microbiol. Biotechnol. 14:133-138. [Google Scholar]
- 32.Teeraphatpornchai, T., T. Nakajima-Kambe, Y. Shigeno-Akutsu, M. Nakayama, N. Nomura, T. Nakahara, and H. Uchiyama. 2003. Isolation and characterization of a bacterium that degrades various polyester-based biodegradable plastics. Biotechnol. Lett. 25:23-28. [DOI] [PubMed] [Google Scholar]
- 33.Tomita, K., Y. Kuroki, and K. Nagai. 1999. Isolation of thermophiles degrading poly(l-lactic acid). J. Biosci. Bioeng. 87:752-755. [DOI] [PubMed] [Google Scholar]
- 34.Vandamme, E., K. H. Schanck-Brodrueck, C. Colson, and J. D. V. Hanotier. October 1987. DNA segment coding for a specific lipase, vectors for the expression thereof, microorganisms transformed by these vectors and use of these microorganisms for the production of the lipase. Patent EP0243338-A/1.
- 35.van Pouderoyen, G., T. Eggert, K.-E. Jaeger, and B. W. Dijkstra. 2001. The crystal structure of Bacillus subtilis lipase: a minimal α/β hydrolase fold enzyme. J. Mol. Biol. 309:215-226. [DOI] [PubMed] [Google Scholar]
- 36.Williams, D. F. 1981. Enzymic hydrolysis of polylactic acid. Eng. Med. 10:5-7. [Google Scholar]
- 37.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]