Abstract
Terminal restriction fragment length polymorphism (T-RFLP) analysis of PCR-amplified genes is a widely used fingerprinting technique in molecular microbial ecology. In this study, we show that besides expected terminal restriction fragments (T-RFs), additional secondary T-RFs occur in T-RFLP analysis of amplicons from cloned 16S rRNA genes at high frequency. A total of 50% of 109 bacterial and 78% of 68 archaeal clones from the guts of cetoniid beetle larvae, using MspI and AluI as restriction enzymes, respectively, were affected by the presence of these additional T-RFs. These peaks were called “pseudo-T-RFs” since they can be detected as terminal fluorescently labeled fragments in T-RFLP analysis but do not represent the primary terminal restriction site as indicated by sequence data analysis. Pseudo-T-RFs were also identified in T-RFLP profiles of pure culture and environmental DNA extracts. Digestion of amplicons with the single-strand-specific mung bean nuclease prior to T-RFLP analysis completely eliminated pseudo-T-RFs. This clearly indicates that single-stranded amplicons are the reason for the formation of pseudo-T-RFs, most probably because single-stranded restriction sites cannot be cleaved by restriction enzymes. The strong dependence of pseudo-T-RF formation on the number of cycles used in PCR indicates that (partly) single-stranded amplicons can be formed during amplification of 16S rRNA genes. In a model, we explain how transiently formed secondary structures of single-stranded amplicons may render single-stranded amplicons accessible to restriction enzymes. The occurrence of pseudo-T-RFs has consequences for the interpretation of T-RFLP profiles from environmental samples, since pseudo-T-RFs may lead to an overestimation of microbial diversity. Therefore, it is advisable to establish 16S rRNA gene sequence clone libraries in parallel with T-RFLP analysis from the same sample and to check clones for their in vitro digestion T-RF pattern to facilitate the detection of pseudo-T-RFs.
One of the most active fields in microbial ecology is the study of microbial communities in their natural habitats. Cultivation-independent molecular methods have become indispensable tools for this type of research, among which PCR is a core technique. Despite its known limitations (for a review, see reference 30), PCR amplification of 16S rRNA genes is an integral part of the so-called full-cycle rRNA analysis approach to community structure analysis (1), which involves cloning of amplified gene products, comparative sequence analysis of individual clones, and, subsequently, probe design and application of probes to environmental samples.
The caveats of the cloning approach (30), namely, the lack of analysis of a statistically significant number of clones required for complex communities, has encouraged the use of molecular techniques, which map the diversity of the community structure by PCR-based fingerprinting. In contrast to cloning analysis, fingerprinting techniques such as denaturing/thermal gradient gel electrophoresis (DGGE/TGGE) (for a review, see reference 19), single-stranded site conformational polymorphism (SSCP) (12, 25), and terminal restriction fragment length polymorphism (T-RFLP) (4, 13) (for reviews, see references 11 and 17) analyses allow the physical separation of the total pool of amplified community gene products.
Typically, T-RFLP analysis involves amplification of target genes from whole-community DNA extracts by using specific primer pairs, one of which is fluorescently labeled. Subsequently, amplicons are digested with restriction enzymes (usually tetranucleotide recognizing) and fragments are size separated via gel electrophoresis on automated sequencers, whereby only the labeled terminal fragments (T-RFs) are detected and quantified. Individual T-RFs can be assigned presumptively to operational taxonomic units, which ideally correspond to phylogenetically related microorganisms, based on in silico search for matching restriction sites in sequences from clone libraries established in parallel from the same sample. The 16S rRNA gene has been used extensively as marker gene for T-RFLP analysis (for reviews, see references 11 and 17).
In general, the T-RFLP technique has proven to be a reproducible and accurate tool for community fingerprinting (4, 13, 18, 22). Since T-RFLP analysis is based on PCR amplification, all biases related to this technique apply (30) and a number of important parameters related to PCR have been identified; it has been found that initial DNA template concentration, number of PCR cycles, annealing temperature, and the choice of Taq DNA polymerase from different manufacturers may affect the composition of T-RFLP profiles (4, 22).
T-RFLP-based gene ratios were found to be influenced by preferential gene amplification of specific templates (PCR drift [23, 28, 29]) when degenerated primers for the amplification of the mcrA (methyl coenzyme M reductase) gene were used (14, 16). On the other hand, Lueders and Friedrich (16) demonstrated that PCR-T-RFLP analysis can accurately reflect template ratios of archaeal 16S rRNA genes in a model community with defined amounts of 16S rRNA gene copies from five different methanogens.
In addition to PCR factors, the composition of T-RFLP profiles can be influenced by factors related to the restriction digestion, such as partially digested PCR products observed in T-RFLP profiles of pure cultures (3, 4) or environmental samples (22, 27). Additional restriction fragments (RFs) in T-RFLP profiles of pure cultures were attributed to either incomplete digestion of the amplicons or sequence heterogeneity of the template, i.e., multiple copies of 16S rRNA genes in single species with different terminal restriction sites (4). If the occurrence of additional peaks originates from incomplete digestion, this may be revealed under limiting restriction enzyme concentration (22). At any rate, incompletely digested PCR products from a complex microbial community may result in additional T-RFs and, consequently, an overestimation of diversity (22).
The present study was initiated to systematically examine the frequent occurrence of unexpected RFs in addition to the expected T-RFs after in vitro digestion of individual environmental 16S rRNA gene clones. These additional, nonterminal RFs in T-RFLP profiles were designated pseudo-T-RFs. Our results indicate that partially single-stranded amplicons are involved in the formation of pseudo-T-RFs.
MATERIALS AND METHODS
DNA extracts.
DNA extracts and bacterial and archaeal 16S rRNA gene clones from gut compartments of cetoniid beetle (Pachnoda ephippiata) larvae are described elsewhere (M. Egert, T. Lemke, B. Wagner, A. Brune, and M. W. Friedrich, unpublished data). Accession numbers of clones used in this study are AJ538350 (clone PeM 75), AJ538351 (PeH59), and AJ538352 (PeMAr04). Pure-culture DNA extracts of Methanococcus jannaschii (DSM 2661T), Methanobacterium bryantii (DSM 863T), Methanospirillum hungatei (DSM 864T), and Methanosaeta concilii (DSM 3671T) were kindly provided by T. Lueders (MaxPlanck Institute, Marburg, Germany).
T-RFLP analysis.
16S rRNA genes were specifically amplified using the primer combination of 6-carboxyfluorescein (FAM)-labeled primers 27f (5′-AGA-GTT-TGA-TCC-TGG-CTC-AG-3′) (5) and 907r (5′-CCG-TCA-ATT-CCT-TTR-AGT-TT-3′) (20) for Bacteria and Ar109f (5′-ACK-GCT-CAG-TAA-CAC-GT-3′) (7) and FAM-labeled Ar912r (5′-CTC-CCC-CGC-CAA-TTC-CTT-TA-3′) (15) for Archaea. The standard reaction mixture contained, in a total volume of 50 μl, 1× PCR buffer II (Applied Biosystems, Weiterstadt, Germany), 1.5 mM MgCl2, a 50 μM concentration of each of the four deoxynucleoside triphosphates (Amersham Pharmacia Biotech, Freiburg, Germany), a 0.5 μM concentration of each primer (MWG Biotech, Ebersberg, Germany), and 1.25 U of Ampli Taq DNA polymerase (Applied Biosystems). In addition, 1 μl of a 1:30 dilution of Pachnoda gut DNA extract, 0.5 μl of a 1:10 dilution of clonal M13 product (including 16S rRNA gene sequence inserts), or 1 μl of pure-culture DNA extract was added as the template. All reaction mixtures were prepared at 4°C in 0.2-ml reaction tubes to avoid nonspecific priming. Amplification was started by placing the reaction tubes immediately into the preheated (94°C) block of a GeneAmp 9700 thermocycler (Applied Biosystems). The standard thermal profiles for the amplification of bacterial 16S rRNA genes were as follows: initial denaturation (94°C for 3 min) followed by 16 (clonal DNA templates) or 32 (environmental DNA templates) cycles of denaturation (94°C for 30 s), annealing (52°C for 30 s), and extension (72°C for 60 s). Thermal profiles for the amplification of archaeal 16S rRNA genes started with an initial denaturation (94°C for 3 min) followed by 16 (clonal and pure-culture DNA templates) or 35 to 38 (environmental templates) cycles of denaturation (94°C for 45 s), annealing (52°C for 45 s), and extension (72°C for 90 s). After terminal extension (72°C for 5 to 7 min), samples were stored at 4°C until further analysis. Aliquots (5 μl) of 16S rRNA amplicons were analyzed by gel electrophoresis on 1% agarose gels and visualized after being stained with ethidium bromide. PCR products were purified with the MinElute PCR purification kit (Qiagen, Hilden, Germany).
Prior to digestion, amplicon concentrations were determined photometrically. DNA (75 ng for amplicons from the gut DNA extract, 50 ng for clonal amplicons), 2.5 U of restriction enzymes (MspI, TaqI, and AluI [Promega, Mannheim, Germany]; MspI, HpaII, HhaI, HaeIII, and BstUI [New England Biolabs, Frankfurt am Main, Germany]; and BsiSI [Minotech Biotechnology, Heraklion, Crete, Greece]), 1 μl of 10× incubation buffer, and 1 μg of bovine serum albumin (if recommended) were combined in a total volume of 10 μl and digested for 3 h at 37°C (MspI, AluI, HpaII, HhaI, and HaeIII), 55°C (BsiSI), 60°C (BstUI), 65°C (TaqI), or 70°C (BsiSI). Fluorescently labeled T-RFs were size separated on an ABI 373A automated sequencer (Applied Biosystems) using an internal size standard (GeneScan-1000 ROX; Applied Biosystems). T-RFLP electropherograms were analyzed with GeneScan 2.1 software (Applied Biosystems) (15).
Mung bean nuclease digest.
The single-stranded DNA parts of 16S rRNA gene amplicons were digested using mung bean nuclease. Approximately 1,000 ng of PCR product was incubated for 1 h at 30°C with 5 U of mung bean nuclease (New England Biolabs) and 10 μl of 10× reaction buffer in a total volume of 100 μl. The digestion was stopped by phenol-chloroform-isoamyl alcohol (25:24:1) extraction, and DNA was recovered by ethanol precipitation. Digested amplicons were purified using the MinElute PCR purification kit.
RESULTS AND DISCUSSION
Occurrence of pseudo-T-RFs in T-RFLP analysis.
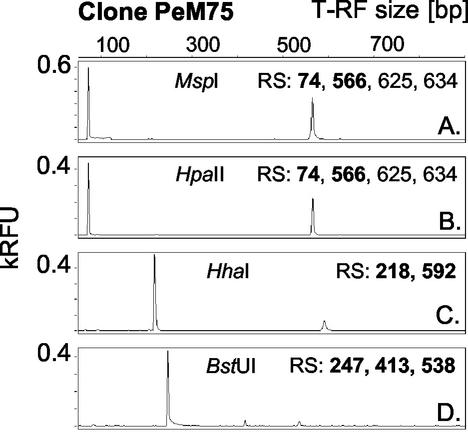
Clones from two bacterial (109 clones) and two archaeal (68 clones) 16S rRNA gene clone libraries (established from DNA extracts of the midgut and hindgut of cetoniid beetle [Pachnoda ephippiata] larvae [Egert et al., unpublished]) were analyzed by PCR-T-RFLP using MspI as the restriction endonuclease for the bacterial clones and AluI as the restriction endonuclease for the archaeal clones. Theoretically, each clone was expected to display a single T-RF with a fragment length which is predictable from the respective sequence data. However, 50% of the bacterial and 78% of the archaeal clones reproducibly showed unexpected RFs in addition to the expected T-RF (examples are given in Fig. 1A and 2B). Moreover, additional RFs were observed for amplicons of two of four archaeal pure cultures (Methanobacterium bryantii and Methanospirillum hungatei) digested with AluI as the restriction enzyme (Fig. 2C). Depending on the restriction enzyme used, single clones displayed either no, one, or (rarely) two or more additional RFs. Additional RFs occurred with a variety of restriction enzymes with different or identical recognition sites when multiple clones were analyzed, i.e., MspI (from two different suppliers), HpaII (an isoschizomer of MspI), HhaI, BstUI (e.g., clone PeM75 [Fig. 1]), AluI (e.g., clones PeMAr04 [Fig. 2B] and PeH59 [Fig. 3B ]), and HaeIII (data not shown). In silico analysis of clonal and pure-culture sequence data revealed that all additional RFs corresponded to restriction sites downstream of the primary site (e.g., clone PeM75 [Fig. 1]). Thus, additional RFs most probably originated from a partial digestion of the amplicons in which the terminal restriction site was not cleaved.
FIG. 1.
Occurrence of pseudo-T-RFs in T-RFLP profiles of a single clone depending on the restriction enzyme used. 16S rRNA gene T-RFLP electropherograms were derived from clone PeM75 (affiliated with Lactobacillales). Numbers indicate restriction sites (RS) for the respective enzyme detected in the clonal sequence between bases 1 and ∼900 (length of the PCR product), counted from the labeled 5′ end. Bold numbers indicate restriction sites with corresponding T-RFs in the electropherogram. RFU, relative fluorescence units.
FIG. 2.
Effect of mung bean nuclease digestion on the occurrence of pseudo-T-RFs in T-RFLP profiles (AluI digests) of environmental, clonal, and pure-culture samples. Insets show the T-RFLP profile after mung bean nuclease digestion. The number of PCR cycles used to produce the amplicons is indicated. Fragment lengths of pseudo-T-RFs are shown in bold. Clone PeMAr04 is affiliated with the kingdom Crenarchaeota. MB, Methanobactericeae; CR, Crenarchaeota; RFU, relative fluorescence units.
FIG. 3.
(A to C) T-RFLP analysis of clone PeH59 (affiliated with the CFB phylum) amplicons after restriction digestion with different enzymes, resulting in the expected T-RFs only (MspI [A]) or in the formation of pseudo-T-RFs (AluI [B] and HhaI [C]). (D) 16S rRNA gene secondary structure of clone PeH59 as predicted by the mfold software including the sequence stretches around detected pseudo-T-RFs. RS, restriction sites. Bold numbers indicate restriction sites with corresponding T-RFs in the electropherogram. RFU, relative fluorescence units.
For these additional, unexpected RFs, we introduce the name “pseudo-T-RFs” (Gr. adj. pseudos, meaning false; i.e., a false T-RF), because they are detectable as terminal, fluorescently labeled fragments in T-RFLP analysis; however, pseudo-T-RFs do not represent the actual (“real”) terminal restriction fragment as predicted from sequence data and therefore have to be regarded as false T-RFs (hence, pseudo-T-RFs).
The frequent occurrence of pseudo-T-RFs in T-RFLP profiles of clones suggested their likely occurrence also in T-RFLP profiles of complex microbial communities. In fact, potential pseudo-T-RFs were identified in T-RFLP profiles of environmental samples, i.e., gut DNA extracts of P. ephippiata larvae and soil which was used for feeding the larvae (Egert et al., unpublished), by comparing predicted T-RFs of clones to those present in the mixed-community T-RFLP profile.
For example, the T-RFLP profile of archaeon-specific 16S rRNA gene amplicons from midgut DNA extracts with AluI digestions was characterized by three T-RFs, two of which could be presumptively assigned to clonal sequences affiliated with the Methanobacteriaceae (T-RF of 64 bp; 6 clones) and Crenarchaeota (125 bp; 12 clones) (Fig. 2A). However, the prominent peak at 165 bp in the electropherogram was not reflected by any clone sequence; i.e., no clone sequence showed a primary, real terminal AluI restriction site of 165 bp. In vitro digestion of clonal PCR amplicons revealed that all clones related to Methanobacteriaceae and Crenarchaeota displayed an additional 165-bp RF (shown in Fig. 2B for the crenarchaeotal clone PeMAr04). Therefore, it was assumed that the 165-bp T-RF in the midgut T-RFLP profile was a pseudo-T-RF.
Involvement of partly single-stranded amplicons in pseudo-T-RF formation.
The occurrence of multiple RFs in T-RFLP profiles from single species has been reported for pure cultures (4, 6) and clonal PCR amplicons (6, 27), which were explained by 16S rRNA gene sequence heterogeneity, e.g., multiple rRNA operons in a single species (4), or partial digestion of the PCR products (4, 22, 27). Sequence heterogeneity of 16S rRNA genes can be excluded as a reason for the formation of pseudo-T-RFs, because we used amplicons of clonal origin. Nevertheless, a characteristic of all clones with pseudo-T-RFs was that the primary terminal restriction site was cleaved by the restriction enzyme for only a fraction of the amplicon pool; i.e., they were only partially digested (Fig. 1, 2B, and 3).
All efforts to overcome a bias related to partial digestion of amplicons were not successful. Use of twice as much enzyme (5 U) as in a typical digest (22) and extension of the digestion time (6 and 24 h) did not relieve the occurrence or the intensity of pseudo-T-RF peaks. It is noteworthy, though, that peaks with a size corresponding to full-length amplicons (∼900 bp) were not present in T-RFLP profiles of clones with and without pseudo-T-RFs (e.g., Fig. 1, 3, and 4), which would have been indicative of incomplete digestion because of limiting enzyme concentration or suboptimal reaction conditions (22).
FIG. 4.
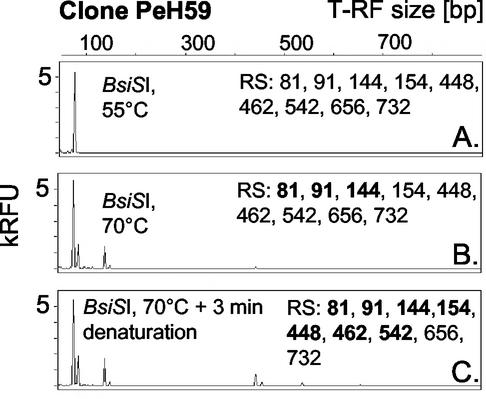
Effect of restriction digest temperature on the formation of pseudo-T-RFs of clone PeH59. Restriction digests were performed using BsiSI at 55°C (A) and 70°C (B) and by using a 3-min denaturation of the PCR amplicon prior to the addition of enzyme and incubation at 70°C (C). Bold numbers indicate restriction sites with corresponding T-RFs in the electropherogram. RFU, relative fluorescence units.
Since restriction endonucleases require double-stranded DNA at the restriction site (21), the presence of single-stranded amplicons in the range of the terminal restriction site was checked for by using mung bean nuclease, which degrades single-stranded DNA (10).
After mung bean nuclease digestion, pseudo-T-RFs were not detectable in environmental (Pachnoda gut), clonal, and pure-culture-derived T-RFLP profiles (Fig. 2). These data indicate clearly that the formation of pseudo-T-RFs results from the presence of at least partly single-stranded DNA amplicons. Single-stranded DNA is not a substrate for type II restriction endonucleases (21), and so the presence of single-stranded 5′-DNA ends of part of the amplicon pool—on otherwise double-stranded PCR products—provides an explanation of why the terminal restriction site was not cut. Similarly, mung bean nuclease treatment was used to remove single-stranded DNA artifacts prior to SSCP (10) and DGGE analysis (26).
By comparing T-RFLP patterns of individual clones with different restriction endonucleases, it became evident that the amplicon pool contains PCR products which are single stranded to different degrees. For example, BstUI digestions of clone PeM75 amplicons yielded pseudo-T-RFs of 413 and 538 bp (Fig. 1D), which suggests that a small part of the amplicon pool is single stranded, at least up to the second BstUI restriction site of PeM75 at bp 413.
The secondary structure of 16S rRNA gene sequences influences restriction digests. Some clones displayed pseudo-T-RFs with one enzyme but not with the other when amplicons from the same PCR batch were analyzed by T-RFLP. For example, clone PeH59 had a primary MspI restriction site at 81 bp and eight subsequent restriction sites as revealed by sequence data analysis (Fig. 3A), but pseudo-T-RFs were not formed. Digests with AluI (Fig. 3B), HhaI (Fig. 3C), and BstUI (data not shown) revealed pseudo-T-RFs up to 638 bp (AluI [Fig. 3B]), which suggests that some amplicons were single stranded at least up to bp 241. According to a model which involves the formation of transiently formed secondary structures composed of recognition sequences with twofold rotational symmetry (“canonical structures”), many type II restriction endonucleases cleave single-stranded DNA (21). Inspection of possible secondary structures as calculated with the program mfold (24) (M. Zuker; http://www.bioinfo.rpi.edu/applications/mfold/old/dna/) showed that the primary MspI restriction site of clone PeH59 was able to form a canonical structure (i.e., a local secondary structure) by folding back with an upstream single-stranded sequence (Fig. 3D). Although the secondary structures did not form a perfect palindrome, it is likely that the primary restriction site of single-stranded amplicons was indeed cleaved by MspI, since pseudo-T-RFs downstream from the primary restriction site were not detected. In contrast, the primary AluI restriction site of clone PeH59 most probably did not form a sterically sufficient secondary structure from single-stranded DNA, and thus AluI did not cleave single-stranded amplicons at the primary recognition site, which corroborates the presence of a pseudo-T-RF at 638 bp. It should be noted, however, that the predicted secondary structures represent the most thermodynamically stable structures according to the underlying model (24) as implemented in mfold; thus, these structures may actually not exist in the reaction mixture of the T-RFLP digest. However, restriction digests conducted at different temperatures provide experimental evidence that canonical structures in single strands might be the reason why some clonal amplicons do not show pseudo-T-RF formation with certain nucleases. Clone PeH59 did not show pseudo-T-RFs when digested with MspI at 37°C (Fig. 3A) or BsiSI at 55°C (Fig. 4A); BsiSI is an isoschizomer of MspI which is not inactivated by heat. However, at 70°C (Fig. 4B), pseudo-T-RFs occurred when BsiSI was used and were even more pronounced when the amplicons were denatured for 3 min at 94°C prior to digestion (Fig. 4C). At increased digestion temperature, canonical structures in single-stranded amplicons are likely to become unstable, rendering the restriction sites inaccessible to the nuclease, which in turn leads to the formation of pseudo-T-RFs. Interestingly, pseudo-T-RFs were not detectable using TaqI as the restriction endonuclease at a digestion temperature of 65°C, even with clones that possessed multiple TaqI restriction sites and displayed pseudo-T-RFs with other nucleases (tested only for archaeal clones [data not shown]). Possibly, TaqI cleaves single-stranded amplicons not involved in canonical structures at a higher rate than the other restriction enzymes analyzed, making TaqI suitable as an endonuclease that avoids formation of pseudo-T-RFs in T-RFLP analysis.
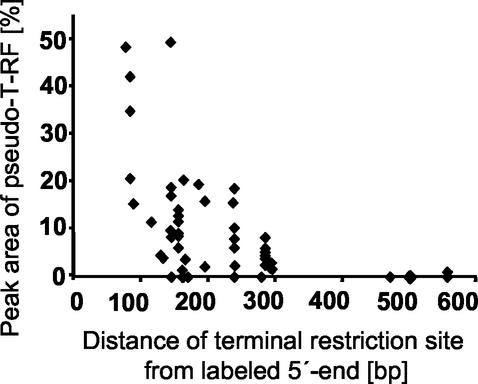
In general, the extent of pseudo-T-RF formation (for all restriction endonucleases tested) decreased with increasing distance of the terminal restriction site from the 5′ labeled end of the amplicon (Fig. 5), which supports the hypothesis that amplicons are partly single stranded.
FIG. 5.
Effect of the position of the terminal restriction site on the extent of pseudo-T-RF formation, based on in vitro T-RF formation of 56 bacterial clones with MspI as the restriction endonuclease. The peak area of the pseudo-T-RF is compared to the peak area of the primary T-RF and given as a percentage. Clones were obtained from a 16S rRNA gene clone library derived from the midgut of cetoniid beetle larvae (Egert et al., unpublished).
Influence of PCR on the formation of pseudo-T-RFs.
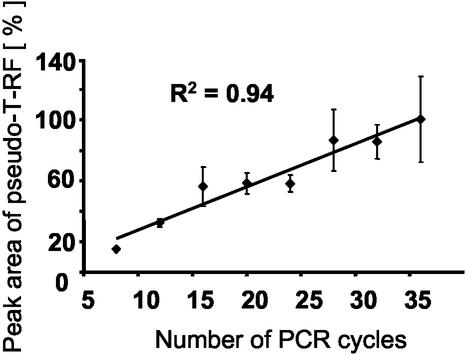
We found that the height and area of a pseudo-T-RF in relation to those of the primary, expected RF increased with the number of PCR cycles used to produce the amplicon (Fig. 6). This shows clearly that the occurrence of pseudo-T-RFs is a PCR artifact and that a PCR bias is apparently involved in the formation of partly single-stranded amplicons. Therefore, we tried to optimize the PCR protocols, but none of the following modifications, tested with selected bacterial clones, significantly affected pseudo-T-RF formation: (i) prolonged extension (1 min 30 s or 2 min) or final extension time (10 or 15 min), (ii) increased concentration of Ampli Taq DNA polymerase (3 U) or addition of fresh polymerase (3 U) before the final extension step to exclude polymerase limitation, (iii) addition of Pfu DNA polymerase (Promega) with proofreading activity, (iv) increased concentration of primers (1 μM each), (v) decreased initial template concentration (∼100 to 10−3 ng μl−1), or (vi) higher (i.e., more stringent) annealing temperatures (55, 57, 59, 61, or 64°C) to determine whether the formation of single-stranded DNA could result from incorrectly annealed primers. Unexpectedly, at annealing temperatures of 61 and 64°C, the number of pseudo-T-RFs even increased. Lower annealing temperatures (50, 48, and 46°C) did not affect the formation of pseudo-T-RFs.
FIG. 6.
Effect of PCR cycle number on the extent of pseudo-T-RF formation observed with amplicons of clone PeM75 after MspI digestion. The peak area of the pseudo-T-RF is compared to the peak area of the primary T-RF and given as a percentage. Error bars (which represent standard deviation) are based on three replicates.
The formation of single-stranded amplicons can be favored by a differential, asymmetric utilization of primers in the PCR amplification due to differences in priming efficiencies, which can result from differences in the G+C content of the primers used (10). Therefore, primer concentrations were varied at ratios of 1:8 to 8:1 (27f versus 907r; G+C content, 50 and 37.5%, respectively) to overcome a possible bias related to asymmetric primer utilization in the PCR, but the formation of pseudo-T-RFs was unaffected.
The formation of partly single-stranded 16S rRNA gene amplicons during PCR may result from template secondary structures (10), which causes the polymerase to pause or fall off the template (23). However, use of the PCR enhancer betaine at various concentrations, which had been shown to be effective in improving the amplification yield and the specificity of templates with high G+C content or secondary structures (9), did not prevent the formation of pseudo-T-RFs.
Model for the formation of pseudo-T-RFs.
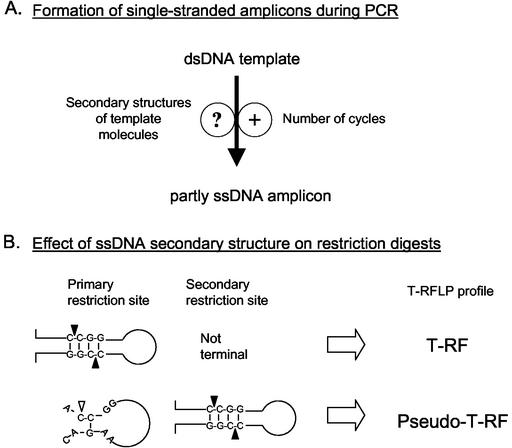
Based on the above results, we propose the following model for the formation of pseudo-T-RFs during T-RFLP analysis of 16S rRNA genes. During PCR of 16S rRNA genes from clonal, pure-culture, and environmental DNA extracts, some of the amplicons formed are at least partly single stranded (as proven by mung bean nuclease digests [Fig. 2]). Since single-stranded terminal restriction sites cannot be cleaved by restriction endonucleases, “pseudo”-terminal restriction sites downstream from the expected primary restriction site can be detected by T-RFLP analysis. The ability of the 16S RNA molecule to backfold with itself (8) may result in an incomplete synthesis of a fraction of 16S rRNA gene amplicons during PCR (Fig. 7A). The involvement of PCR in the generation of (partly) single-stranded amplicons is corroborated by the strong dependence on the number of PCR cycles (Fig. 6). Similarly, the number of PCR cycles has been implicated as a controlling factor in a kinetic model which describes the reannealing of single-stranded templates as a source of the PCR bias (28). Accordingly, the formation of single-stranded amplicons may be viewed as an extension of the original kinetic model of template reannealing: when the amplicon concentration increases at greater PCR cycle numbers, the rate of interaction between single-stranded template molecules increases, which may result not only in interstrand reannealing as described by Suzuki and Giovannoni (28) but also in intrastrand annealing, hence the formation of local secondary structures. In turn, these temporary secondary structures of template molecules may cause the DNA polymerase molecules to fall off with higher frequency (23), thereby leaving the template strands (partially) unamplified. Furthermore, we hypothesize that single-stranded 16S rRNA gene amplicons can form local palindromic secondary structures, which in turn allow restriction enzymes to cut “single-stranded” DNA (21). This hypothesis helps explain why T-RFLP analyses with certain enzymes yield pseudo-T-RFs whereas others from the same PCR amplification do not (Fig. 7B): a secondary restriction site will be detected in T-RFLP analysis only if the primary restriction is not part of a canonical structure. We could show that higher temperatures (70°C, [Fig. 4B and C]) during restriction digestion resulted in the formation of pseudo-T-RFs, most probably because local secondary structures were unstable under these conditions and consequently were no longer substrates for the restriction enzyme. Thus, the sequence context around the primary restriction site most probably will determine whether even a single-stranded amplicon can be digested at its primary, real terminal restriction site.
FIG. 7.
Schematic model of pseudo-T-RF formation. (A) PCR-related parameters influencing the formation of partly single-stranded amplicons. (B) Involvement of the secondary structure of partly single-stranded amplicons in the formation of pseudo-T-RFs. dsDNA, double-stranded DNA; ssDNA, single-stranded DNA; solid triangles, restriction site cut (MspI); open triangle, restriction site not cut.
Regardless of the underlying mechanism for secondary-structure formation, pseudo-T-RFs occurred in 16S rRNA gene clones from diverse phylogenetic lineages (i.e., Lactobacillales, Bacillales, Clostridia, high-G+C gram-positive Bacteria, Crenarchaeota, and Euryarchaeota [Egert et al., unpublished]), which shows that most 16S rRNA gene amplicons may be affected. Interestingly, pseudo-T-RFs were not detected in T-RFLP analysis of cloned nirK genes (copper-containing nitrite reductase [2; G. Braker, personal communication]) or mcrA genes (methyl-coenzyme M reductase [T. Lueders, personal communication]); apparently, PCR products of these genes do not form secondary structures which are as thermodynamically stable as those formed in 16S rRNA molecules.
Conclusions and recommendations.
The occurrence of pseudo-T-RFs in T-RFLP profiles has consequences for the interpretation of the underlying microbial diversity. First of all, if pseudo-T-RFs are not identified, diversity may be overestimated because of the larger number of peaks in T-RFLP profiles. For example, in the AluI-based archaeon-specific T-RFLP analysis of the Pachnoda midgut, the prominent T-RF of 165 bp (Fig. 2A) did not represent additional diversity which was overlooked by clone library analysis but, rather, could be clearly identified as a pseudo-T-RF originating from clone sequences related to Methanobacteriaceae and Crenarchaeota. Second, curing of pseudo-T-RFs by simple elimination through mung bean nuclease digestion (Fig. 2A) will result in an underestimation of the relative gene frequency of amplicons which are affected by the formation of pseudo-T-RFs; in the case of the prominent archaeal pseudo-T-RF of 165 bp, 26% of the total 16S RNA gene frequency was represented by the pseudo-T-RF. When the in vitro digestion pattern of clones was tested, the number of assignable T-RFs in a Bacteria-specific T-RFLP profile from the hindgut of Pachnoda larvae (Egert et al., unpublished) increased from 18 to 27. To this end, it should be kept in mind that the restriction enzyme for T-RFLP analysis which produces the largest number of peaks from a given amplicon pool may not be the most suitable one, because the increase in the number of T-RF peaks may be a reflection of an increased number of pseudo-T-RFs only.
Since the extent of pseudo-T-RF formation is likely to be dependent on the species (gene) composition of the system under investigation and the chosen restriction endonuclease(s), it is advisable to perform T-RFLP analysis and cloning in parallel. Although this results in increased effort, the T-RF patterns of clones should be determined by in vitro T-RFLP analysis under the applied PCR and T-RFLP conditions, in particular when T-RFs are supposed to be quantitatively assigned to species or phylogenetic groups. Assigning T-RFs solely on the basis of in silico or database search is insufficient because of the potential occurrence of pseudo-T-RFs in T- RFLP profiles. In agreement with several other studies (22, 23, 28, 29), the number of PCR cycles should be limited to a minimum because pseudo-T-RF formation increases linearly with the cycle number. Beyond T-RFLP fingerprinting, the formation of (partly) single-stranded 16S rRNA gene amplicons during PCR may also affect other core techniques in microbial ecology, e.g., 16S rRNA gene cloning. In 16S rRNA gene clone libraries, sequences with a strong tendency to produce single-stranded amplicons are likely to be underrepresented because the single-stranded fraction of the amplicons cannot be ligated into the cloning vector.
Acknowledgments
This study was supported by a grant by the Deutsche Forschungsgemeinschaft (DFG) and by the Max Planck Society.
We thank Bianca Wagner for excellent technical assistance, and we thank Gesche Braker and Tillmann Lueders for data on the in vitro T-RF formation pattern of cloned functional genes.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avrahami, S., R. Conrad, and G. Braker. 2002. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl. Environ. Microbiol. 68:5685-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruce, K. D. 1997. Analysis of mer gene subclasses within bacterial communities in soils and sediments resolved by fluorescent-PCR-restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 63:4914-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clement, B. G., L. E. Kehl, K. L. Debord, and C. L. Kitts. 1998. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J. Microbiol. Methods 31:135-142. [Google Scholar]
- 5.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes—characterization of a gene coding for 16S-ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez, J. M., R. Simo, R. Massana, J. S. Covert, E. O. Casamayor, C. Pedros-Alio, and M. A. Moran. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grosskopf, R., S. Stubner, and W. Liesack. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutell, R. R. 1994. Collection of small-subunit (16S and 16S-like) ribosomal-RNA structures—1994. Nucleic Acids Res. 22:3502-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henke, W., K. Herdel, K. Jung, D. Schnorr, and S. A. Loening. 1997. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 25:3957-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen, M. A., and N. Straus. 1993. Effect of PCR conditions on the formation of heteroduplex and single-stranded DNA products in the amplification of bacterial ribosomal DNA spacer regions. PCR Methods Appl. 3:186-194. [DOI] [PubMed] [Google Scholar]
- 11.Kitts, C. L. 2001. Terminal restriction fragment patterns: a tool for comparing microbial communities and assessing community dynamics. Curr. Issues Intest. Microbiol. 2:17-25. [PubMed] [Google Scholar]
- 12.Lee, D. H., Y. G. Zo, and S. J. Kim. 1996. Nonradioactive method to study genetic profiles of natural bacterial communities by PCR-single-strand-conformation polymorphism. Appl. Environ. Microbiol. 62:3112-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lueders, T., K. J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme M reductase-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 15.Lueders, T., and M. Friedrich. 2000. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lueders, T., and M. W. Friedrich. 2003. Evaluation of PCR amplification bias by T-RFLP analysis of SSU rRNA and mcrA genes using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2:323-327. [DOI] [PubMed] [Google Scholar]
- 18.Moeseneder, M. M., J. M. Arrieta, G. Muyzer, C. Winter, and G. J. Herndl. 1999. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:3518-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muyzer, G. 1999. DGGE/TGGE a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 2:317-322. [DOI] [PubMed] [Google Scholar]
- 20.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationship of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 21.Nishigaki, K., Y. Kaneko, H. Wakuda, Y. Husimi, and T. Tanaka. 1985. Type-II restriction endonucleases cleave single-stranded DNAs in general. Nucleic Acids Res. 13:5747-5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 23.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.SantaLucia, J. 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 95:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson, J. M., V. J. McCracken, B. A. White, H. R. Gaskins, and R. I. Mackie. 1999. Application of denaturant gradient gel electrophoresis for the analysis of the porcine gastrointestinal microbiota. J. Microbiol. Methods 36:167-179. [DOI] [PubMed] [Google Scholar]
- 27.Song, B., L. J. Kerkhof, and M. M. Haggblom. 2002. Characterization of bacterial consortia capable of degrading 4-chlorobenzoate and 4-bromobenzoate under denitrifying conditions. FEMS Microbiol. Lett. 213:183-188. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki, M. T., M. S. Rappe, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Wintzingerode, F., U. B. Goebel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples—pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]