Abstract
Phylogenetic relationships among members of the marine Synechococcus genus were determined following sequencing of the 16S ribosomal DNA (rDNA) from 31 novel cultured isolates from the Red Sea and several other oceanic environments. This revealed a large genetic diversity within the marine Synechococcus cluster consistent with earlier work but also identified three novel clades not previously recognized. Phylogenetic analyses showed one clade, containing halotolerant isolates lacking phycoerythrin (PE) and including strains capable, or not, of utilizing nitrate as the sole N source, which clustered within the MC-A (Synechococcus subcluster 5.1) lineage. Two copies of the 16S rRNA gene are present in marine Synechococcus genomes, and cloning and sequencing of these copies from Synechococcus sp. strain WH 7803 and genomic information from Synechococcus sp. strain WH 8102 reveal these to be identical. Based on the 16S rDNA sequence information, clade-specific oligonucleotides for the marine Synechococcus genus were designed and their specificity was optimized. Using dot blot hybridization technology, these probes were used to determine the in situ community structure of marine Synechococcus populations in the Red Sea at the time of a Synechococcus maximum during April 1999. A predominance of genotypes representative of a single clade was found, and these genotypes were common among strains isolated into culture. Conversely, strains lacking PE, which were also relatively easily isolated into culture, represented only a minor component of the Synechococcus population. Genotypes corresponding to well-studied laboratory strains also appeared to be poorly represented in this stratified water column in the Red Sea.
Phycobilisome-containing unicellular cyanobacteria of the genus Synechococcus have a ubiquitous distribution in oceanic waters (see reference 52 for a review) responsible for around a quarter of the primary production in some regions (37). The now well-recognized importance of these marine Synechococcus organisms in global carbon cycling has led to increased interest in the physiology and molecular biology of the genus (see references 15, 26, 64, and 73 for reviews), focusing on aspects of nutrient acquisition (18, 24, 38, 39, 41, 61, 62), photosynthesis (23, 25, 28, 33, 34, 66, 76), motility (10), and cell cycle behavior (3, 7, 8, 32). Generally though, such work has focused on a few relatively well-studied strains, and there is little known of the specific environmental niches occupied by such isolates or, indeed, the dominant genotypes found in situ.
Marine Synechococcus isolates are taxonomically most closely related to Prochlorococcus isolates (70), which lack discrete phycobilisomes but contain divinyl derivatives of chlorophyll a and b as their major light-harvesting pigments (17, 53). The genus Synechococcus includes both marine and freshwater strains and is clearly polyphyletic (31, 58). It has thus been suggested that since these organisms are phylogenetically diverse they should be reclassified into several independent taxonomic units (31). The marine Synechococcus lineage has been split into clusters based on the composition of the major light-harvesting pigments of these organisms, an ability to perform a novel swimming motility, whether there is an elevated salt requirement for growth, and G+C content (75). These clusters, originally defined as marine clusters A, B, and C (MC-A, MC-B, and MC-C) (75), have recently been reclassified (29). MC-A and MC-B are now combined into two subclusters within Synechococcus cluster 5. MC-B (Synechococcus subcluster 5.2 but with strain PCC7001 removed, molecular percentage of G+C = 63 to 66) contains mostly halotolerant strains isolated from coastal waters that possess phycocyanin but lack phycoerythrin (PE). However, one strain, WH 8007, does have elevated salt requirements for growth. It is the MC-A group (Synechococcus subcluster 5.1, molecular percentage of G+C = 55 to 62), however, which is the dominant Synechococcus group within the euphotic zone of both open-ocean and coastal waters (20, 52, 71, 80, 84). All members of this subcluster have elevated salt (Na+, Cl−, Mg2+, and Ca2+) requirements for growth and contain PE as their major light-harvesting pigment. There is considerable spectral diversity in the PEs possessed by subcluster 5.1 strains, however, largely determined by the presence or absence of the chromophore phycourobilin (PUB), as well as the ratio of PUB to phycoerythrobilin (PEB) chromophores (83, 85; reviewed by Glazer in reference 23). In addition, it is now known that some members of Synechococcus subcluster 5.1 are capable of chromatic adaptation, being able to increase their PUB/PEB ratio under blue light compared with growth under white light (51). This subtle tuning capacity of the phycobilisome content of specific cultures to the light environment is reflected in the distribution of natural marine Synechococcus populations. Thus, strains containing PUB-lacking PEs have been shown to dominate in turbid waters (classified optically as case 2 waters), whereas PUB-containing PE strains are more dominant in highly transparent waters (case 1 waters) (14, 48, 49, 84). However, since such analyses were performed prior to the discovery that some strains are capable of chromatic adaptation, it is difficult to interpret whether such distributions reflect the presence of genetically similar populations that possess this property or of genetically dissimilar ones that lack the trait. Certainly, there is good evidence for a wide genetic diversity among members of the marine Synechococcus cluster 5 lineage (reviewed in reference 63), through studies based on the DNA-dependent RNA polymerase, rpoC1, sequence data (20, 50, 67, 69) as well as sequences for the 16S-23S ribosomal DNA (rDNA) internal transcribed spacer (ITS) region (59). This latter study showed six distinct clades of Synechococcus subcluster 5.1 to be well supported phylogenetically based on sequences of the ITS. Knowledge of the specific environments or niches occupied by members of each clade is poor, however. In addition, little is known of the genetic structure of marine Synechococcus populations down a water column.
In order to better define the relationships between diversity and physiology among marine Synechococcus spp., we have characterized here the genetic signature and some physiological traits (swimming capacity, PUB/PEB ratio, and nitrogen and salt requirements) of a wide range of marine Synechococcus strains, including 31 novel isolates from the Red Sea, Pacific and Atlantic Oceans, Mediterranean Sea, and North Sea. Furthermore, to address the issue of the distribution of the different clades in the field, we have developed 16S rDNA oligonucleotides specific for individual Synechococcus cluster 5 clades. Such probes can be used in dot blot hybridization (21, 22, 27, 77, 87) or fluorescent in situ hybridization techniques (reviewed in reference 2) to analyze the abundance, distribution, and activity of specific genotypes in situ. Here, we have applied the former method and our newly designed probes to study a natural Synechococcus community in the Gulf of Aqaba, Red Sea, during a spring maximum. We demonstrate that in these conditions one Synechococcus clade predominates over all others throughout the water column.
MATERIALS AND METHODS
Sampling.
Water samples for DNA extraction were collected at station A (29° 28′ N 34° 55′ E) in the Gulf of Aqaba, Red Sea, on 27 April 1999, and strains were isolated from surface and deep waters at specific times during the 1999 calendar year aboard the RV Sea Surveyor. Samples were obtained from discrete depths on 27 April by using 30-liter Niskin bottles on a hydrographic cable. Conductivity, temperature, and barometric pressure were measured simultaneously with a CTD (conductivity-temperature-depth meter; model STD-plus; Applied Microsystems). Seawater (5 liters) was filtered from each depth onto 47-mm diameter, 0.45-μm-pore-size polysulfone filters (Supor-450, 0.45-μm pore size; Gelman Sciences, Inc., Ann Arbor, Mich.) under gentle vacuum (10 mm Hg). The filter was cut into four pieces, placed in a 5-ml cryovial with 3 ml of DNA lysis buffer (0.75 M sucrose, 400 mM NaCl, 20 mM EDTA, 50 mM Tris-HCl [pH 9.0]), and stored at −20°C until extraction.
Chlorophyll a measurement.
Phytoplankton were collected on a 25-mm diameter GF/F filter from 250 ml of water. Chlorophyll a was then extracted with 90% acetone and measured with a Turner Designs 10-000R fluorometer equipped with a 10-040 filter set, as previously described (40).
Flow cytometric analysis.
Duplicate 1-ml samples were placed in 2-ml cryovials and fixed with 1% (wt/vol) paraformaldehyde and 0.05% (vol/vol) glutaraldehyde for 15 min at room temperature before freezing in liquid nitrogen and subsequent storage at −80°C until analysis. Picophytoplankton were enumerated by using a FACSort flow cytometer (Becton Dickinson) equipped with a 15-mW argon ion laser exciting at 488 nm as described previously (43).
Culture isolation and growth.
Seawater samples were filtered through 25-mm diameter, 0.8-μm-pore-size cellulose acetate filters (Whatman) without refrigeration. Nutrients based on SN medium (74) were added at a 1:10 dilution, containing either 100 μM NaNO3 or 100 μM NH4Cl as the sole N source. Cycloheximide (final concentration, 0.5 mg ml−1) was then added to inhibit the growth of eukaryotic algae, and the samples were incubated at 25°C and 10 microeinsteins m−2 s−1 until Synechococcus cell pellets were observed (typically 1 to 2 weeks). Cell pellets were then transferred to full-strength SN medium (1 mM N source) with cycloheximide for maintenance. Clonal Synechococcus cultures were obtained by successively plating isolates on solid SN medium 3 times, as described by Brahamsha (9, 11). Diluted cells were mixed at 37°C with SN medium containing 0.6% (wt/vol) washed Bacto agar (Difco) and containing 1 mM of the appropriate N source. The mixture was then poured immediately into petri dishes before incubation at 25°C and 5 microeinsteins m−2 s−1 for 24 h then at 10 microeinsteins m−2 s−1 until colonies appeared (typically 1 to 4 weeks). Subsequent clonal Synechococcus sp. strains were maintained in SN (with 1 mM N source) at 25°C and 10 microeinsteins m−2 s−1 and transferred to fresh medium every 2 to 4 weeks. Cultures isolated from locations other than the Red Sea were maintained in similar growth conditions but with the medium PCRS-11 (57) instead of SN. Novel strains (Table 1) are available upon request from the Roscoff culture collection (http://www.sb-roscoff.fr/Phyto/collect.html).
TABLE 1.
Origin and characteristics of Synechococcus strains used in this study
| Clade no. | RCC no.h | Strain | Site of isolation | Depth (m) | Isolation datel | PUB/PEB ratio | Motil- ityi | N require- ment(s) | Obligately marinej | Isolating scientist(s) |
|---|---|---|---|---|---|---|---|---|---|---|
| I | 43 | Almo3 | Mediterranean Sea 36°11′N, 1°51′W | 0 | 1-5-91 | 0.4 | − | + | F. Partensky, this study | |
| I | CC9311a,d | California current | 95 | 1993 | 1.16 | NO3− | B. Palenik | |||
| I | WH 8016a,e | Woods Hole, Mass. | 6-80 | 0.40 | NO3−, NH4+ | J. Waterbury | ||||
| I | WH 8020a,e | Sargasso Sea 38°41′N, 69°19′W | 50 | 26-6-80 | 0.78 | NO3−, NH4+ | + | J. Waterbury | ||
| II | CC9605a,f | California current | 1996 | NO3− | B. Palenik | |||||
| II | 541 | RS9902a | Gulf of Aqaba 29°28′N, 34°55′E | 1 | 29-3-99 | 1.9 | − | NO3−, NH4+ | + | N. Fuller, this study |
| II | 542 | RS9903a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 11-5-99 | 2.3 | − | NO3−, NH4+ | + | N. Fuller, this study |
| II | 543 | RS9904a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 14-6-99 | 2.1 | − | NO3−, NH4+ | + | N. Fuller, this study |
| II | 546 | RS9907a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 23-8-99 | 0.5 | − | NO3−, NH4+ | + | N. Fuller, this study |
| II | 547 | RS9908a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 7-9-99 | 0.5 | − | NO3−, NH4+ | + | N. Fuller, this study |
| II | 549 | RS9910a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 7-9-99 | 0.5 | − | NO3−, NH4+ | + | N. Fuller, this study |
| II | 550 | RS9911a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 11-5-99 | 1.0 | − | NO3−, NH4+ | + | N. Fuller, this study |
| II | 551 | RS9912a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 23-8-99 | 0.5 | − | NO3−, NH4+ | + | N. Fuller, this study |
| II | 558 | RS9919a | Gulf of Aqaba 29°28′N, 34°55′E | 50 | 22-11-99 | 1.9 | − | NO3−, NH4+ | + | N. Fuller, this study |
| II | WH 8002a,e | Gulf of Mexico 19°45′N, 92°25′W | 15-4-80 | 0.48 | NO3−, NH4+ | + | J. Waterbury | |||
| II | WH 8109a,e | Sargasso Sea 39°29′N, 70°28′W | 6-81 | 0.89 | NO3−, NH4+ | J. Waterbury | ||||
| III | C129a,c | Gulf of Aqaba 29°28′N, 34°55′E | 20 | 9-93 | 0.9g | + | D. Lindell and A. Post | |||
| III | 30 | Max42 | Sargasso Sea 26°18′N, 63°26′W | 120 | 9-10-87 | 1.4 | − | + | D. Vaulot and C. Courties, this study | |
| III | 62 | Minos12 | Mediterranean Sea 34°0′N, 18°0′E | 20 | 19-6-96 | 1.8 | − | + | D. Vaulot, this study | |
| III | 311 | Minos02a | Mediterranean Sea 39°10′N, 6°10′E | 15 | 26-5-96 | 1.6 | − | + | F. Partensky, this study | |
| III | 544 | RS9905a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 18-7-99 | 1.3 | − | NO3−, NH4+ | + | N. Fuller, this study |
| III | 554 | RS9915a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 18-10-99 | 0.8 | − | NO3−, NH4+ | + | N. Fuller, this study |
| III | WH 8102a,b,e | Tropical Atlantic 22°30′N, 65°36′W | 15-3-81 | 1.90 | + | NO3−, NH4+ | + | J. Waterbury | ||
| III | WH 8103a,b,e | Sargasso Sea 28°30′N, 67°24′N | 17-3-81 | 1.3 | + | NO3−, NH4+ | + | J. Waterbury | ||
| V | UW01a | 0.5 | − | NO3− | + | K. Peeters | ||||
| V | WH 7803a,b,e | Sargasso Sea 33°45′N, 67°30′W | 1-7-78 | 0.5 | − | NO3−, NH4+ | J. Waterbury | |||
| VI | WH 7805a,b,e | Sargasso Sea 33°45′N, 67°30′W | 30-6-78 | No PUB | NO3−, NH4+ | + | J. Waterbury | |||
| VI | WH8018a,b,e | Woods Hole, Mass. | 6-80 | No PUB | − | NO3−, NH4+ | + | J. Waterbury | ||
| VII | 37 | Eum14 | Tropical Atlantic 21°2′N, 31°8′W | 105 | 1-10-91 | 2.2 | − | + | F. Partensky, this study | |
| VII | 44 | Oli31 | Equatorial Pacific 5°60′S, 150°0′W | 70 | 11-11-94 | 1.7 | − | + | D. Vaulot, this study | |
| VII | RS9920a | Gulf of Aqaba 29°28′N, 34°55′E | 150 | 22-11-99 | 1.7 | − | NO3−, NH4+ | N. Fuller, this study | ||
| VIII | 545 | RS9906a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 23-8-99 | NAk | − | NO3−, NH4+ | − | N. Fuller, this study |
| VIII | 548 | RS9909a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 7-9-99 | NA | − | NO3−, NH4+ | − | N. Fuller, this study |
| VIII | 552 | RS9913a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 7-9-99 | NA | − | NH4+ | − | N. Fuller, this study |
| VIII | 553 | RS9914a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 18-10-99 | NA | − | NH4+ | − | N. Fuller, this study |
| VIII | 556 | RS9917a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 22-11-99 | NA | − | NH4+ | − | N. Fuller, this study |
| VIII | 557 | RS9918a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 22-11-99 | NA | − | NO3−, NH4+ | − | N. Fuller, this study |
| VIII | WH 8101a,b,e | Woods Hole, Mass. | 1981 | NA | NO3−, NH4+ | − | J. Waterbury | |||
| IX | 540 | RS9901a | Gulf of Aqaba 29°28′N, 34°55′E | 1 | 29-3-99 | 0.7 | − | NO3−, NH4+ | + | N. Fuller, this study |
| IX | 555 | RS9916a | Gulf of Aqaba 29°28′N, 34°55′E | 10 | 22-11-99 | 0.7 | − | NO3−, NH4+ | + | N. Fuller, this study |
| IX | 559 | RS9921a | Gulf of Aqaba 29°28′N, 34°55′E | 150 | 22-11-99 | 0.7 | − | NO3−, NH4+ | + | N. Fuller, this study |
| X | 61 | Minos11 | Mediterranean Sea 34°0′N, 18°0′E | 20 | 19-6-96 | 0.7 | − | + | D. Vaulot, this study | |
| X | 309 | Minos01a | Mediterranean Sea 39°10′N, 6°10′E | 15 | 26-5-96 | 0.7 | − | + | F. Partensky, this study | |
| NS01a | Central North Sea | NA | − | NO3− | − | G. Nieuwland | ||||
| WH 5701a,e | Long Island Sound | 1957 | NA | NO3−, NH4+ | − | J. Waterbury |
Clonal strain.
16S rDNA sequence obtained from database.
Strain information and 16S rDNA sequence from D. Lindell (GenBank accession no. AY210409).
Reference 67.
Reference 73.
Reference 68.
Reference 59.
RCC, Roscoff culture collection.
+, motile; −, nonmotile.
+, obligately marine; −, halotolerant (growth in both ASW and BG-11 medium).
NA, not applicable.
Dates are given as year, day-month-year, or month-year.
Strain characterization.
Strain motility was determined by colony morphology (motile strains causing larger, diffuse, spherical colonies) and by microscopic observation with a Nikon Axiophot epifluorescence microscope. Ratios of PUB to PEB were calculated from the excitation spectra from cultures diluted 1:30, exciting at 400 to 570 nm, measuring emission at 585 nm, with 2.5-nm excitation and emission slits, by using a Perkin Elmer scanning spectrofluorimeter model LS-5. An excitation wavelength range up to 570 nm was used since it was recently shown that some Synechococcus strains possess a form of PE in which the PEB has an excitation maximum around 566 nm (71). PUB/PEB ratios were averaged from triplicate profiles. For the purpose of calculating PUB/PEB ratios, all strains were grown at 10 microeinsteins m−2 s−1 in ASW medium (81). Strains were also tested for their ability to grow on nitrate and ammonium, in ASW-based medium, and in the freshwater medium BG-11 (56) (Table 1).
Environmental DNA isolation.
DNA was extracted from the filters in lysis buffer largely following the method described by Gordon and Giovannoni (27), except that 5-ml cryovials were used for the lysis, and the filter was transferred with the lysate to four Eppendorf tubes for phenol extraction. Phenol dissolves the Supor filters, increasing the efficiency of DNA recovery (results not shown). Nucleic acids were extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1) before being precipitated with 1 volume of isopropanol and 0.4 volume of 7.5 M ammonium acetate at room temperature. Nucleic acids were recovered by centrifugation, washed once with 70% (vol/vol) ethanol, and resuspended in TE2 (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA) prior to storage at −80°C.
Chromosomal DNA isolation.
DNA was prepared from Synechococcus strains by resuspending pelleted cells in lysis buffer (0.25 M Tris-HCl [pH 8.0], 25% [wt/vol] sucrose, 10 mg ml−1 lysozyme [Sigma]) and incubated at 37°C for 1 h. Sarkosyl (1% [vol/vol]) and proteinase K (final concentration, 200 μg ml−1) were then added, and the cell lysate was incubated at 65°C for 1 h. Proteins were extracted once with phenol-chloroform (25:24) and once with chloroform-isoamyl alcohol (24:1) before precipitating nucleic acids with 1 volume of isopropanol and 0.4 volume of 7.5 M ammonium acetate at room temperature. Nucleic acids were recovered by centrifugation, washed once with 70% (vol/vol) ethanol, and resuspended in TE (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA) prior to storage at −20°C.
PCR amplification.
16S rDNA sequences were amplified from environmental DNA and cultured strains by using the primers shown in Table 2. The 27F-OXY1313R and OXY107F-1522R primer pairs were used to amplify the near full-length 16S rDNA from most of the cultured strains. 16S rDNA from strains isolated by F. Partensky and D. Vaulot, plus strains UW01 and CC9311, were amplified with 27F and 1522R and cloned into the TA vector pCR2.1-TOPO (Invitrogen). Subsequent clones were putatively identified as Synechococcus sp., rather than as a heterotrophic bacterium, by PCR screening with the OXY359F and OXY1313R primer pair prior to confirmation by sequencing. Control DNAs for use in dot blot hybridization experiments were prepared by amplification of 16S rDNA sequences from cultured Prochlorococcus and Synechococcus strains by using the OXY107F and OXY1313R primers. The same primer pair was used to amplify the Red Sea environmental DNA samples.
TABLE 2.
16S rDNA and ntcA PCR primers and oligonucleotide probes used in this study
| Primer or probe | Td(°C) | Sequence (5′-3′) | Target organisms | Reference |
|---|---|---|---|---|
| PCR primers | ||||
| OXY107F | GGA CGG GTG AGT AAC GCG TG | Oxygenic phototrophs | 78 | |
| OXY359F | GGG GAA TYT TCC GCA ATG GG | Oxygenic phototrophs | 47 | |
| OXY1313R | CTT CAY GYA GGC GAG TTG CAG C | Oxygenic phototrophs | 78 | |
| SYN1017R | TCC CGA AGG CAC CCT CTC G | Synechococcus clades III, IV, V, and VI | This study | |
| 27F | AGA GTT TGA TCM TGG CTC AG | Eubacteria | 21 | |
| 1522R | AAG GAG GTG ATC CAN CCR CA | Eubacteria | 21 | |
| G15Fa | GAR TCN GGB GAA GAG ATC ACY GT | All PE-containing MC-A Synechococcus & Prochlorococcus strains tested | 41 | |
| G16Fa | GAR TCW GGW GAA GAR ATW ACW GT | All PE-containing MC-A Synechococcus & Prochlorococcus strains tested | 41 | |
| 4ARa | ATG GCY TCG GCK ATG GCY TGR T | All PE-containing MC-A Synechococcus & Prochlorococcus strains tested | 41 | |
| Oligonucleotide probes | ||||
| EUB338 | 46 | GCT GCC TCC CGT AGG AGT | Eubacteria | 2 |
| SYN1006 | 44 | CTC TCA AGT TTC CAA GAG | Synechococcus clade I | This study |
| SYN1006RS | 42 | CTC TCC TGT TTC CAA GAG | Synechococcus clade II | This study |
| SYN262 | 53 | GAT GCC TTG GTA GGC CTT | Synechococcus clade III | This study |
| SYN635 | 50 | AAG CCC CTC AGT TTC CAC | Synechococcus clade IV | This study |
| SYN1280 | 50 | GAG CCA CGG TTT ATG AGA | Synechococcus clades V, VI, and VII | This study |
| SYN620 | 51 | CAC TGC CAC GAT GGA GTT | Synechococcus clade VIII | This study |
| SYN1000 | 45 | GGT TTC CCA GAA ATT CGC | Synechococcus clade IX | This study |
| SYN1007 | 48 | ACC CTC CGG TTT CCC AGA | Synechococcus clade X | This study |
| SYN1258 | 47 | TTG TCC TCG CGA ACT TGC | Synechococcus clades I-VII and X, Prochlorococcus sp. strain MIT9303 | This study |
ntcA primer.
PCR amplification of 16S rDNA from strains was carried out in a total reaction volume of 100 μl containing 2 μl of cells (preheated at 95°C for 5 min for lysis), 200 μM concentrations of deoxynucleoside triphosphates, 1.5 mM MgCl2, 0.2 μM concentrations of primers, and 2.5 U of Taq polymerase in 1× enzyme buffer (GIBCO BRL, Life Technologies Ltd., Paisley, Scotland). Amplification of environmental DNA also included bovine serum albumin (1 mg ml−1; Sigma). When OXY107F, OXY359F, SYN1017R, or OXY1313R was used, the amplification conditions comprised 95°C for 5 min and 80°C for 1 min, at which time Taq polymerase was added, followed by 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 6 min. Amplification with the primer pair 27F and 1522R was identical, except that the annealing temperature was 50°C. Environmental DNA was amplified from each depth in duplicate, and the reaction mixtures were pooled prior to further use.
PCR amplicons of ntcA for restriction fragment length polymorphism (RFLP) analysis were similarly amplified from preheated cells. The primers G15-16F and 4AR (41) were used to amplify approximately 370-bp products. Primer G15-16F is a mixture of G15F and G16F at a ratio of 1:1.3. Reaction mixtures of 50 μl contained 1 μl of cells, 200 μM concentrations of deoxynucleoside triphosphates, 1 mM MgCl2, 0.5 μM concentrations of primers, and 2.5 U of Taq polymerase in 1× enzyme buffer. Amplification conditions comprised 95°C for 5 min and 80°C for 1 min, at which time Taq polymerase was added, followed by 35 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 6 min.
RFLP analysis.
ntcA PCR amplicons were subject to restriction digestion in 20-μl reaction mixtures with 2 U (each) of BstUI and HaeIII in 1× NEBuffer 2 (New England BioLabs Ltd., Hitchin, England), with incubation at 37°C for 2 h and then at 60°C for 2 h. Fragments were then resolved by gel electrophoresis on a 10% (vol/vol) polyacrylamide gel at 60 mA for 2 h.
Cloning of 16S rDNA operons from Synechococcus sp. strain WH 7803.
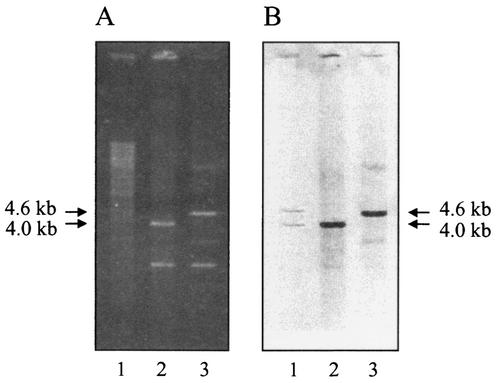
Chromosomal DNA extracted from Synechococcus sp. strain WH 7803 was digested to completion by overnight digestion with various restriction enzymes before gel electrophoresis and Southern blotting with standard protocols (60). Prehybridization and hybridization were performed in 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 5× Denhardt's solution, and 0.5% (wt/vol) sodium dodecyl sulfate (SDS) (60). A 16S rDNA PCR fragment from Synechococcus sp. strain PCC7942 was used as a probe following random priming with the Prime-a-gene labeling system (Promega). The blot was washed at room temperature in 2× SSPE and 0.1% (wt/vol) SDS for 1 h and then at 61°C in 2× SSPE and 0.1% (wt/vol) SDS for 25 min. Hybridizing 16S rDNA fragments (4.0- and 4.6-kb HindIII fragments) were then cloned from a 3.5- to 5-kb HindIII partial library of Synechococcus sp. strain WH 7803 genomic DNA cloned into pUC19. Escherichia coli strain TG1 transformants were screened by PCR with primers OXY359F and SYN1017R followed by plasmid DNA extraction and restriction digestion with HindIII to identify each 16S rDNA copy. Confirmation that each 16S rDNA copy had been cloned was also determined by Southern hybridization (Fig. 1).
FIG. 1.
Agarose gel (A) and corresponding Southern blot (B) showing the two 16S rDNA copies from Synechococcus sp. strain WH 7803 cloned into pUC19, probed with digoxigenin-labeled 16S rRNA PCR product amplified from the same strain. Lanes: 1, strain WH 7803 genomic DNA; 2 and 3, pUC19 containing a 16S rDNA copy on a 4.0- and 4.6-kb HindIII fragment, respectively.
DNA sequencing.
Double-stranded plasmid DNAs, containing the two 16S rDNA copies from Synechococcus strain WH 7803 or a single 16S rDNA copy from the other strains, were sequenced bidirectionally by using an ABI 373A automated sequencer (Applied Biosystems, Foster City, Calif.). 16S rDNA from the Red Sea strains was sequenced bidirectionally from PCR products by using a commercial sequencing company (MWG-Biotech).
Phylogenetic analysis.
Sequences were checked for the presence of chimeric artifacts by using the Ribosomal Database Project program CHECK_CHIMERA (42) and by the construction of phylogenetic trees by using each half of the sequence separately. Sequence alignment and phylogenetic analysis were performed by using the ARB program (http://www.mikro.biologie.tu-muenchen.de/pub/ARB/).
Oligonucleotide probes.
16S rDNA oligonucleotide probes were designed and tested for specificity by using the probe design and probe match tools from the ARB program. Probes (30 ng) were end labeled with γ-32P by using T4 polynucleotide kinase in 1× forward kinase buffer (GIBCO BRL) at 37°C for 60 min. Unincorporated nucleotides were removed by purification through a Sephadex G-25 column (60).
Dot blot hybridization of Red Sea environmental DNA vertical profile.
16S rDNA amplicons were purified by using the QIAquick PCR purification kit (Qiagen Ltd.) and quantified spectrophotometrically before denaturation and blotting onto nylon membranes (Zetaprobe; Bio-Rad Laboratories Ltd., Hemel Hempstead, Herts, United Kingdom). For the control DNAs (see “PCR amplification” above), a dilution series of 5 to 50 ng of PCR products was blotted, and for the natural samples, 30 ng of PCR products was blotted in triplicate. Blots were prehybridized in 10 ml of Z-hyb buffer (1 mM EDTA, 0.5 M Na2HPO4 [pH 7.2], 7% [wt/vol] SDS) for 30 min at 30°C and hybridized in 10 ml of fresh Z-hyb buffer containing labeled oligonucleotide overnight at 30°C as described previously (27). Membranes were washed in 0.2× SSPE and 0.1% (wt/vol) SDS for three 15-min washes at 30°C followed by a 10-min wash at the stringency temperature. The stringency wash temperatures for each probe were assessed empirically by producing wash curves at 3 to 5°C increments from 30 to 65°C as described previously (87). However, the wash temperatures which were finally used for each probe were usually slightly higher and determined by specificity blots with a range of 16S rDNA amplicons from other Synechococcus and Prochlorococcus strains. The temperature of the stringency wash was increased in subsequent hybridization experiments until the appropriate specificity for the probe was reached. Final wash (or dissociation) temperatures (Td) are shown in Table 2. For EUB338, 46°C was the wash temperature based on experiments described previously (27). Hybridization was quantified by using a phosphorimager and Image Quant software (Molecular Dynamics). The relative hybridization of the Synechococcus genotype-specific probes to the total oxygenic phototroph 16S rDNA sequences was calculated according to the equation:
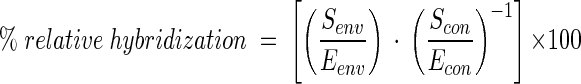 |
where Senv and Eenv represent hybridization to environmental DNA of the specific and eubacterial probes, respectively, and Scon and Econ are the slopes of the specific and eubacterial probe-binding curves, respectively, calculated by hybridizing each probe to a dilution series of homogenous control DNAs. The relative hybridization of a given specific probe compared with that of the eubacterial probe to the control DNAs was averaged where more than one control DNA was used.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database under the following accession numbers: AY172800 to AY172837.
RESULTS
Two copies of 16S rDNA in marine Synechococcus.
Southern analysis of the axenic marine Synechococcus sp. strain WH 7803 revealed the presence of two copies of the 16S rRNA gene. These two copies were cloned from a HindIII partial library of genomic DNA (Fig. 1) on specific 4.0- and 4.6-kb fragments. DNA sequencing revealed the two 16S rRNA genes to be identical over their entire length. Similarly, using sequence information from the recently completed Synechococcus sp. strain WH 8102 genome (http://bahama.jgi-psf.org/prod/bin/microbes/syn/home.syn.cgi), the two 16S rDNA copies from this strain show identity over their entire length apart from two 1-bp deletions in conserved regions of one copy. This initial analysis thus showed no significant microheterogeneity within the 16S rRNA gene of at least two Synechococcus strains and suggested that this molecule would be an appropriate tool for the construction of phylogenetic relationships and clade-specific oligonucleotides for this genus.
Strain isolation and 16S rDNA sequences.
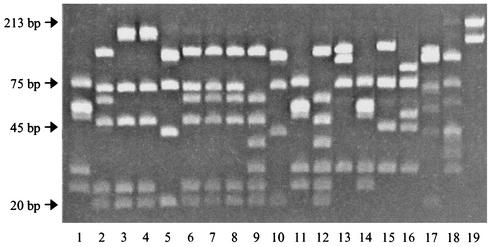
Thirty-one Synechococcus strains were isolated or obtained from several oceanic regions (Table 1), including 21 clones isolated during a monthly sampling program in 1999 at station A in the Gulf of Aqaba. These Red Sea strains were derived from 257 initial isolates, obtained by using pour plate techniques from either surface (1 or 10 m) or deep (50 or 150 m) waters. An RFLP analysis was developed for rapid genetic screening of these initial isolates by using the ntcA gene, which encodes a global N regulator, amplified with general marine picocyanobacterial primers that had been developed elsewhere (41). ntcA RFLP profiles from these Red Sea Synechococcus strains and from those strains isolated from other oceanic environments, representing different phylogenetic clades (see below), are shown in Fig. 2. This simple and rapid RFLP method for screening marine Synechococcus strains affords high resolution; some strains with different ntcA RFLP patterns have identical 16S rDNA sequences, e.g., strains RS9905 and WH 8103 or strains RS9902, RS9903, and RS9911. For the Red Sea isolates, HaeIII- and BstUI-digested ntcA products identified six different RFLP types (Fig. 2). Using this ntcA RFLP information, as well as pigment composition (see below) and date of isolation, 21 clonal isolates were identified as being potentially distinct and thus suitable for 16S rDNA sequence analysis. These strains were named RS9901 to RS9921.
FIG. 2.
RFLP analysis of Synechococcus sp. isolates from the Red Sea. PCR amplicons of ntcA were digested with HaeIII and BstUI and separated by electrophoresis on a 10% polyacrylamide gel, revealing six different RFLP patterns. Patterns from other Synechococcus strains are shown for comparison. Lanes 1 to 14: Synechococcus sp. strains RS9901, RS9902, RS9903, RS9904, RS9905, RS9907, RS9908, RS9910, RS9911, RS9915, RS9916, RS9919, RS9920, and RS9921, respectively. Lanes 15 to 19: Synechococcus sp. strains WH 7803, WH 8018, WH 8103, Minos01, and CC9311, respectively.
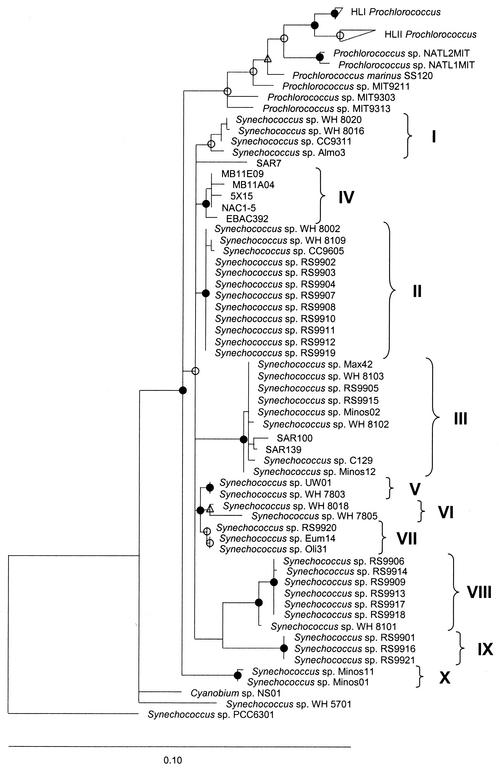
Near complete 16S rRNA gene sequences were obtained for all marine Synechococcus isolates (Table 1). Phylogenetic relationships between these strains and other marine Synechococcus and Prochlorococcus sequences already in the database are shown in Fig. 3. The Synechococcus strains fall into ten distinct clades, which are well supported by high bootstrap values. Clade designation is based on data from Ferris and Palenik (20) and Rocap et al. (59). Clades I to VI have been previously described (59) while clade VIII has been assigned to what was previously just strain WH 8101 (a member of MC-B) (73) but now contains 6 new Red Sea isolates. Clades VII, IX, and X are novel. Interestingly, sequences from members of clade X, in phylogenetic analyses, lie outside of the main MC-A Synechococcus (subcluster 5.1) group, branching at the same point as Prochlorococcus.
FIG. 3.
Neighbor-joining phylogenetic tree of 16S rDNA sequences from marine Synechococcus and Prochlorococcus strains, with Jukes-Cantor correction. Partial sequences (<1,390 nucleotides) were added to the tree by using a maximum-parsimony option within ARB. The freshwater strain PCC6301 was used as the root. The tree was constructed with a filter of 1,345 nucleotides that excluded alignment positions with sequence ambiguity or missing data. The confidence of branch points was determined by three separate analyses (maximum likelihood, neighbor joining, and maximum parsimony), with multifurcations indicating branch points that were collapsed until supported in all three analyses by using a strict consensus rule. Bootstrap values (100 replicates) are shown from neighbor-joining analysis with Jukes-Cantor correction. Closed symbols represent values of >95%, open symbols represent values of 70 to 95%, and values of <70% are not shown. Circles represent values where full-length sequences (1,345 nucleotides) were available for the analysis; triangles represent values where shorter sequences (681 nucleotides) were used.
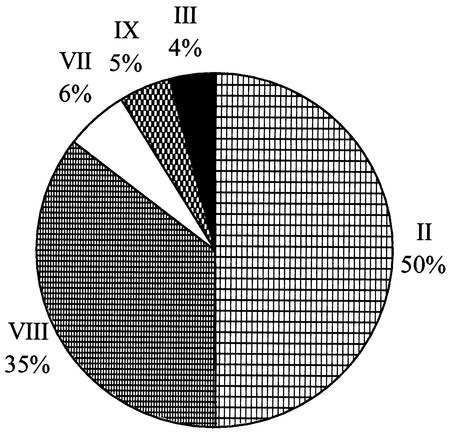
Phylogenetic relationships between representatives of these marine Synechococcus isolates and other Synechococcus lineages are shown in Fig. 4. Strains PCC7001 and PCC7009, which were previously recognized as Synechococcus sp. have recently been reclassified as members of the genus Cyanobium (29). Hence, we have renamed all members of this clade Cyanobium sp. (Fig. 4). The Synechococcus MC-A (subcluster 5.1) strains, together with the MC-B (subcluster 5.2) clade VIII isolates (see Fig. 3) and Prochlorococcus strains, form a monophyletic group with good bootstrap support, distinct from other MC-B (subcluster 5.2) (e.g., WH 5701) and Cyanobium strains. The MC-A (subcluster 5.1) and Prochlorococcus group, together with the WH 5701 MC-B (subcluster 5.2) and the Cyanobium clusters, form a distinct lineage, supported by a high bootstrap value, clearly separate from freshwater Synechococcus (clusters 1 and 2) (e.g., PCC7942 and PCC6716) and MC-C (clusters 3 and 4) (e.g., PCC7002 and PCC7335) groups. Strain NS01 falls within the Cyanobium cluster, but all the other PE-lacking halotolerant strains form a distinct clade, VIII, which lies within what is recognized as MC-A (Synechococcus subcluster 5.1). Using ntcA RFLP profile information, the 257 cultured Synechococcus isolates from the Red Sea contain representatives from 5 of the 10 16S rDNA clades described above. Figure 5 details the proportion of these 257 isolates represented by each of these five 16S rDNA clades. The large majority (85%) of isolates fell into just two clades, II and VIII, with clade VIII comprising the green, PE-lacking, halotolerant strains.
FIG. 4.
Neighbor-joining tree of 16S rDNA sequences from cyanobacterial strains, with Jukes-Cantor correction. The positions of Synechococcus MC-A, MC-B, and MC-C and their current phylogenetic assignments are indicated by brackets. *, strain WH 8101 was previously designated a member of MC-B (subcluster 5.2) but is designated here a member of Synechococcus subcluster 5.1. For Synechococcus sp. strain G2.1, only a 951-bp sequence was available and this was added to the tree by using a maximum-parsimony option within ARB. Escherichia coli was used as the root. The tree was constructed with a filter of 1,263 nucleotides that excluded alignment positions with sequence ambiguity or missing data. The confidence of branch points was determined by three separate analyses (maximum likelihood, neighbor joining, and maximum parsimony), with multifurcations indicating branch points that were collapsed until supported in all three analyses by using a majority consensus rule. Bootstrap values (100 replicates) are shown from neighbor-joining analysis with Jukes-Cantor correction. Closed symbols represent values of >95%, open symbols represent values of 70 to 95%, and values of <70% are not shown. Circles represent values where full-length sequences (1,263 nucleotides) were available for the analysis; triangles represent values where shorter sequences (877 nucleotides) were used.
FIG. 5.
Pie chart of the proportion of cultured isolates from the Red Sea belonging to particular Synechococcus clades.
The extent of microheterogeneity (i.e., the fractional identity of the 16S rDNA sequences) among the marine Synechococcus sp. strains analyzed (data not shown), reveals that clade VI contains the greatest within-clade diversity. This clade has a minimum identity between sequences of 99.1%. Among all the MC-A sequences analyzed here, WH 7805 and RS9901 from clades VI and IX, respectively, show the lowest degree of identity, 96.2%. Some of the other clades are much more closely related, e.g., sequences of clades II and IV are ∼99.4% identical while those of clades V, VI, and VII are around 99.7% identical to each other.
Strain characterization.
The 31 novel Synechococcus sp. isolates were subjected to preliminary physiological characterization, assessing cell motility, PUB/PEB ratio, N requirements, and salt requirements, as shown in Table 1. Interestingly, no motile strains were obtained from the Red Sea in this study, even though from phylogenetic inference, two of them, RS9905 and RS9915, lie within the motile clade (clade III) (Fig. 3) described by Toledo et al. (69). Three other isolates, Max42, Minos2, and Minos12, from the Sargasso and Mediterranean Seas also lie within clade III yet appear to be nonmotile.
The new isolates exhibited a wide range in PUB/PEB ratio, from 0.4 to 2.3, though there appeared to be little correlation with depth of isolation, with surface isolates possessing either high or low ratios. From the Red Sea, low-PUB, high-PUB, and PE-lacking isolates were obtained. All of the Red Sea isolates from 1999 grew on both nitrate and ammonium as the sole N source, except strains RS9913, RS9914, and RS9917, which appeared to be unable to grow on nitrate (Table 1). These latter three strains fall within clade VIII (MC-B [73] or Synechococcus subcluster 5.2 [29]), along with the nitrate-utilizing strains RS9906, RS9909, and RS9918. Despite the different phenotype, strains RS9913 and RS9914 and strains RS9906 and RS9909 show identical 16S rDNA sequences over a 1,370-bp PCR product, while all six strains are identical over the length of the shortest sequence, that of RS9917, which is 870 bp (Fig. 3). Moreover, strains RS9913 and RS9917 were isolated from the same water samples as strains RS9909 and RS9918, respectively.
All of the new marine Synechococcus isolates, except the PE-lacking strains, appear to be obligately marine, since none would grow in the freshwater medium BG-11. The PE-lacking strains, i.e., strain NS01 and all those new isolates clustering within clade VIII, all grew in this medium (for those strains from clade VIII which could not utilize nitrate, growth was observed in BG-11 medium in which sodium nitrate was replaced with ammonium chloride).
Probe development.
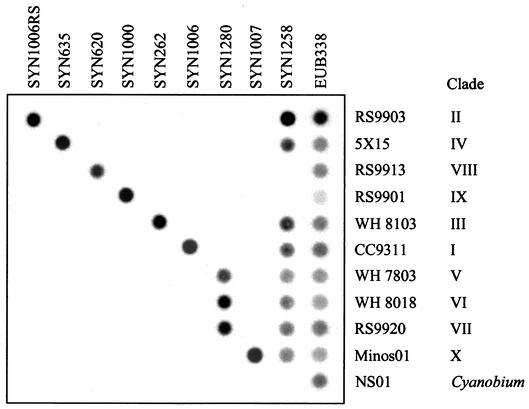
By comparing the alignments of all available Synechococcus 16S rDNA sequences, variable regions were identified that allowed oligonucleotide probes to be designed to specifically target the different clades (Table 2). Probe SYN1258 has more general specificity, recognizing all marine MC-A (subcluster 5.1) Synechococcus strains known so far except clades VIII and IX and no Prochlorococcus strains except MIT9303. Probe SYN1280 recognizes clades V, VI, and VII, since specific probes capable of recognizing these clades separately could not be established. The specificities of all probes were checked by using the ARB sequence database tools and by performing BLAST searches (1). Specificity was then confirmed experimentally by hybridization of the probes to dot blots containing 16S rDNA amplicons from different nontarget Synechococcus and Prochlorococcus strains. None of the probes hybridized to nontarget strains (Fig. 6). Only environmental 16S rDNA sequences (clade IV) (5, 65, 80) are targeted by the SYN635 probe for which, at present, a clonal culture remains to be isolated.
FIG. 6.
Representative dot blots showing the specificity of hybridization of each Synechococcus clade-specific oligonucleotide (SYNn) to arrays of control 16S rDNA amplicons (Synechococcus sp. strain RS9903, Synechococcus 16S rDNA clone 5X15, Synechococcus sp. strains RS9913, RS9901, WH 8103, CC9311, WH 7803, WH 8018, RS9920, and Minos01 and Cyanobium sp. strain NS01). EUB338, eubacterial probe.
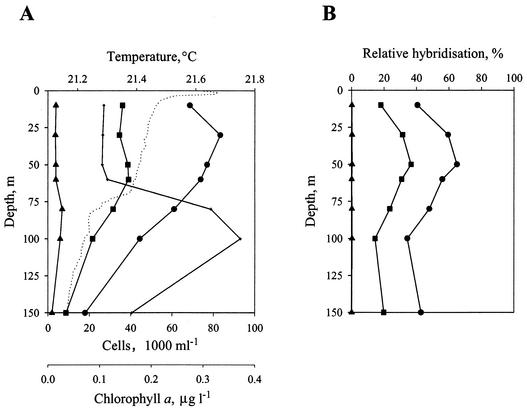
Hydrological characteristics of station A, Gulf of Aqaba, Red Sea.
Profiles of temperature (Fig. 7A) and salinity (data not shown) revealed a stratified water column at station A in the northern tip of the Gulf of Aqaba on 27 April 1999. The temperature profile showed shallow surface warming down to about 6 m then a surface mixed layer (SML) down to about 70 m, and a slight thermocline from 70 m to about 160 m. Salinity data confirmed the shallow surface layer, the SML, and the water below the SML as distinct water masses. Chlorophyll a measurements detected a subsurface maximum at around 80 to 100 m, just below the SML (Fig. 7A). Based on the orange fluorescence of their PE, flow cytometry data revealed high Synechococcus numbers throughout the SML (3.5 × 104 to 3.9 × 104 cells ml−1), decreasing down the thermocline to 0.9 × 104 cells ml−1 at 150 m (Fig. 7A). However, Prochlorococcus was more abundant in the water column, with numbers ranging between 1.8 × 104 and 8.3 × 104 cells ml−1.
FIG. 7.
Ancillary CTD, flow cytometry, and dot blot hybridization data from a depth profile at station A, Gulf of Aqaba, Red Sea on 27 April 1999. (A) Temperature (· · · ·), chlorophyll a concentration (in micrograms liter−1) (+), and Synechococcus (▪), Prochlorococcus (•), and photosynthetic picoeukaryote (▴) cell numbers. (B) Relative hybridization abundance of Synechococcus clades detected by 16S rDNA oligonucleotide probes SYN1006RS (clade II) (▪), SYN1258 (clades I to VII and X) (•), and SYN262 (clade III) (▴), quantified from dot blots.
Dot blot hybridization of the Red Sea environmental DNA depth profile.
The distribution of Synechococcus genotypes in the water column at station A on 27 April 1999 was examined by dot blot hybridization. The specific oligonucleotide probes (Table 2) were hybridized to PCR amplicons of oxygenic phototroph (cyanobacteria and chloroplasts) 16S rDNA from each depth. Probes were always hybridized simultaneously to blots containing control DNAs for subsequent quantitation and to check the specificity of the probes. No cross-hybridization of the probes to nonhomologous sequences was observed (Fig. 6). All of the marine Synechococcus probes showed negligible hybridization to DNA amplified from the depth profile (data not shown) except for the probe specific for clade II (SYN1006RS) and the general Synechococcus probe, SYN1258 (Fig. 7B). This suggested that these other genotypes were absent or were a minor component of the Synechococcus population at this time and that the Synechococcus population largely consisted of genotypes of a single clade, clade II, throughout the depth profile. Note that the relative hybridization values for the general Synechococcus probe (SYN1258) are less than 100% (Fig. 7B) since Prochlorococcus cell density exceeds Synechococcus in this water column (Fig. 7A) and both cell types are amplified by the PCR primers used to generate target DNA for hybridization studies. Although probe SYN1258 does not detect members of clades VIII and IX (Table 2), members of these two clades were shown by the specific probes SYN620 and SYN1000, respectively, to be virtually absent.
While the hybridization data clearly indicate a predominance of genotypes of a single phylogenetic clade at station A, the observed difference in relative hybridization values between the clade II-specific probe and the general MC-A Synechococcus probe, as well as no hybridization with probe 181LL (78) (data not shown) recognizing Prochlorococcus MIT9303, suggests one or more other MC-A clades await identification in this water column. Such clades may be represented by the rpoC1 clades (20) for which 16S rDNA sequence information is as yet unavailable or may be other novel clades for which cultured representatives await isolation.
Other marine Synechococcus lineages for which specific probes have yet to be designed include MC-B and MC-C strains as well as the recently described isolate G2.1 (82). However, given the high relative hybridization signal for Synechococcus clade II and the SYN1258 probe, as well as the high Prochlorococcus cell density in the water column, these would likely be a minor component of this picocyanobacterial community.
DISCUSSION
Phylogenetic analysis of 16S rDNA sequences from the several new marine Synechococcus isolates reported here clearly support and extend previous sequence and RFLP data of a high genetic diversity within this group of organisms (19, 35, 50, 67, 69, 70, 86). The large number (10) of MC-A (subcluster 5.1) clades found in this study also highlights that the resolution capacity of this molecule to distinguish genetic diversity within the genus, i.e., in being able to resolve specific clades, is comparable to that obtained with the 16S-23S rDNA ITS (59) and rpoC1 (67, 69) gene loci. In fact, thus far, there is very good congruence between the clades identified by 16S rDNA sequencing and those identified by these other gene markers. The 16S rDNA clade designations reported here follow the Roman numeral ITS nomenclature of Rocap et al. (59) for the six clades which are common in the two studies. With reference to the eight marine Synechococcus clades identified by rpoC1 sequences (20), clades 1 to 3 of that study correspond to clades I to III resolved by sequencing the ITS and 16S rDNA. Clades 4 and 8 contain rpoC1 sequences from environmental clone libraries and as yet cannot be matched with ITS and 16S rDNA data. Similarly, clades 5 and 6 of Ferris and Palenik (20), although containing sequences from cultured isolates, await further ITS and 16S rDNA sequence information before a specific comparison can be made. Clade 7 recognized from rpoC1 sequencing, and consisting of strains WH 7803 and WH 7805, was divided into clades V and VI based on ITS sequencing and since physiologically members of clade VI lack PUB (59). The three novel clades identified from 16S rDNA sequences reported here include one clade, clade VII, which is phylogenetically closely related to clades V and VI. rpoC1 sequencing of selected isolates of clade VII, and indeed clades IX and X, confirm the novelty of these designations (M. Mühling, N. J. Fuller, D. J. Scanlan, and N. H. Mann, unpublished data). Clade IV is another phylogenetically well-defined clade, with strong bootstrap support with both 16S rDNA and ITS sequence data, but as yet contains no cultured counterpart, it being represented only by sequence information from environmental BAC or clone libraries (5, 65, 80).
Recently, the ITS sequence from a marine Synechococcus strain, MIT S9220, was determined (46). Phylogenetic analysis supported its designation as a novel clade and different from the six clades already designated by Rocap et al. (59). This ITS sequence has around 280 bp in common with the 16S rDNA sequences in our study. Unfortunately, over this region the MIT S9220 16S rDNA sequence is identical to sequences from both clade VI and VII. However, since the complete ITS sequence shows MIT S9220 to be only 84% identical to other members of clade VI (46), it is possible that MIT S9220 represents a member of clade VII designated in this report. Further sequencing will be needed to clarify this. Physiologically, MIT S9220 is certainly distinct from the clade VII isolate RS9920, since the former cannot utilize nitrate as a sole N source for growth, nor can it be rescued from N limitation by nitrate (46), while nitrate does support the growth of RS9920.
The phylogenetic position of 16S rDNA sequences of isolates from clade X is interesting since they branch outside the main marine Synechococcus subcluster 5.1 but at the same branch node as Prochlorococcus. Certainly, the dominant accessory light-harvesting pigment in members of this clade is low-PUB PE. However, it is intriguing whether these strains might possess chlorophyll b or Prochlorococcus-like photosynthetic antenna (pcb) genes.
Synechococcus sp. strain WH 8101, a member of clade VIII, is recognized as a member of MC-B, lacking PE and with a high DNA base composition (molecular percentage of G+C content) (73). However, the identification of multiple new clade VIII sequences, and their phylogenetic analysis in the work presented here, as well as preliminary indications in previous studies (59, 70, 78), clearly show WH 8101 to fall within the MC-A (subcluster 5.1) group. The reference strain for MC-B (subcluster 5.2) (29), is WH 5701. Since strain WH 5701 is phylogenetically distinct from members of clade VIII, we suggest the latter clade represents a halotolerant, PE-lacking clade within the MC-A (subcluster 5.1) group.
Phenotypically, motility is one of the few marine Synechococcus characters that appears to show monophyletic clustering (69). However, strains isolated in this study whose 16S rDNA sequences fall within clade III, the motile cluster, appear to be nonmotile (Table 1). Since nonmotile mutants have been observed to occur relatively commonly in motile strains (B. Brahamsha, personal communication), it is possible that these are motile strains that have lost the ability to swim. Certainly, motile strains belonging to clade III have previously been isolated from the same location, station A, in the Red Sea (strains C8015 and C129) (59, 69). Moreover, the oligotrophic nature of this body of water is consistent with the water type from which other motile strains have been isolated, where it is thought that motility plays an important role in translocating these organisms towards microscale nutrient patches (79). However, the possibility now exists that clade III comprises both motile and nonmotile strains. An absence of any motile strains isolated in this study does, however, confirm that the motility phenotype appears not to be found outside this clade.
With regard to the phenotypic character of the Red Sea isolates, it is perhaps surprising that any halotolerant PE-lacking strains were isolated, given the high salinity in the north of the Gulf (ca. 41 ppt), higher even than most other open-ocean environments (ca. 37 ppt). Certainly these halotolerant strains comprised a relatively high percentage (35%) of the total isolates obtained from the station A site. However, from the dot blot hybridization data with a clade-specific 16S rDNA oligonucleotide, members of this clade seem to represent a very minor component of the Synechococcus community at this site. Such data highlight the caution required when extrapolating in situ abundance data from the frequency at which a strain appears in culture.
Three of the isolates from clade VIII are unusual in that they are incapable of growth on nitrate as the sole N source. Although there is a recent report of one other marine Synechococcus strain (MIT S9220), isolated from surface waters of the equatorial Pacific, that cannot utilize nitrate for growth (46), this trait appears to be generally uncommon in marine Synechococcus isolates. This is in contrast to all current isolates of the Prochlorococcus genus, none of which can utilize nitrate for growth (46), and several freshwater thermophilic cyanobacteria (for an example, see reference 44). The high N demand of the N-rich phycobilisome has been proposed as an important driving force for maintaining the nitrate utilization capacity of marine Synechococcus isolates (46) in an environment where N availability is potentially limiting. Since Synechococcus strain MIT S9220 is a low-PUB isolate and appears to be phylogenetically distinct from the three PE-lacking clade VIII isolates (see above), this lack of growth on nitrate is correlated with neither phylogenetic position nor pigment composition. Furthermore, the fact that four clade VIII isolates are capable of growth on nitrate although having identical 16S rDNA sequences to those of the three clade VIII isolates lacking this capacity illustrates the potential phenotypic diversity among members of the same clade.
Dot blot hybridization data using the Synechococcus clade-specific probes developed in this study showed the prevalence of a single clade, clade II (probe SYN1006RS), throughout the euphotic zone down to 150 m at station A, with all other MC-A clades virtually undetectable (Fig. 7B). The abundance of clade II genotypes in this oligotrophic water column concurs with recent immunological data on the distribution of CC9605 (a member of clade II) serotypes in waters of the California current and which also appeared more numerous at oligotrophic offshore sites (68). Interestingly, clades represented by well-studied laboratory strains, such as WH 7803 (clade V) and WH 8103 (clade III), were virtually absent from this Red Sea water column. This has potentially important implications, since ecologically it is of obvious importance to focus laboratory work on those strains known to be abundant in situ and which likely make the greatest contribution to oceanic primary production. Further work is required though, to assess whether these distribution and abundance patterns are true of other oceanic regions and also to corroborate these dot blot hybridization data with PCR-independent approaches, such as fluorescent in situ hybridization, which lack any associated problems of PCR bias.
The absence of the clade V genotypes is perhaps not so surprising given the fact that high PUB/PEB ratio cells tend to dominate open-ocean systems (36, 49, 85) and indeed supports earlier immunological data that suggested that the WH 7803 serogroup was not typical of Synechococcus populations in the open ocean (14).
The availability of both genetic (this study) and immunological probes (12-14, 68) now allows a comparative approach to be taken for identifying clade-specific Synechococcus niches, though care needs to be taken when comparing these data, since the two approaches may have different resolutions. Thus, to date it has been difficult to assess whether specific serotypes are monophyletic or are polyphyletic but share common antigens. The phylogenetic data reported here clarify this problem, being able to partly resolve these serotypes in a genetic context while also highlighting the need to extend cross-reactivity studies of antisera to isolates from all known genetically defined Synechococcus clades.
The underlying rationale for the development of 16S rDNA oligonucleotides capable of recognizing phylogenetically defined Synechococcus clades is to establish the specific niches that these distinct genetic packets occupy. Given that the oceanic environment comprises strong vertical and horizontal gradients of light, temperature, and nutrients, and with the vertical light gradient having both a light intensity and spectral quality component, it is likely that there has been strong evolutionary pressure to adapt to a specific water column environment. At the phylogenetic level this manifests itself in 16S rDNA microheterogeneity and likely correlates with distinct Synechococcus populations that change in time and space as conditions fluctuate and, hence, as different niches appear and disappear. This microheterogeneity thus likely serves to maintain the function of the specific microbial guild over an array of different microhabitats, as has been hypothesized elsewhere for freshwater bacterioplankton populations (16).
Even given the caveat that overlying the adaptation to these abiotic factors are controls on the Synechococcus community structure by viral infection and/or grazing, the availability of a suite of genetic probes to map clade distribution allows us to address more specifically genotype-phenotype-environment relationships within the genus. Certainly the idea that specific bacterial clades or ecotypes are partitioned into specific niches in situ is now well demonstrated both in marine and freshwater environments (4, 6, 21, 22, 45, 54, 55, 72, 77, 78).
However, although recent genomic information gives insights into the underlying genetic basis for Prochlorococcus ecotypes that occupy distinct surface or deep niches in situ (30), it is possible that the greater genetic diversity of marine Synechococcus lineages defines a more complicated suite of niches and underlying physiologies. This may also explain the lack of distinct physiological characteristics of specific clades. Certainly, the 10 MC-A clades described here show few common within-clade phenotypic characteristics. Even if specific Synechococcus clades cannot be defined by a common phenotypic characteristic, and/or physiological traits are shared between clades, this is perhaps to be expected since it is the total suite of traits that correlates with the environmental niche occupied. At the genome level this would be reflected in a genomic core found in all strains and an auxiliary set of genes found in different strain subsets (a phylogenetic clade) required for occupation of that specific niche. Certainly we expect that further application of the clade-specific oligonucleotide probes developed here will permit interrogation of the distribution of the different clades in situ and ultimately allow correlation of such data with environmental parameters that will help to elucidate the underlying nature of the phenotypic fingerprints belonging to each marine Synechococcus clade.
Acknowledgments
We are grateful to John Waterbury, Brian Palenik, Gabrielle Rocap, Annick Wilmotte, and Anna Noordeloos for providing Synechococcus DNA or cells of some of the strains used in this study. S. Boulben and F. Le Gall are warmly thanked for purifying and maintaining the Roscoff strains over several years. We also thank Asaph Rivlin for supplying CTD data from the Red Sea depth profile and Nyree West for designing primer SYN1017R.
This work was supported by NERC grant GR3/11606 and by the EU program PICODIV (EVK3-CT-1999-00021).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armbrust, E. V., J. D. Bowen, R. J. Olson, and S. W. Chisholm. 1989. Effect of light on the cell cycle of a marine Synechococcus strain. Appl. Environ. Microbiol. 55:425-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, S., M. Fahrbach, P. Böger, and A. Ernst. 2002. Quantitative tracing, by Taq nuclease assays, of a Synechococcus ecotype in a highly diversified natural population. Appl. Environ. Microbiol. 68:4486-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. [DOI] [PubMed] [Google Scholar]
- 6.Béjà, O., E. N. Spudich, J. L. Spudich, M. Leclerc, and E. F. DeLong. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786-789. [DOI] [PubMed] [Google Scholar]
- 7.Binder, B. 2000. Cell cycle regulation and the timing of chromosome replication in a marine Synechococcus (Cyanobacteria) during light- and nitrogen-limited growth. J. Phycol. 36:120-126. [Google Scholar]
- 8.Binder, B. J., and S. W. Chisholm. 1995. Cell cycle regulation in marine Synechococcus sp. strains. Appl. Environ. Microbiol. 61:708-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brahamsha, B. 1996. A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 62:1747-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brahamsha, B. 1999. Non-flagellar swimming in marine Synechococcus. J. Mol. Microbiol. Biotechnol. 1:59-62. [PubMed] [Google Scholar]
- 11.Brahamsha, B. 1999. Genetic manipulations in Synechococcus spp. of marine cluster A. Bull. Inst. Oceanogr. 19:517-527.
- 12.Campbell, L., E. J. Carpenter, and V. J. Iacono. 1983. Identification and enumeration of marine chroococcoid cyanobacteria by immunofluorescence. Appl. Environ. Microbiol. 46:553-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell, L., and E. J. Carpenter. 1987. Characterisation of phycoerythrin-containing Synechococcus spp. populations by immunofluorescence. J. Plankton Res. 9:1167-1181. [Google Scholar]
- 14.Campbell, L., and R. Iturriaga. 1988. Identification of Synechococcus spp. in the Sargasso Sea by immunofluorescence and fluorescence excitation spectroscopy performed on individual cells. Limnol. Oceanogr. 33:1196-1201. [Google Scholar]
- 15.Carr, N. G., and N. H. Mann. 1994. The oceanic cyanobacterial picoplankton, p. 27-48. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 16.Casamayor, E. O., C. Pedrós-Alió, G. Muyzer, and R. Amann. 2002. Microheterogeneity in 16S ribosomal DNA-defined bacterial populations from a stratified planktonic environment is related to temporal changes and to ecological adaptations. Appl. Environ. Microbiol. 68:1706-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chisholm, S. W., R. J. Olson, E. R. Zettler, R. Goericke, J. B. Waterbury, and N. A. Welschmeyer. 1988. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 334:340-343. [Google Scholar]
- 18.Collier, J. L., B. Brahamsha, and B. Palenik. 1999. The marine cyanobacterium Synechococcus sp. WH 7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology 145:447-459. [DOI] [PubMed] [Google Scholar]
- 19.Douglas, S. E., and N. G. Carr. 1988. Examination of genetic relatedness of marine Synechococcus spp. using restriction fragment length polymorphisms. Appl. Environ. Microbiol. 54:3071-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferris, M. J., and B. Palenik. 1998. Niche adaptation in ocean cyanobacteria. Nature 396:226-228. [Google Scholar]
- 21.Field, K. G., D. Gordon, T. Wright, M. Rappé, E. Urbach, K. Vergin, and S. J. Giovannoni. 1997. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl. Environ. Microbiol. 63:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giovannoni, S. J., M. S. Rappé, K. L. Vergin, and N. L. Adair. 1996. 16S ribosomal RNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc. Natl. Acad. Sci. USA 93:7979-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glazer, A. N. 1999. Cyanobacterial photosynthetic apparatus: an overview. Bull. Inst. Oceanogr. 19:419-434. [Google Scholar]
- 24.Glibert, P. M., and R. T. Ray. 1990. Different patterns of growth and nitrogen uptake in two clones of marine Synechococcus spp. Mar. Biol. 107:273-280. [Google Scholar]
- 25.Glibert, P. M., T. M. Kana, R. J. Olson, D. L. Kirchman, and R. S. Alberte. 1986. Clonal comparisons of growth and photosynthetic responses to nitrogen availability in marine Synechococcus spp. J. Exp. Mar. Biol. Ecol. 101:199-208. [Google Scholar]
- 26.Glover, H. E. 1985. The physiology and ecology of the marine cyanobacterial genus Synechococcus. Adv. Aquat. Microbiol. 3:49-107. [Google Scholar]
- 27.Gordon, D. A., and S. J. Giovannoni. 1996. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic Ocean and Pacific Ocean. Appl. Environ. Microbiol. 62:1171-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hassidim, M., N. Keren, I. Ohad, L. Reinhold, and A. Kaplan. 1997. Acclimation of Synechococcus strain WH 7803 to ambient CO2 concentration and to elevated light intensity. J. Phycol. 33:811-817. [Google Scholar]
- 29.Herdman, M., R. W. Castenholz, I. Iteman, J. B. Waterbury, and R. Rippka. 2001. Subsection I (Formerly Chroococcales Wettstein 1924, emend. Rippka, Deruelles, Waterbury, Herdman and Stanier 1979), p. 493-514. In D. R. Boone, R. W. Castenholz, and G. M. Garrity (ed.), Bergey's Manual of Systematic Bacteriology, 2nd ed., vol. 1. The archaea and the deeply branching and phototrophic bacteria. Springer Publishers, New York, N.Y.
- 30.Hess, W. R., G. Rocap, C. S. Ting, F. Larimer, S. Stilwagen, and S. W. Chisholm. 2001. The photosynthetic apparatus of Prochlorococcus: insights through comparative genomics. Photosyn. Res. 70:53-72. [DOI] [PubMed] [Google Scholar]
- 31.Honda, D., A. Yokota, and J. Sugiyama. 1999. Detection of seven major evolutionary lineages in cyanobacteria based on 16S rRNA gene sequence analysis with new sequences of five marine Synechococcus strains. J. Mol. Evol. 48:723-739. [DOI] [PubMed] [Google Scholar]
- 32.Jacquet, S., F. Partensky, J.-F. Lennon, and D. Vaulot. 2001. Diel patterns of growth and division in marine picoplankton in culture. J. Phycol. 37:357-369. [Google Scholar]
- 33.Kana, T. M., and P. M. Glibert. 1987. Effects of irradiances up to 2000 μE m−2 s−1 on marine Synechococcus WH 7803. I. Growth, pigmentation and cell composition. Deep Sea Res. 34:479-495. [Google Scholar]
- 34.Kana, T. M., and P. M. Glibert. 1987. Effects of irradiances up to 2000 μE m−2 s−1 on marine Synechococcus WH 7803. II. Photosynthetic responses and mechanisms. Deep Sea Res. 34:497-516. [Google Scholar]
- 35.Laloui, W., K. A. Palinska, R. Rippka, F. Partensky, N. Tandeau de Marsac, M. Herdman, and I. Iteman. 2002. Genotyping of axenic and non-axenic isolates of the genus Prochlorococcus and the OMF-‘Synechococcus’ clade by size, sequence analysis of RFLP of the internal transcribed spacer of the ribosomal operon. Microbiology 148:453-465. [DOI] [PubMed] [Google Scholar]
- 36.Lantoine, F., and J. Neveux. 1997. Spatial and seasonal variations in abundance and spectral characteristics of phycoerythrins in the tropical northeastern Atlantic Ocean. Deep Sea Res. I 44:223-246. [Google Scholar]
- 37.Li, W. K. W. 1994. Primary production of prochlorophytes, cyanobacteria, and eukaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol. Oceanogr. 39:169-175. [Google Scholar]
- 38.Lindell, D., E. Padan, and A. F. Post. 1998. Regulation of ntcA expression and nitrite uptake in the marine Synechococcus sp. strain WH 7803. J. Bacteriol. 180:1878-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindell, D., E. Padan, and A. F. Post. 1999. Effect of ammonium on nitrate/nitrite uptake and ntcA expression in Synechococcus sp. strain WH 7803. Bull. Inst. Oceanogr. 19:273-278. [Google Scholar]
- 40.Lindell, D., and A. F. Post. 1995. Ultraphytoplankton succession is triggered by deep winter mixing in the Gulf of Aqaba (Eilat), Red Sea. Limnol. Oceanogr. 40:1130-1141. [Google Scholar]
- 41.Lindell, D., and A. F. Post. 2001. Ecological aspects of ntcA gene expression and its use as an indicator of the nitrogen status of marine Synechococcus spp. Appl. Environ. Microbiol. 67:3340-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1996. The ribosomal database project. Nucleic Acids Res. 24:82-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marie, D., C. Brussaard, F. Partensky, and D. Vaulot. 1999. Flow cytometric analysis of phytoplankton, bacteria and viruses, p. 11.11.1-11.11.15. In J. P. Robinson et al. (ed.), Current protocols in cytometry. John Wiley & Sons, Inc., New York, N.Y.
- 44.Miller, S. R., and R. W. Castenholz. 2001. Ecological physiology of Synechococcus strain SH-94-5, a naturally occurring cyanobacterium deficient in nitrate assimilation. Appl. Environ. Microbiol. 67:3002-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 46.Moore, L. R., A. F. Post, G. Rocap, and S. W. Chisholm. 2002. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol. Oceanogr. 47:989-996. [Google Scholar]
- 47.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR Primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olson, R. J., S. W. Chisholm, E. R. Zettler, and E. V. Armbrust. 1988. Analysis of Synechococcus pigment types in the sea using single and dual beam flow cytometry. Deep Sea Res. 35:425-440. [Google Scholar]
- 49.Olson, R. J., S. W. Chisholm, E. R. Zettler, and E. V. Armbrust. 1990. Pigments, size, and distribution of Synechococcus in the North Atlantic and Pacific Oceans. Limnol. Oceanogr. 35:45-58. [Google Scholar]
- 50.Palenik, B. 1994. Cyanobacterial community structure as seen from RNA polymerase gene sequence analysis. Appl. Environ. Microbiol. 60:3212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palenik, B. 2001. Chromatic adaptation in marine Synechococcus strains. Appl. Environ. Microbiol. 67:991-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Partensky, F., J. Blanchot, and D. Vaulot. 1999. Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bull. Inst. Oceanogr. 19:457-475. [Google Scholar]
- 53.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Postius, C., U. Kenter, A. Wacker, A. Ernst, P. Böger. 1998. Light causes selection among two phycoerythrin-rich Synechococcus isolates from Lake Constance. FEMS Microbiol. Ecol. 25:171-178. [Google Scholar]
- 55.Ramsing, N. B., M. J. Ferris, and D. M. Ward. 2000. Highly ordered vertical structure of Synechococcus populations within the one-millimetre-thick photic zone of a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 66:1038-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 57.Rippka, R., T. Coursin, W. R. Hess, C. Lichtlé, D. J. Scanlan, K. Palinska, I. Iteman, F. Partensky, J. Houmard, and M. Herdman. 2000. Prochlorococcus marinus Chisholm et al. 1992, subsp. nov. pastoris, strain PCC 9511, the first axenic chlorophyll a2/b2-containing cyanobacterium. Int. J. Syst. E vol. Microbiol. 50:1833-1847. [DOI] [PubMed] [Google Scholar]
- 58.Robertson, B. R., N. Tezuka, and M. M. Watanabe. 2001. Phylogenetic analyses of Synechococcus strains (cyanobacteria) using sequences of 16S rDNA and part of the phycocyanin operon reveal multiple evolutionary lines and reflect phycobilin content. Int. J. Syst. E vol. Microbiol. 51:861-871. [DOI] [PubMed] [Google Scholar]
- 59.Rocap, G., D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68:1180-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 61.Scanlan, D. J., N. H. Mann, and N. G. Carr. 1993. The response of the picoplanktonic marine cyanobacterium Synechococcus species WH 7803 to phosphate starvation involves a protein homologous to the periplasmic-binding protein of Escherichia coli. Mol. Microbiol. 10:181-191. [DOI] [PubMed] [Google Scholar]
- 62.Scanlan, D. J., N. J. Silman, K. M. Donald, W. H. Wilson, N. G. Carr, I. Joint, and N. H. Mann. 1997. An immunological approach to detect phosphate stress in populations and single cells of photosynthetic picoplankton. Appl. Environ. Microbiol. 63:2411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scanlan, D. J., and N. J. West. 2002. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol. Ecol. 40:1-12. [DOI] [PubMed] [Google Scholar]
- 64.Scanlan, D. J. Physiological diversity and niche adaptation in marine Synechococcus. Adv. Microb. Phys., in press. [DOI] [PubMed]
- 65.Suzuki, M. T., O. Béjà, L. T. Taylor, and E. F. DeLong. 2001. Phylogenetic analysis of ribosomal operons from uncultivated coastal marine bacterioplankton. Environ. Microbiol. 3:323-331. [DOI] [PubMed] [Google Scholar]
- 66.Tchernov, D., M. Hassidim, B. Luz, A. Sukenik, L. Reinhold, and A. Kaplan. 1997. Sustained net CO2 evolution during photosynthesis by marine microorganisms. Curr. Biol. 7:723-728. [DOI] [PubMed] [Google Scholar]
- 67.Toledo, G., and B. Palenik. 1997. Synechococcus diversity in the California current as seen by RNA polymerase (rpoC1) gene sequences of isolated strains. Appl. Environ. Microbiol. 63:4298-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toledo, G., and B. Palenik. A Synechococcus serotype is found preferentially in surface marine waters. Limnol. Oceanogr., in press.
- 69.Toledo, G., B. Palenik, and B. Brahamsha. 1999. Swimming marine Synechococcus strains with widely different photosynthetic pigment ratios form a monophyletic group. Appl. Environ. Microbiol. 65:5247-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Urbach, E., D. J. Scanlan, D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 1998. Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria). J. Mol. Evol. 46:188-201. [DOI] [PubMed] [Google Scholar]
- 71.Uysal, Z. 2000. Pigments, size and distribution of Synechococcus spp. in the Black Sea. J. Mar. Syst. 24:313-326. [Google Scholar]
- 72.Ward, D. M., M. J. Ferris, S. C. Nold, and M. M. Bateson. 1998. A natural view of microbial diversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62:1353-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Waterbury, J. B., S. W. Watson, F. W. Valois, and D. G. Franks. 1986. Biological and ecological characterisation of the marine unicellular cyanobacterium Synechococcus. Can. Bull. Fish. Aquat. Sci. 214:71-120. [Google Scholar]
- 74.Waterbury, J. B., and J. M. Willey. 1988. Isolation and growth of marine planktonic cyanobacteria. Methods Enzymol. 167:100-105. [Google Scholar]
- 75.Waterbury, J. B., and R. Rippka. 1989. Subsection 1. Order Croococcales Wettsten 1924, emend. Rippka et al., 1979, p. 1728-1746. In J. T. Staley, M. P. Bryant, N. Pfenning, and J. G. Holt (ed.), Bergey's Manual of Systematic Bacteriology, vol. 3. Williams and Wilkins, Baltimore, Md.
- 76.Watson, G. M. F., and F. R. Tabita. 1996. Regulation, unique gene organisation, and unusual primary structure of carbon fixation genes from a marine phycoerythrin-containing cyanobacterium. Plant Mol. Biol. 32:1103-1115. [DOI] [PubMed] [Google Scholar]
- 77.West, N. J., and D. J. Scanlan. 1999. Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl. Environ. Microbiol. 65:2585-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.West, N. J., W. A. Schönhuber, N. J. Fuller, R. I. Amann, R. Rippka, A. F. Post, and D. J. Scanlan. 2001. Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology 147:1731-1744. [DOI] [PubMed] [Google Scholar]
- 79.Willey, J. M., and J. B. Waterbury. 1989. Chemotaxis towards nitrogenous compounds by swimming strains of marine Synechococcus spp. Appl. Environ. Microbiol. 55:1888-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilmotte, A., C. Demonceau, A. Goffart, J.-H. Hecq, V. Demoulin, and A. C. Crossley. 2002. Molecular and pigment studies of the picophytoplankton in a region of the Southern Ocean (42-54°S, 141-144°E) in March 1998. Deep Sea Res. II 49:3351-3363. [Google Scholar]
- 81.Wilson, W. H., N. G. Carr, and N. H. Mann. 1996. The effect of phosphate status on the kinetics of cyanophage infection in the oceanic cyanobacterium Synechococcus sp. WH 7803. J. Phycol. 32:506-516. [Google Scholar]
- 82.Wingard, L. L., S. R. Miller, J. M. L. Sellker, E. Stenn, M. M. Allen, and A. M. Wood. 2002. Cyanophycin production in a phycoerythrin-containing marine Synechococcus strain of unusual phylogenetic affinity. Appl. Environ. Microbiol. 68:1772-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wood, A. M., P. K. Horan, K. Muirhead, D. A. Phinney, C. M. Yentsch, and J. B. Waterbury. 1985. Discrimination between types of pigments in marine Synechococcus spp. by scanning spectroscopy, epifluorescence microscopy, and flow cytometry. Limnol. Oceanogr. 30:1303-1315. [Google Scholar]
- 84.Wood, A. M., D. A. Phinney, and C. S. Yentsch. 1998. Water column transparency and the distribution of spectrally distinct forms of phycoerythrin-containing organisms. Mar. Ecol. Prog. Ser. 162:25-31. [Google Scholar]
- 85.Wood, A. M., M. Lipsen, and P. Coble. 1999. Fluorescence-based characterisation of phycoerythrin-containing cyanobacterial communities in the Arabian Sea during the northeast and early southwest monsoon (1994-1995). Deep Sea Res. II 46:1769-1790. [Google Scholar]
- 86.Wood, A. M., and D. Townsend. 1990. DNA polymorphism within the WH 7803 serogroup of marine Synechococcus spp. (cyanobacteria). J. Phycol. 26:576-585. [Google Scholar]
- 87.Zheng, D. D., E. W. Alm, D. A. Stahl, and L. Raskin. 1996. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl. Environ. Microbiol. 62:4504-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]