Abstract
Microeukaryotes in oxygen-depleted environments are among the most diverse, as well as the least studied, organisms. We conducted a cultivation-independent, small-subunit (SSU) rRNA-based survey of microeukaryotes in suboxic waters and anoxic sediments in the great Sippewisset salt marsh, Cape Cod, Mass. We generated two clone libraries and analyzed approximately 300 clones, which contained a large diversity of microeukaryotic SSU rRNA signatures. Only a few of these signatures were closely related (sequence similarity of >97%) to the sequences reported earlier. The bulk of our sequences represented deep novel branches within green algae, fungi, cercozoa, stramenopiles, alveolates, euglenozoa and unclassified flagellates. In addition, a significant number of detected rRNA sequences exhibited no affiliation to known organisms and sequences and thus represent novel lineages of the highest taxonomical order, most of them branching off the base of the global phylogenetic tree. This suggests that oxygen-depleted environments harbor diverse communities of novel organisms, which may provide an interesting window into the early evolution of eukaryotes.
The extent of protistan diversity on this planet represents a hotly debated issue. The concept of limited diversity (everything is everywhere) (17) competes with a view that the number of protistan species exceeds that of any other eukaryotic group (35).
This debate is a natural reflection of how limited our knowledge is on microeukaryotic diversity. The data available are derived primarily from cultivation studies and manual collection of cells from environmental samples. The proportion of cultivable microorganisms is likely to be under 1% (4, 24, 26), and this fact alone suggests that there may be an enormous reservoir of protistan diversity yet to be discovered (35).
The rRNA approach bypasses the problems of microbial uncultivability and has revolutionized the diversity studies. Its application to prokaryotic diversity resulted in some of the most interesting discoveries in environmental microbiology of the past decade (2, 6, 9, 23, 25, 28, 30, 37, 40, 41). In contrast, there are surprisingly few works exploring eukaryotic diversity in a similar manner (5, 7, 31, 34, 36). These studies reported a number of novel protistan lineages, some at the kingdom level. They also abundantly showed that the study of real microeukaryotic diversity is still in its infancy. Apart from identifying substantial gaps in our knowledge, this has an important implication for molecular phylogeny. Dawson and Pace (5) showed that the currently available collection of rRNA sequences is heavily biased toward only a few phylogenetic kingdoms. The accuracy with which both known as well as novel lines of descent are resolved can only be improved through a substantial growth of the sequence collection. This paper is a further contribution to the survey of marine microeukaryotic diversity and improvement of the resolution of the eukaryotic tree. We applied the 18S rRNA approach to samples from several suboxic and anoxic marine systems. We chose these environments because some of the most deeply divergent known eukaryotic lineages contain microaerobic and anaerobic organisms (15). Anoxic environments have occurred continuously throughout the Earth's history and promise to harbor a diversity of novel sequences potentially relevant to the early evolution of eukaryotes.
MATERIALS AND METHODS
Study site and sampling.
Our study site was located in the Great Sippewisset salt marsh on Cape Cod, Mass. We sampled a well-protected intertidal pool approximately 1.50 m deep and 10 m in diameter. The pool is connected to the marsh by a narrow channel, which limits the input of fresh oxygenated water and results in suboxic conditions in the water column and anoxia in sediments (Table 1) (oxygen measurements were made using the YSI 556 multiprobe system [Marion, Mass.]). Both the water column and sediments were sampled to generate small-subunit (SSU) rRNA clone libraries.
TABLE 1.
Physicochemical characteristics of samples used to generate clone libraries of eukaryotic 18S rRNA genes
| Parameter (unit) | Result for library
|
||
|---|---|---|---|
| CCW | CCI | CCA | |
| Temperature (°C) | 10 | 9.2 | 8.1 |
| Salinity (‰) | 31.6 | 33.5 | 34.4 |
| Oxygen (mg/ml) | 0.06 | NDa | ND |
| Oxygen saturation (%) | 0.8 | ND | ND |
| Eh (mV) | −151 | −189 | −237 |
| Characteristicb | Microaerobic | Anoxic | Anoxic |
ND, not detectable.
According to reference 15, characteristics were based on oxygen concentration and saturation.
Organisms from the water samples were first fractionated based on their size. To collect larger organisms, 1 to 2 liters of seawater was filtered through a 5-μm-pore-size Durapore membrane filter (47-mm diameter; Millipore, Bedford, Mass.) by using a peristaltic pump at the rate of 50 ml min−1. Smaller organisms from the filtrate were then concentrated on 0.2-μm-pore-size Durapore membrane filters. Both filtration steps were carried out in the field immediately following sampling. Filters of the same sample set were put into a cryovial with lysis buffer (100 mM Tris-HCl [pH 8], 100 mM Na2EDTA [pH 8], 100 mM NaPO4 [pH 8], 1.5 M NaCl, 1% cetyltrimethylammonium bromide, proteinase K [100 μg ml−1 final concentration]), immediately frozen in liquid nitrogen, and stored at −80°C until further analyses. Sediment samples were taken by coring the sediments to an approximately 20-cm depth by using a 5-cm diameter plastic core and further subsampled. The uppermost 1 cm of the sediments, which included an approximately 0.5-cm-thick layer of bottom water, and a 1-cm-thick layer at a depth of 9 to 10 cm, were obtained by slicing the original cores. These subsamples are hereafter referred to as water-sediment interface and sediment samples, respectively. Three subsamples of each horizon (10 ml each, taken with a syringe) were pooled to prevent selective sampling. Samples were immediately frozen in liquid nitrogen and stored at −80°C until further processing.
Nucleic acid isolation.
High-molecular-weight DNA was obtained from the frozen filters and 5-g aliquots of sediment samples as described by Zhou et al. (43). In short, the samples were heated to 65°C for 2 h in a high-salt extraction buffer (see above) with sodium dodecyl sulfate (20%), cetyltrimethylammonium bromide (1%), and proteinase K (0.1 mg ml−1 final concentration). The lysates were purified twice by extraction with an equal volume of chloroform-isoamyl alcohol (24:1) and precipitated with a 0.6 volume of isopropanol. Humic acids and other potential inhibitors of downstream applications were removed by DNA purification with the resin-based Wizard DNA clean up system (Promega, Madison, Wis.). The integrity of the total DNA was checked by agarose gel electrophoresis (0.8%), and the DNA yield was quantified photometrically (Biophotometer; Eppendorf, Hamburg, Germany). The molecular size of extracted genomic DNA was between 11 and 13 kb (as determined by comparison to a molecular size standard). On average, we extracted 4.5 μg of crude DNA from a filter (1-liter sample). The benthic DNA yield was 9.5 and 52 μg of DNA per g of interface and sediment samples, respectively. After Wizard purification, we recovered approximately 40% of the crude DNA. Humic materials in crude extracts are assumed to interfere with DNA binding to the resin (43), resulting in these low recovery rates.
PCR amplification of eukaryotic rRNAs.
The nearly full-length 18S rRNA genes were PCR amplified by using a universal eukaryotic primer set (EukA 1 to 21 and EukB 1795 to 1772) (33). Each PCR mixture contained 10 to 20 ng of DNA template, 2.5 U of HotStar Taq DNA polymerase (Qiagen, Valencia, Calif.) in the manufacturer-provided reaction buffer [Tris-HCl [pH 8.7], KCl, and (NH4)2SO4], 1.5 mM MgCl2, 200 μM concentrations of each deoxynucleoside triphosphate, and 0.5 μM concentrations of each oligonucleotide primer. The final volume was adjusted to 50 μl with sterile water. The PCR protocol for 18S rRNA gene amplification consisted of an initial hot start incubation (15 min at 95°C) followed by 30 identical amplification cycles (denaturation at 95°C for 45 s, annealing at 55°C for 1 min, and extension at 72°C for 2.5 min) and a final extension at 72°C for 7 min. Negative control reactions included bacterial (Escherichia coli) or archaeal (Halobacterium salinarium ATCC 19700) DNA as a template.
Cloning, ARDRA, and sequencing.
Fresh PCR products were used to construct a clone library by using the TA cloning kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Positive clones were identified by blue-white screening. Plasmids were isolated from overnight cultures by using the Qiaprep Spin Miniprep kit (Qiagen). The presence of an 18S rRNA gene insert was confirmed by PCR reamplification with the primer set and PCR protocol described above. Between 200 and 400 ng of positive amplification products of the target size were digested with 7.5 U of the restriction endonuclease HaeIII (New England Biolabs, Beverly, Mass.) for 60 min at 37°C, followed by an inactivation step for 20 min at 80°C. The resulting bands were separated by electrophoresis in a 2.5% low-melting-point agarose gel at 80 V for 2 to 3 h. Clones with identical amplified ribosomal DNA restriction fragment analysis (ARDRA) patterns were considered members of the same operational taxonomic unit (OTU). At least one clone of each OTU was sequenced at the University of Maine DNA Sequencing Facility with an Applied Biosystems (ABI) 373 DNA stretch sequencer, with the XL Upgrade and the ABI Prism BigDye, Terminator version 3.0, cycle sequencing ready reaction kit. Approximately 300 clones with unique ARDRA patterns were sequenced for the Sippewisset clone library. Low-quality sequence reads (7 sequences) and nontarget metazoan sequences (<10% in the water body and about 50% in each of both sediment layers) were excluded from the phylogenetic analyses. The clones from the water body are referred to as CCW (Cape Cod, water column), the clones from the sediment-water interface are referred to as CCI, and the clones from the deep anoxic layer are referred to as CCA.
Phylogenetic analyses.
The environmental 18S rRNA gene sequences obtained were compared to those in a GenBank by using gapped BLAST analysis (1) and to over 5,000 prealigned eukaryotic SSU rRNAs in the Ribosomal Database Project (32). The closest matches were aligned together with the Sippewisset clone sequences by using ClustalX (39), and the resulting multiple alignments were manually refined by using phylogenetically conserved secondary structures. The RNA secondary structure prediction and comparison tools of the Vienna RNA Package (27) were used to construct secondary structure models, which were verified against known conserved secondary structure regions (3). Environmental gene sequences were tested by using the RDP CHECK_CHIMERA program (32) to detect potential chimeric gene artifacts (38). Additionally, we tested 50 random 18S rRNA gene sequences by constructing sets of phylogenetic trees by using different regions of the same gene and examining the identity of the resulting trees' topology (16, 38). Further phylogenetic analyses included only conserved nucleotides at unambiguously aligned positions. Evolutionary trees were constructed by a distance-based (evolutionary distance) and by a character-based (maximum parsimony) method by using the PAUP* software package, version 4.0b10 (D. L. Swofford, Sinauer Associates, Sunderland, Mass.). All heuristic searches were performed by using random, stepwise addition of taxa with the tree bisection reconnection (TBR) branch-swapping algorithm. For each evolutionary distance tree we evaluated several models of character change: a Tamura-Nei minimum evolutionary distance model, a Kimura 2-Parameter model, an HKY85 model, and a general time-reversible substitution model under maximum-likelihood conditions (with two substitution models, a Ti/tv ratio of 2 or a six-category estimated general time-reversible model with individual base frequencies determined empirically). We assessed the relative stability of tree topologies by using 1,000 bootstrap replicates. Heuristic searches for bootstrap analyses employed stepwise addition, starting trees with simple addition of sequences and TBR branch swapping. Relative similarities between two different clones are based on ClustalX alignments of homologous regions of these two clones.
Nucleotide sequence accession number.
The gene sequences reported in this study have been deposited in the GenBank database and assigned GenBank accession numbers AY179969 to AY180047 and AY230211.
RESULTS
Microaerobic water column.
The 100-clone CCW library contained 69 unique ARDRA patterns, 57 of which were represented only once. Sequence homogeneity within each repetitive pattern was confirmed by sequencing 40% of all the repeats. Sequences exhibiting identical ARDRA patterns proved similar at or above the 99.8% level, and those of different patterns diverged by at least 2.5%. Therefore, we consider all unique ARDRA patterns as separate OTUs.
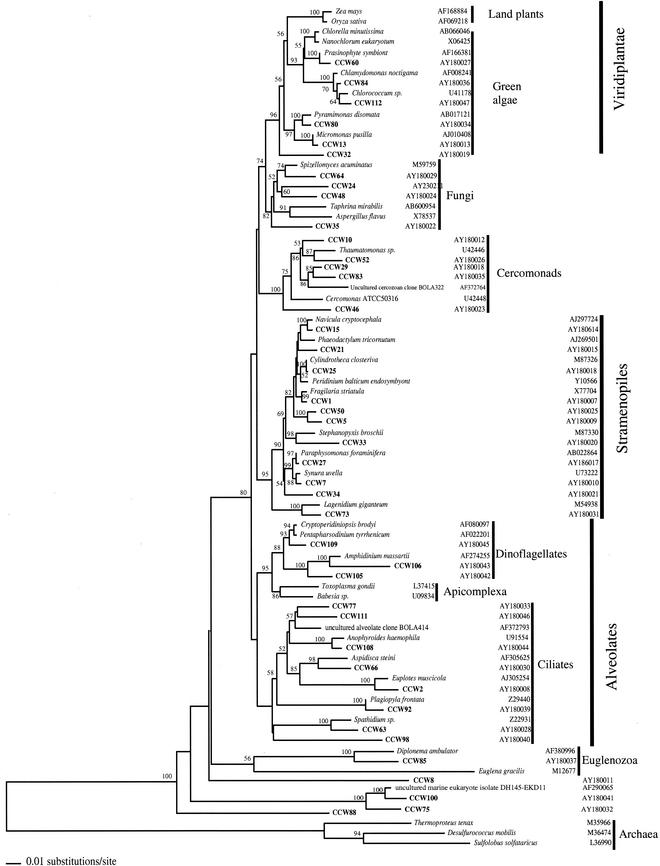
Several OTUs were excluded from analyses either because they represented nontarget organisms (2 crustaceans, 2 nematodes, and 2 polychaetes) or due to low quality or chimeric nature of their 18S rRNA gene sequences (21 OTUs). The final phylogenetic analyses thus included sequences of 42 unique protistan and fungal OTUs. This analysis showed (Fig. 1) that the CCW library was dominated by stramenopiles and alveolates (26% of the total number of unique OTUs each), followed by green algae (14%) and cercozoans (12%), fungi (10%), and euglenoids (2%). Four sequences showed no clear affiliation. Only 17% of sequences were closely related (>97% similarity) to sequences reported previously, mainly restricted to stramenopiles (e.g., CCW25 and CCW27) and green algae (CCW13). Most alveolate, cercozoan, and fungal sequences obtained were divergent from the closest known relatives by at least 5%. Several sequences could not be identified with any established protistan group, i.e., the three independent clades CCW8, CCW100, and CCW75, as well as CCW88, all of which branched in the base of the tree. Some of the deeply rooted sequences showed affiliation with uncultured clones from previous surveys of microeukaryotic SSU rRNA in aquatic environments. These included an independent clade CCW100/CCW75, a alveolate clade CCW77/CCW111, and a cercozoan clade CCW29/CCW83.
FIG. 1.
Phylogenetic tree for nearly complete SSU rRNA sequences of environmental clones from oxygen-depleted water in the Great Sippewisset salt marsh, Cape Cod, Mass., and the most closely related cultured organisms and environmental 18S rRNA signatures. Environmental clones are indicated by boldface type, and each clone is designated by the library designation (CCW) followed by a number. The tree topology was obtained by a heuristic search with stepwise addition of taxa (TBR branch swapping) under a Tamura Nei minimum evolutionary distance model, with gapped and ambiguously aligned positions excluded. Distance bootstrap values over 50% from an analysis of 1,000 bootstrap replicates are given at respective nodes. Sequences are followed by GenBank accession numbers.
Anoxic sediment.
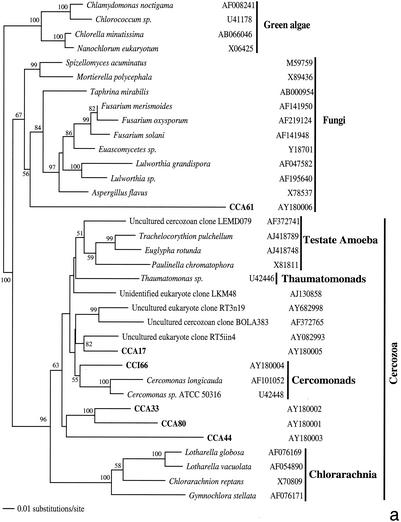
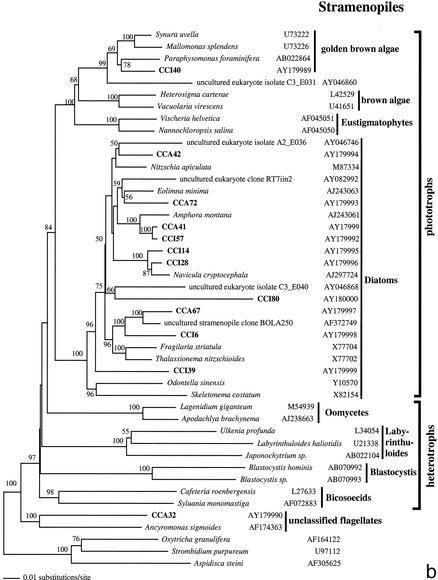
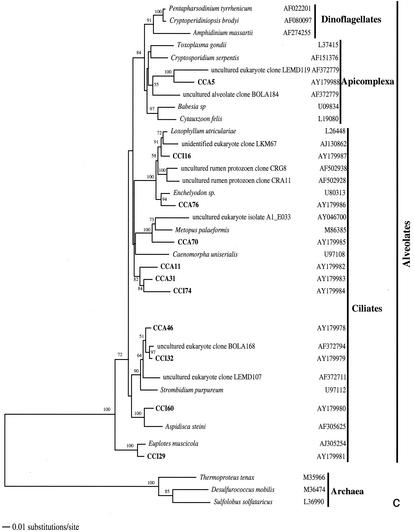
From the two clone sublibraries from anoxic sediments (CCI and CCA), we analyzed approximately 200 clones, 48 of which proved to have unique ARDRA patterns. The level of sequence similarity within and between ARDRA patterns paralleled that in the CCW clone library, and we consider these unique patterns as separate OTUs. Seven highly repetitive OTUs belonged to the nematode phylum; additionally, we detected one bivalve and two polychaete sequences. These were excluded from phylogenetic analyses, as were 2 foraminiferan sequences (CCI5 and CCA68, both 97% similar to Ammonia beccarii) due to their destabilizing effect on the phylogenetic trees. Therefore, the final phylogenetic analysis of OTUs from anoxic sediments included 36 rRNA sequences, of which 34 were protistan and 2 were archaeal (Fig. 2).
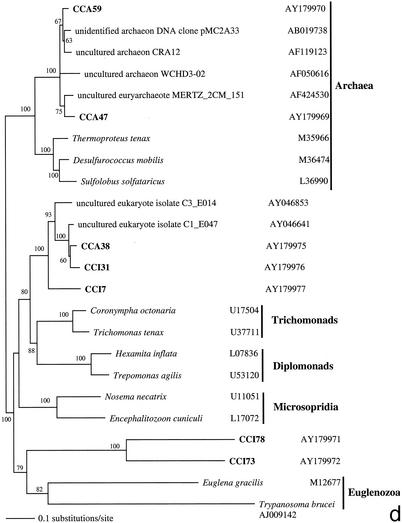
FIG. 2.
Phylogenetic trees for nearly complete SSU rRNA sequences of environmental clones in the Great Sippewisset salt marsh, Cape Cod, Mass., and the most closely related cultured organisms and environmental 18S rRNA signatures. Environmental clones are indicated by boldface type, and each clone is designated by the library designation (CCI for water-sediment interface; CCA for subsurface sediment [10 cm]) followed by a number. The tree topologies were obtained by a heuristic search with stepwise addition of taxa (TBR branch swapping) under a Tamura Nei minimum evolutionary distance model (trees a, b, and c) and maximum-likelihood (tree d) trees, with gapped and ambiguously aligned positions excluded. Distance bootstrap values over 50% from an analysis of 1,000 bootstrap replicates are given at the respective nodes. Sequences are followed by GenBank accession numbers.
The protistan sequences were dominated by phototrophic stramenopiles and alveolates, primarily ciliates (32% each) (Fig. 2b and c), and followed by cercozoans (15%) (Fig. 2a). Four cercozoan OTUs, all from the deep anoxic layer (CCA 17, CCA33, CCA44, and CCA80), four stramenopile OTUs, mainly from the sediment-water interface (CCI6, CCI39, CCI80, and CCA67), and three alveolate OTUs (mainly ciliates; CCA5, CCA11, CCA31, and CCI74) appear as clades independent of any sequence from known species within the respective groups. Among other sequences detected, there were one fungal (CCA61) (Fig. 2a) and one unclassified heterotrophic flagellate (CCA32) (Fig. 2b). Unassignable lineages represented 15% of the clones. These branch off as independent clades at the base of the tree (Fig. 2d), some in affiliation with previously detected environmental rRNA signatures (CCA38, CCI31, and CCI7). In contrast to the CCW clone library, in anoxic sediments we detected no green algae OTUs but amplified two euryarchaeote sequences (CCA59 and CCA47) (Fig. 2d).
DISCUSSION
The two main goals of this research are to survey protistan diversity by using an 18S rRNA approach and to compare the sequence diversity with that of organisms known to inhabit the environments of choice. We chose the Sippewisset salt marsh as a test environment because marine soft sediments with adjacent near-bottom waters represent one of the most diverse habitats on this planet. Intertidal and shallow water systems harbor especially abundant populations of microalgae, flagellates, ciliates, fungi, and other microeukaryotes (14).
As expected, the sequences obtained from the clone libraries encompass all major protistan lineages, including alveolates, stramenopiles, cercomonads, euglenoids, and green algae, as well as fungi and nontargeted metazoan groups, such as invertebrates. Although PCR and cloning reactions tend to produce a biased outcome, screening of only ca. 300 clones yielded representatives of all these major groups expected in the environment under study.
The sequences of our SSU rRNA libraries (76 unique protistan OTUs) fall into three different categories, according to their relatedness to the ribosomal sequences reported earlier. The first category (29%) is the sequences closely related to the named organisms known from prior taxonomic surveys and/or available in culture collections. In general, the sequences from this group are 95 to 99% similar to those of known organisms, which indicates relatedness at the specific to generic levels (R. Droste and S. Epstein, submitted for publication). The closest matches (up to 99% similarity and above) included widely distributed ciliates (e.g., Plagiopyla, and Euplotes), green algae (e.g., Micromonas, Chlorococcum, and Chlamydomonas), and stramenopiles (e.g., Navicula, Amphora, Fragilaria, Synura, and Paraphysomonas).
The second category of sequences (59%) represents sequences that are not closely related to any named organisms but are branching within well-established protistan clades. As a rule, the depth of branching of these sequences is comparable to class level differences. This group contains the novel golden-brown algae CCI40 and the diatom clone CCA72. We also include in this category clones CCA42, CCA67, CCI32, CCW77, and CCW111, which have unclear taxonomic affiliations but are related to the A2- and BOLA-type sequences obtained in molecular surveys of other marine oxygen-depleted environments (5, 11).
The third group of sequences (12%) cannot be assigned to any established clade and seem to represent novel groups at the kingdom level. Four of the clones (CCW8, CCW88, CCI78, and CCI73) form early branches on the eukaryotic tree that precede the euglenozoa. To the same category belong several clones that are equally divergent from the known organisms but are affiliated with previously reported novel 18S rRNA-based kingdom level clades. Among these are clones CCA38, CCI7, and CCI31, which are related to two sequences recovered from a deep-sea hydrothermal vent site (11). These clones support and increase the diversity in what appears to be a novel protistan kingdom branching off the eukaryotic tree preceding trichomonads. Additionally, clones CCW75 and CCW100 are affiliated with a recently reported sequence DH145-EKD11 (31). Together, these three clones form a tight protistan clade whose origin seems to precede the crown radiation of eukaryotes. Interestingly, the CCW and DH clones originated from samples collected in different hemispheres and drastically different environments, yet their sequences are similar at the level approaching specific (>97%). This indicates that the novel clade may be ubiquitous and probably contains cosmopolitan representatives. Collectively, these findings illustrate how decidedly incomplete is the eukaryotic evolutionary tree and our view on protistan diversity.
It is important to know how well the sequence diversity in the clone libraries represents the organism diversity in the target community. Ciliated Protozoa may serve as a convenient model to address this issue because they have been surveyed by classical taxonomists for over a century (8, 10, 12, 19, 20, 21, 22, 29). This makes it possible to conduct a direct comparison between the molecular and classical inventories.
Taxonomic and ecological research of the past indicate that suboxic and anoxic sediments and waters are characterized by a range of facultative and obligate anaerobes, such as Plagiopyla, Metopus, Caenomorpha, and others (13, 14, 15, 18). Using the rRNA approach, we detected sequences related to all of the above genera (e.g., clone CCW92 reveals 99% similarity to Plagiopyla frontata) as well as to other typical benthic ciliates such as Euplotes, Aspidisca, and Spathidium. In addition, we detected a large number of lineages within the ciliate clade whose phylogenetic position suggest relatedness to anaerobic and microaerophilic ciliates (e.g., CCA11, CCA31, CCI74, CCI32, and CCA46). Therefore, even limited clone libraries such as ours with only ca. 300 clones processed contain the signatures of those genera that are expected to dominate the sampling site. This identifies the 18S rRNA approach as useful in identifying at least some of the key elements in the ciliate assemblage in question. The latter is undoubtedly more diverse than the collection of ciliate-related sequences we obtained. However, the ciliate sequence diversity should increase dramatically as the clone library is sampled more systematically, perhaps reaching, and likely exceeding, the level of ciliate diversity known from classical research. The relatively good coverage of the ciliate dominant taxa exhibited by our clone libraries tentatively indicates that these libraries may contain a reasonable diversity record of other protistan taxa from the Sippewisset salt marsh.
In conclusion, the 18S rRNA approach seems to be a valuable tool in assessing the global diversity of aquatic eukaryotes. The first applications of this approach to microeukaryotes in aquatic environments (5, 7, 11, 31, 34, 36, 42) uncovered a substantial number of novel clades from the specific to the kingdom level. This work supports some of the previously reported clades and identifies novel deeply branching lineages in the eukaryotic evolutionary tree. The oxygen-depleted habitats seem to be particularly promising in a search for such new lineages, which calls for an extended SSU RNA sequence collection in this and other extreme environments as part of surveying the diversity of life and in an attempt to develop models of the early Precambrian microbial communities.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft grant STO414/21 to T.S., NSF grant DEB-0103599 to S.E., and several Northeastern University grants to S.E.
We are indebted to Edward Jarroll (Marine Science Center, Northeastern University) for invaluable support throughout this study in too many areas to list. We thank Ted Maney (Marine Science Center, Northeastern University) for lending us a hand in many challenging situations. Our acknowledgments extend to Rita Droste and Victoria Alla (Marine Science Center, Northeastern University) for help in the field as well as in the lab. We are grateful to Virginia Edgcomb (Woods Hole Oceanographic Institution) for stimulating discussions on protistan phylogeny.
Footnotes
This is contribution no. 245 of the Marine Science Center of Northeastern University.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The Comparative RNA Web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. [Correction, 3:15.]. [DOI] [PMC free article] [PubMed]
- 4.Colwell, R. R., and D. J. Grimes. 2000. Semantics and strategies. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment, ASM Press, Washington D.C.
- 5.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99:8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dièz, B., C. Pedros-Alio, and R. Massana. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 67:2932-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doflein, F. 1901. Die Protozoen als Parasiten und Krankheitserreger Nach Biologischen Gesichtspunkten Argestellt. G. Fisher, Jena, Germany.
- 9.Dojka, M. A., J. K. Harris, and N. R. Pace. 2000. Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria. Appl. Environ. Microbiol. 66:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dragesco, J. 1960. Cilies mesopsammiques littoraux. Trav. Stat. Biol. Roscoff 12:1-365. [Google Scholar]
- 11.Edgcomb, V. P., D. T. Kysela, A. Teske, A. de Vera Gomez, and M. L. Sogin. 2002. Benthic eukaryotic diversity in the Guyamas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. USA 99:7658-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrenberg, C. C. 1838. Die Infusionstheirchen als Vollkommene Organismen. Leopold Voss, Leipzig, Germany.
- 13.Esteban, G., B. J. Finlay, and T. M. Embley. 1993. New species double the diversity of anaerobic ciliates in a Spanish lake. FEMS Microbiol. Lett. 109:93-100. [Google Scholar]
- 14.Fenchel, T. 1992. What can ecologists learn from microbes-life beneath a square centimeter of sediment surface. Funct. Ecol. 6:499-507. [Google Scholar]
- 15.Fenchel, T., and B. J. Finlay. 1995. Ecology and evolution in anoxic worlds. Oxford University Press, Oxford, United Kingdom.
- 16.Field, K. G., D. Gordon, T. Wright, M. Rappé, E. Urbach, K. Vergin, and S. J. Giovannoni. 1997. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl. Environ. Microbiol. 63:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finlay, B. J. 2002. Global dispersal of free-living microbial species. Science 296:1061-1063. [DOI] [PubMed] [Google Scholar]
- 18.Finlay, B. J., K. J. Clarke, E. Vicente, and M. R. Miracle. 1991. Anaerobic ciliates from sulphidic-rich solution lake in Spain. Eur. J. Protistol. 27:148-159. [DOI] [PubMed] [Google Scholar]
- 19.Foissner, W., H. Blatterer, H. Berger, and F. Kohmann. 1991. Taxonomische und oekologische Revision der Ciliaten des Saprobiensystems-Band I: Crytophorida, Oligotrichida, Hypotrichia, Colpodea. Informationsberichte des Bayer. Landesamtes fuer Wasserwirtschaft. Bartels und Wernitz Druck, Munich, Germany.
- 20.Foissner, W., H. Berger, and F. Kohmann. 1992. Taxonomische und oekologische Revision der Ciliaten des Saprobiensystems-Band II: Peritrichia, Heterotrichida, Odontomastida. Informationsberichte des Bayer. Landesamtes fuer Wasserwirtschaft. Bartels und Wernitz Druck, Munich, Germany.
- 21.Foissner, W., H. Berger, and F. Kohmann. 1994. Taxonomische und oekologische Revision der Ciliaten des Saprobiensystems-Band III: Hymenostomata, Protomatida, Nassulida. Informationsberichte des Bayer. Landesamtes fuer Wasserwirtschaft. Bartels und Wernitz Druck, Munich, Germany.
- 22.Foissner, W., H. Berger, and F. Kohmann. 1995. Taxonomische und oekologische Revision der Ciliaten des Saprobiensystems-Band IV: Gymnostomatea, Loxodes, Suctoria. Informationsberichte des Bayer. Landesamtes fuer Wasserwirtschaft. Bartels und Wernitz Druck, Munich, Germany.
- 23.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1992. Novel major archae-bacterial group from marine plankton. Nature 356:148-149. [DOI] [PubMed] [Google Scholar]
- 24.Giovannoni, S. G., and M. Rappé. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, Inc., New York, N.Y.
- 25.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 26.Grimes, D. J., A. L. Mills, and K. H. Nealson. 2000. The importance of viable but nonculturable bacteria in biogeochemistry. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, D.C.
- 27.Hofacker, I. L., W. Fontana, P. F. Stadler, L. S. Bonhoeffer, M. Tacker, and M. Schuster. 1994. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 125:167-188. [Google Scholar]
- 28.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kahl, D. M. 1930-1935. Urtiere oder Protozoa. I: Wimpertiere oder Ciliata (Infusoria), eine Bearbeitung der freilebenden und ectocomenslaen Infusorien der Erde, unter Ausschluss der marinen Tintinnidae. Tierwelt Dtl. Gustav Fischer, Jena, Germany.
- 30.Liesack, W., and E. Stackebrandt. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 174:5072-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez-Garcia, P., F. Rodriguez-Valera, C. Pedros-Alio, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 32.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acid Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medlin, L., H. J. Elwood, S. Stickel, and M. L. Sogin. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491-499. [DOI] [PubMed] [Google Scholar]
- 34.Moon-van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 35.Patterson, D. J. 1999. The diversity of eukaryotes. Am. Nat. 154:96-124. [DOI] [PubMed] [Google Scholar]
- 36.Rappé, M. S., M. T. Suzuki, K. L. Vergin, and S. J. Giovannoni. 1998. Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the Pacific and Atlantic coasts of the United States. Appl. Environ. Microbiol. 64:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold, marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robison-Cox, J. F., M. M. Bateson, and D. M. Ward. 1995. Evaluation of nearest-neighbor methods for detection of chimeric small-subunit rRNA sequences. Appl. Environ. Microbiol. 61:1240-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal-W-improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:82-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 42.Zettler, L. A. A., F. Gomez, E. Zettler, B. G. Keenan, R. Amils, and M. L. Sogin. 2002. Eukaryotic diversity in Spain' s River of Fire. Nature 417:137.. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, J., M. A. Brunns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]