Abstract
We studied the growth of six culturable bacterial lineages from coastal North Sea picoplankton in environmental samples under different incubation conditions. The grazing pressure of heterotrophic nanoflagellates (HNF) was reduced either by double prefiltration through 0.8-μm-pore-size filters or by 10-fold dilutions with 0.2-μm (pore-size) prefiltered seawater. We hypothesized that those γ-proteobacterial genera that are rapidly enriched would also be most strongly affected by HNF regrowth. In the absence of HNF, the mean protein content per bacterial cell increased in both treatments compared to environmental samples, whereas the opposite trend was found in incubations of unaltered seawater. Significant responses to the experimental manipulations were observed in Alteromonas, Pseudoalteromonas, and Vibrio populations. No treatment-specific effects could be detected for members of the Roseobacter group, the Cytophaga latercula-C. marinoflava lineage, or the NOR5 clade. Statistical analysis confirmed a transient increase in the proportions of Alteromonas, Pseudoalteromonas, and Vibrio cells at reduced HNF densities only, followed by an overproportional decline during the phase of HNF regrowth. Cells from these genera were significantly larger than the community average in the dilution treatments, and changes in their relative abundances were negatively correlated with HNF densities. Our findings suggest that bacteria affiliated with frequently isolated genera such as Alteromonas, Pseudoalteromonas, and Vibrio might be rare in coastal North Sea picoplankton because their rapid growth response to changing environmental conditions is counterbalanced by a higher grazing mortality.
During the past several years, our understanding of the ecological importance of culturable and uncultured lineages of marine picoplankton has been repeatedly challenged. For more than a decade, direct molecular cloning of microbial 16S ribosomal DNA (rDNA) sequences has revealed a high diversity of uncultured bacteria and archaea (11, 15). These microbial lineages were regarded as more typical for the pelagic environment than members of the traditionally isolated genera. However, cloning of PCR amplificates has been shown to discriminate, e.g., against the Bacteroidetes (2), an important group of coastal marine bacteria (7) with many culturable representatives (34). At the same time, the ecological importance of members from culturable bacterial lineages such as the Roseobacter clade is increasingly recognized (19, 52). Moreover, innovative cultivation approaches have yielded isolates from previously uncultured bacterial lineages that form substantial populations in the marine environment (7, 37). The most extreme positions have been voiced by Pinhassi et al. (34), who argued that the majority of common picoplankton bacteria are able to form colonies on agar plates and rich media, and Garcia-Martínez et al. (13), who claimed that >50% of picoplankton rRNA in deeper Mediterranean waters originates from Alteromonas macleodii-like bacteria.
Nevertheless, some genera of marine bacteria that are frequently isolated by classic ZoBell techniques are apparently rare in the picoplankton of coastal North Sea surface waters, as determined by direct counts after specific fluorescence in situ hybridization (FISH) (5, 6). Representatives of such readily culturable bacterial lineages with conspicuously low in situ population densities in the German Bight are Alteromonas, Colwellia, Pseudoalteromonas, and Vibrio (6). Since members of these genera typically form larger cells than so-called “oligotrophic” marine isolates (1, 37), we hypothesized that these opportunistically growing γ-proteobacteria might be a preferred target of selective predation.
Free-living heterotrophic nanoflagellates (HNF) are ubiquitous bacterial grazers (38, 40). Although the phenotypic prey selectivity of aquatic HNF was first discovered in marine systems (20, 28), its potential effects on the genotypic composition of microbial communities have mainly been studied in freshwaters (22, 25, 33, 44, 45). So far, few investigations have presented evidence that taxon-specific grazing effects might occur in marine picoplankton. Suzuki (48) reported significant differences in the composition of PCR-amplified microbial 16S rDNA from 24-h bottle incubations of filtered and unfiltered seawater, but the available method did not allow a precise phylogenetic affiliation of the rDNA in question. Other investigations have reported qualitative community changes during confinement (27, 39) but could not establish a quantitative relationship between the dynamics of individual bacterial populations, the phenotypic traits of single cells, and HNF abundances.
We set up incubations of North Sea water in which HNF abundances were reduced by filtration or dilution and then monitored the development of the microbial community in these treatments during the initial phase of HNF regrowth. Based on previous observations (5), we presumed that members of different frequently cultured γ-proteobacterial lineages would rapidly establish high densities after HNF removal, so that the effects of predation on these bacteria could be studied as HNF populations recovered without the addition of tracer cells from cultures. The dynamics of control groups were also monitored, i.e., of other bacteria that are also culturable but nevertheless common in the picoplankton. In addition, the cell sizes of three rapidly enriched bacterial genera and of the total community were determined, as well as the changes in mean per-cell protein content of the bacterial assemblages during the incubations.
MATERIALS AND METHODS
Study site and experimental design.
The incubation experiments were conducted at the Biologische Anstalt Helgoland of the Alfred-Wegener Institute for Polar and Marine Research between 29 August and 3 September 2000. The island of Helgoland is located ca. 50 km offshore in the German Bight of the North Sea. Water was collected at the sampling site Helgoland Roads (54.09 N, 7.52 E) from ca. 1 m depth in a prerinsed 50-liter polyethylene container. It was stored at 4°C until further processing (within 6 h after sampling). Three experimental treatments were set up in triplicates of 3-liter volumes in acid-washed, prerinsed 5-liter glass bottles (Schott, Germany). Treatments were categorized as follows: (i) untreated (i.e., bottles were filled with 3,000 ml of the unmanipulated seawater); (ii) dilution (i.e., 300 ml of seawater was 10-fold diluted with 2,700 ml of seawater that had been prefiltered through 0.2-μm-pore-size filters [Sartorius, Göttingen, Germany]); (iii) prefiltration (i.e., seawater was filtered twice through 0.8-μm-pore-size polycarbonate membranes [type ATTP; diameter, 47 mm; Millipore, Eschborn, Germany]). The choice of filter types was done according to the recommendations of Gasol and Morán (14). All bottles were incubated in the dark at 16°C for 5 days. Samples were analyzed daily (days 0 to 5) to determine total bacterial and HNF cell numbers, bacterial per-cell protein content, and the population dynamics and cell sizes of selected bacterial groups. At the sampling date and during the first 3 days of the incubations, additional water samples were collected daily at Helgoland Roads and also analyzed for bacterial numbers and per-cell protein content.
HNF abundances.
Portions of 100 to 300 ml were fixed with alkaline Lugol solution (final concentration, 0.1%), followed by particle-free formaldehyde (1.8%) and sodium thiosulfate (60 μg ml−1) (modified from reference 41). Samples were fixed at 4°C for 1 to 24 h. Portions of 20 to 50 ml from these subsamples were concentrated on black polycarbonate membranes (type ATTP; diameter, 25 mm; pore size, 0.8 μm; Millipore) and double stained with DAPI (4′,6′-diamidino-2-phenylindole; 1 μg ml−1; staining time, 3 min) and fluorescein isothiocyanate (FITC; 33 μg ml−1; staining time, 6 min) (modified from reference 41). Filters were embedded on microscopic slides in Cargille-Type A immersion oil (Cargille Laboratories, Cedar Grove, N.J.). Two parallel filters were produced for every time point and bottle and then stored at −20°C until evaluated. Eukaryotic cells were visualized by epifluorescence microscopy at ×1,000 magnification (Axiophot2; Carl Zeiss, Jena, Germany). HNF cells (cell length, 2 to 20 μm) could be unambiguously identified by a combined inspection at UV and blue excitation (filter sets Zeiss01 and Zeiss09, respectively; Carl Zeiss) to detect both their DAPI-stained nucleus and their FITC-stained body outline and, if visible, the flagella. Pigmented (mixo- and autotrophic) flagellates were distinguished from exclusively heterotrophic forms by the red autofluorescence of chloroplasts. At least 20 microscopic fields of view or 200 HNF cells were counted per polycarbonate membrane.
Bacterial abundances and per-cell protein content.
For bacterial analyses, portions of 40 to 100 ml from each time point and bottle and from environmental samples between days 0 to 3 were fixed with freshly prepared buffered paraformaldehyde solution as described previously (17). For the determination of bacterial abundances and protein content, 4-ml portions of fixed subsamples were stored frozen until further analysis. Total bacterial cell numbers and the average per-cell protein content were determined by flow cytometry (FACStar Plus flow cytometer; Becton Dickinson, Heidelberg, Germany) as described previously (5, 53). The carefully thawed samples were double stained with the DNA specific stain Hoechst 33342 and the protein stain SYPRO red (Molecular Probes, Leiden, The Netherlands). At least 2,000 Hoechst 33342-positive cells were counted per sample. Counts were converted into numbers of bacteria per milliliter of sample from known concentrations of fluorescent beads added to each sample. These beads served as internal standard to normalize SYPRO fluorescence intensity for estimates of the mean cellular protein content. Samples from day 0 of the prefiltered treatment for bacterial counts and protein content determination were lost by accident.
Bacterial population dynamics and cell sizes.
Subsamples of 10 ml from the paraformaldehyde fixed samples (see above) were concentrated on polycarbonate membranes (type GTTP; diameter, 47 mm; pore size, 0.2 μm; Millipore), rinsed with double-distilled water, and stored at −20°C. Two parallel filters were produced for every time point and bottle, and replicate samples from two of the bottles were evaluated per treatment.
North Sea bacterioplankton community structure and population dynamics were analyzed by FISH as described previously (31). Cells on filter sections were hybridized with 5′ Cy3-monolabeled oligonucleotide probes (ThermoHybaid, Ulm, Germany). The probes were targeted to all bacteria (EUB338, EUB338 II, and EUB338 III) (3), γ-proteobacteria (GAM42a) (26), Alteromonas spp. (ALT1413), Pseudoalteromonas spp. (PSA184) (6), Vibrio spp. (GV822), Roseobacter spp. (GRb626) (16), and members of the Cytophaga latercula-C. marinoflava lineage (CYT1448), including two North Sea isolates, and to bacteria affiliated with the NOR5 cluster (NOR5-730) (7). The EUB338 antisense probe NON338 served as a negative control. Hybridized filter sections were counterstained with DAPI (1 μg ml−1) and embedded on microscopic slides in a 1:5 mixture of Vecta Shield (Vecta Laboratories, Burlingame, Calif.) and Citifluor AF1 (Citifluor, Ltd., London, United Kingdom) mounting fluids. Cell counts were performed by epifluorescence microscopy at a ×1,000 magnification. The abundances of FISH-stained cells were determined at green excitation and counted as percentages of all DAPI-stained cells in identical microscopic fields. From 400 to >2,000 DAPI-stained cells were enumerated per sample.
In samples from day 2 of the dilution treatments the cell sizes of bacteria that hybridized with probes EUB338, ALT1413, PSA184, and GV822 were determined. Image pairs of FISH and DAPI-stained cells were captured in 10 microscopic fields with a cooled slow-scan charge-coupled device camera (ORCA; Hamamatsu, Herrsching, Germany). Since DAPI staining may underestimate cell sizes (35), all measurements were performed on FISH-stained preparations. However, objects visible at green excitation without colocalized DAPI fluorescence were excluded from measurements. Cell edges were established by previously described strategies (36), and mean cell volumes were estimated from measured area and perimeter. Cell size data from the two replicate bottles were pooled. Altogether, >1,000 EUB338 stained cells were measured and 150 to 200 cells hybridized with each of the specific probes.
Statistical analyses.
We tested for treatment-specific differences in the relative abundances of individual bacterial populations from the six studied lineages. Changes in the community contribution of the different bacterial lineages over time were approximated by a mixed logistic regression model with fixed effects for time and treatment and random effects for replicates. This model was chosen as the least complex equation that could be fitted to the data, and because it is adaequate for ratio data (range, 0 to 1). As a precondition for the estimation of random effects (i.e., biological variability of replicates), the precision of FISH counts at different relative abundances was established from separate preparations (ca. 5, 25, and 90% of probe-positive cells, five replicates each). This empirical factor was then included in a bootstrap analysis, i.e., the random generation of 2,000 regression curves. The 95% confidence intervals were subsequently calculated from the distribution of the Kolmogorov-Smirnov distances between the newly generated and the original regression curves.
A two-step likelihood ratio test was then applied to determine whether the observed data from the three treatments could be described by a single regression (common model), or if the data could be significantly better approximated by different regression curves for different treatments (separate model). First, the common model was tested against the separated model in all six bacterial groups. The significance level was adjusted to 1/6 alpha (Bonferroni correction, P = 0.0083) to maintain the experimentwise error rate. Subsequent pairwise tests between individual treatments were conducted only for those groups in which the common model (i.e., no treatment effects) could be significantly rejected. Due to the closed test principle for less than three pairwise comparisons, no further alpha adjustment was required (47).
In bacterial groups that were significantly affected by the treatments, as detected by the analysis described above, we then tested whether the responses in community contributions of the individual bacterial groups were significantly related to HNF numbers (Spearman rank sum correlations). Changes were calculated as the differences between the relative abundances of the populations on subsequent days. This analysis was limited to the dilution treatments, because HNF in the prefiltration treatment formed substantial populations only during the last 2 days of the incubation period. All calculations were carried out with the SAS ISTAT package (v.8.2; SAS, Inc., Cary, N.C.).
RESULTS
Total abundances of bacteria and HNF and evaluation of bacterial protein content.
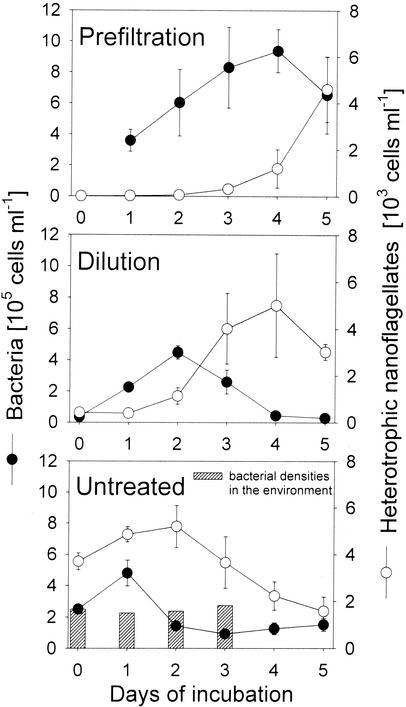
Bacterial total cell numbers transiently increased during the incubations in all three variants and declined thereafter, whereas no change in abundances was detected in environmental samples (Fig. 1). Maximum abundances were found after 24, 48, and 96 h of enrichment in the untreated, dilution, and prefiltration treatments, respectively. In the dilution treatment a bloom of HNF followed the bacterial bloom with a delay of 1 day. HNF were effectively reduced to <10 individuals ml−1 by double filtration but reestablished a substantial population in these treatments during the last 2 days of the experiment. In the untreated variants, high initial HNF cell numbers were maintained during the first 48 h but decreased thereafter.
FIG. 1.
Cell numbers of bacteria and heterotrophic nanoflagellates in the different treatments (mean ± the standard deviation). Hatched bars indicate the daily samples of North Sea picoplankton.
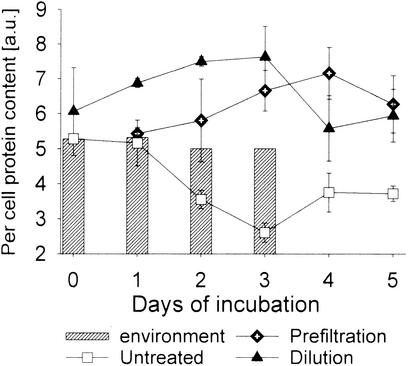
Between days 1 and 3, the mean bacterial per-cell protein content was significantly higher in the dilution treatment than in the environmental samples and decreased during HNF regrowth (Fig. 2). An increase in the average protein content was also observed in the prefiltration treatment between days 2 and 4. In the untreated variants, protein content per cell dropped by >50% during the first 3 days of incubation compared to the environmental values and slightly increased thereafter.
FIG. 2.
Changes in bacterial mean per-cell protein content in the different treatments during the incubation period (mean ± the standard deviation). a.u: arbitrary units. Hatched bars indicate the daily samples of North Sea picoplankton.
Community contribution of bacterial groups.
The majority of bacteria in the incubations were detectable by FISH with monolabeled probes (Table 1). On average, 79% of all DAPI-stained cells were detectable in the dilution and prefiltration treatments, and 74% in the untreated variants, and detection rates changed little over the incubation period. The fraction of γ-proteobacteria decreased by almost 50% in the untreated incubations but almost doubled in the dilution and prefiltration treatments. Between days 1 and 5 of the incubation period, averages of 71 and 77% of all γ-proteobacteria were affiliated with Alteromonas, Pseudoalteromonas, or Vibrio in the dilution and prefiltration treatments, respectively.
TABLE 1.
Relative abundances of cells detectable by FISH with the probes EUB338 I to III (all bacteria) and GAM42a (γ-proteobacteria) in the various treatments during the incubation period
| Phylogenetic group | Day | % All DAPI-stained cells, replicates A and B
|
|||||
|---|---|---|---|---|---|---|---|
| 1:10 Dilution
|
0.8-μm prefiltration
|
Untreated
|
|||||
| A | B | A | B | A | B | ||
| Bacteria (probe EUB338 I to III) | 0 | 70 | 79 | 84 | 89 | 88 | 82 |
| 1 | 90 | 72 | 83 | 85 | 81 | 81 | |
| 2 | 90 | 78 | 73 | 83 | 72 | 69 | |
| 3 | 90 | 81 | 73 | 78 | 69 | 58 | |
| 4 | 82 | 66 | 73 | 78 | 71 | 69 | |
| 5 | 79 | 65 | 75 | 74 | 71 | 78 | |
| γ-Proteobacteria (probe GAM42a) | 0 | 17 | 12 | 13 | 12 | 12 | 19 |
| 1 | 17 | 17 | 14 | 15 | 12 | 12 | |
| 2 | 30 | 28 | 29 | 37 | 8 | 10 | |
| 3 | 32 | 27 | 30 | 39 | 8 | 11 | |
| 4 | 21 | 21 | 40 | 43 | 9 | 9 | |
| 5 | 18 | 20 | 30 | 37 | 7 | 8 | |
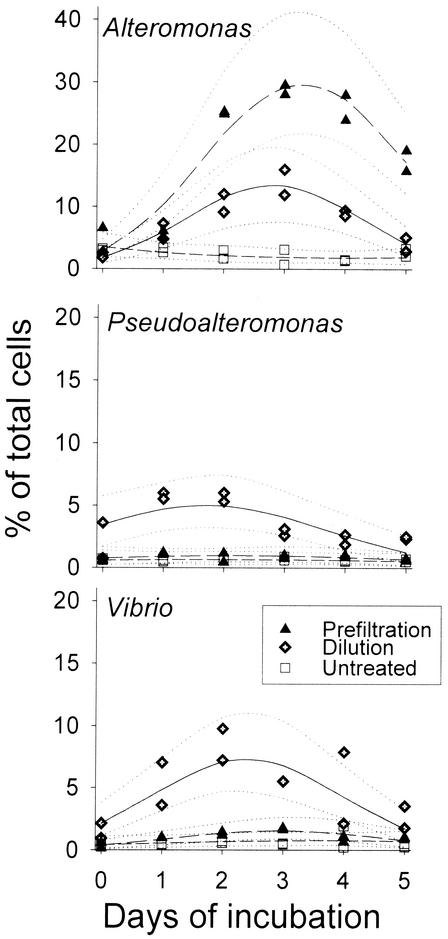
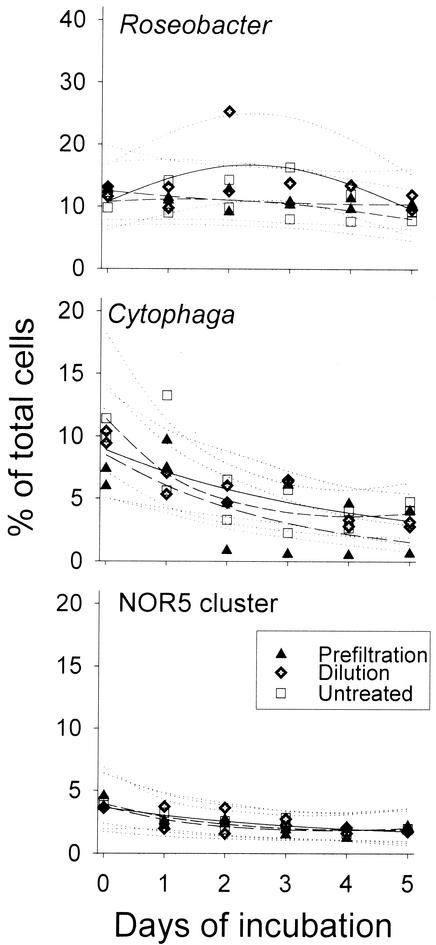
Treatment-specific differences in the temporal development of bacterial abundances were compared by likelihood ratio tests. Of the studied lineages, only bacteria affiliated with Alteromonas significantly responded to both experimental manipulations (P < 0.001; Fig. 3 and Table 2). In the prefiltration treatment this group temporarily constituted almost 30% of all bacteria but declined significantly by the end of the experiment to < 20%. In the dilution treatments, a significant transient rise in community contribution was also observed for populations related to Pseudoalteromonas and Vibrio (Table 2 and Fig. 3). No treatment-specific response was detected for Roseobacter spp., bacteria related to the C. latercula-C. marinoflava lineage, or for members of the NOR5 clade (Fig. 4 and Table 2). The community contribution of these groups either remained constant or gradually declined in all variants.
FIG. 3.
Relative contributions of the genera Alteromonas, Pseudoalteromonas, and Vibrio to the total microbial assemblages in the different treatments. The solid lines represent the population development as modeled by logistic regression and the dotted lines depict the upper and lower limits of the 95% confidence intervals.
TABLE 2.
Results of a two-step likelihood ratio test for significant treatment-specific differences in the studied bacterial groupsa
| Bacterium detected by probe |
Pb
|
|||
|---|---|---|---|---|
| Step 1 (DL=PR=UN) | Step 2
|
|||
| DL=PR | DL=UN | PR=UN | ||
| ALT1413 | <0.0005 | <0.0005 | <0.0005 | <0.0005 |
| PSA184 | <0.0005 | <0.0005 | <0.0005 | 0.2865 |
| GV822 | <0.0005 | <0.0005 | <0.0005 | 0.0350 |
| CYT1448 | 0.0990 | |||
| NOR5-730 | 0.9460 | |||
| GRb626 | 0.1285 | |||
P values above the critical value indicate that the pooled data from several treatments can be equally well described by a single regression curve (i.e., there were no significant differences between treatments). Significant values of P are depicted in boldface, DL, dilution; PR, prefiltration; UN, untreated.
Bonferroni adjustment (critical P value = 0.0083).
FIG. 4.
Relative contributions of bacteria related to Roseobacter spp., C. latercula-C. marinoflava, and to the NOR5 clade to the total microbial assemblages in the different treatments.
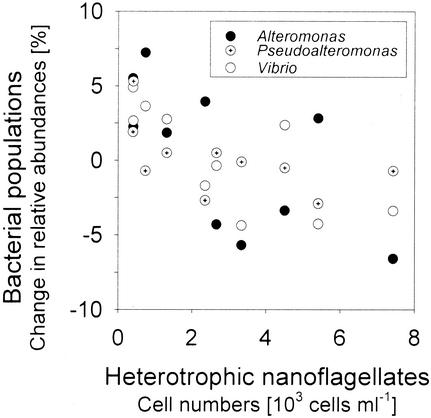
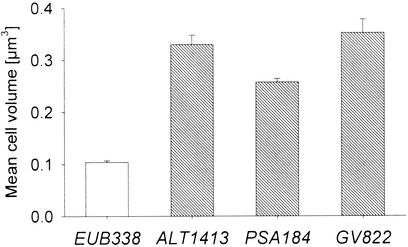
In the dilution treatments, significant negative correlations were found between the abundances of HNF and the changes in community contribution of bacteria of the genera Alteromonas (Spearman rank correlation coefficent rS = −0.66, P < 0.05), Pseudoalteromonas (rS = −0.63, P < 0.05), and Vibrio (rS = −0.81, P < 0.005) (Fig. 5). The mean cell volumes of bacteria related to Alteromonas, Pseudoalteromonas, and Vibrio spp. were significantly larger in these treatments at day 2 than the mean cell volume of bacteria hybridizing with the general probe EUB338 (Fig. 6).
FIG. 5.
Relationship between the cell numbers of heterotophic nanoflagellates and the changes in relative abundances of Alteromonas, Pseudoalteromonas, and Vibrio spp. in the dilution treatments. Negative y-axis values indicate a decrease in community contribution.
FIG. 6.
Mean cell volume (± standard error) of the total FISH-stained bacterial assemblage and of individual populations in the dilution treatments at day 2. The FISH probes were as follows: EUB338 I to III, all bacteria; ALT1413, Alteromonas spp.; PSA143, Pseudoalteromonas spp.; and GV822, Vibrio spp.
DISCUSSION
Incubation of seawater filtrates in bottles.
All six microbial groups studied include culturable strains (6, 7, 19), but there is a conspicuous ecological difference between them. Alteromonas, Pseudoalteromonas, and Vibrio spp. are usually rare (<1% of all cells) in coastal North Sea bacterioplankton (5, 6). In contrast, members of the genus Roseobacter, bacteria related to the C. latercula-C. marinoflava lineage, and members of the recently cultured NOR5 clade can all seasonally form substantial populations in the German Bight (7, 29). This dichotomy was also reflected in the responses of the different groups to our experimental manipulations (Fig. 3 and 4). As predicted by our hypothesis, the rare culturable γ-proteobacteria were only enriched in treatments with reduced predator encounter rates and not in the untreated variants (Fig. 3).
However, we caution against oversimplification of our results. In 1925 Tansley and Adamson (49) described the invasion and dominance of rare plant species in chalk grassland after the fencing out of rabbits and thus discovered the potential importance of grazing to prey (i.e., plant) community diversity. Although our bottle incubations of filtered and diluted seawater share some elements with this classic study, our experiments also differ in several important features. For one, the physicochemical contact with the environment is much more drastically cut in such manipulations than by a fence, which would probably be more adequately modeled by a dialysis membrane (4, 44). This separation in itself potentially causes a substantial change of the nutrient and substrate conditions, e.g., by sedimentation of particulate matter (46). Confinement of unmanipulated seawater may stimulate the growth of aquatic microbial assemblages (39), as was also reflected in the initial increase of bacterial abundances in the untreated variants (Fig. 1). More importantly, filtration is known to increase the overall activity of microbial communities (8, 42) and release dissolved organic matter, e.g., from breaking algal cells (18). A 10-fold dilution with 0.2-μm-pore-size filtrate thus probably caused a substrate input that stimulated the growth of the studied γ-proteobacterial groups in these treatments during the initial days of the incubations.
In fact, both Pseudoalteromonas and Vibrio spp. have been shown to require additional substrates for growth in 0.8-μm filtrates of North Sea water, whereas members of the genus Alteromonas could be readily enriched in unamended incubations (5). Eilers et al. postulated an ecological difference between these three groups of opportunistically growing bacteria because of their contrasting threshold substrate concentrations required for growth. This was confirmed by our observations. Even if no accidental release or transformation of substrates by filtration is assumed, a 10-fold dilution with ambient seawater will necessarily result in reduced interspecific competition between bacterial populations and higher substrate availability per microbial cell. The growth of Pseudoalteromonas and Vibrio spp. in the dilution but not in the prefiltration treatments (Fig. 3) thus provides further evidence that these bacteria indeed form larger populations only at increased substrate concentrations.
Other lines of evidence also indicate that the population increase of Alteromonas, Pseudoalteromonas, and Vibrio spp. after 24 h of bottle incubation must in part be a reaction to a sudden change in substrate availability. In cocultures, members of the genus Pseudoalteromonas outcompeted other marine bacteria at rapidly changing substrate conditions due to its shorter lag phase (30) but were disadvantaged when exposed to flatter substrate gradients. Strains of Alteromonas, Pseudoalteromonas, and Vibrio spp. conserved high levels of ribosomes also during extended periods of nongrowth (5, 30), which is most probably a physiological precondition for an opportunistic, “feast-and-famine” life strategy (9). In summary, the rapid growth response of Alteromonas, Pseudoalteromonas, and Vibrio spp. and other γ-proteobacteria in the dilution or prefiltration treatment cannot be unambiguously assigned to the experimental reduction of grazing mortality, but it is most probably also related to an increased substrate input.
Size-selective HNF predation.
In contrast, there is much clearer evidence for top-down, i.e., predation control of Alteromonas, Pseudoalteromonas, and Vibrio populations during the second half of the incubation period. Our study might provide the first experimental evidence from marine waters for a larger conceptual model about the impact of size-selective grazing on microbial community structure that has been continuously refined during the past few years (21, 22, 24, 25, 32, 33, 44, 45). This concept is based on the observation that there are marked differences in the average cell sizes of various bacterial species at comparable growth rates. Many bacterivorous marine and freshwater HNF are strongly size-selective feeders (20, 43). Bacterial species within a particular cell size range (ca. 1 to 4 μm of the cell length) are thus expected to suffer overproportional loss rates. For example, Hahn and Höfle (21, 22) present a qualitative model that predicts an increase of such “medium-size” bacteria only in the absence of grazing pressure.
Bottle incubations of North Sea water filtrates or dilutions typically induce both an increase in the community contribution of large γ-proteobacteria and an increase of the mean community cell size (protein content) (5, 10) (cf. Table 1 and Fig. 2). Bacteria affiliated with the genera Alteromonas, Pseudoalteromonas, and Vibrio constituted almost three quarters of all γ-proteobacteria in the dilution and prefiltration treatments. Members of these groups in the dilution treatments were significantly larger than the community average at day 2 (Fig. 6) and fall well in the preferred cell size range for many HNF (24). The significantly overproportional decline of the three γ-proteobacterial populations (Fig. 3) in the dilution or prefiltration treatments between days 3 and 5 is, therefore, likely a consequence of size-selective grazing mortality. It is, moreover, clearly paralleled by a decrease in the mean per-cell protein content (a proxy for cell size) of the bacterial community in the dilution treatment during HNF regrowth (Fig. 1 and 2). A connection between the fate of the bacterial groups and the predator population in the dilution treatment is also suggested by the significant negative correlations between changes in the community contributions of Alteromonas, Pseudoalteromonas, and Vibrio spp. and the abundances of HNF (Fig. 5). In these groups high HNF abundances can thus be related to losses that significantly exceed the overall bacterial mortality rate, i.e., declining relative abundances during a phase of declining total cell numbers (Fig. 1 and 3). This indicates that selective mortality was acting upon cells from the three γ-proteobacterial genera in the dilution treatment during the period of high HNF densities and declining total bacterial abundances (Fig. 1).
Our results apparently contradict the findings of Schäfer et al. (39), who reported an increase of A. macleodii-related 16S rDNA sequences in clone libraries after a grazing-induced decline of a bacterial bloom in a mesocosm of Mediterranean coastal waters. However, the bulk of A. macleodii-related sequence types in that study were obtained either before the effects of grazing were visibly affecting bacterial abundances or else after the collapse of the predator population. Moreover, Cottrell and Kirchman (2) caution against a misinterpretation of qualitative diversity information as a measure of population sizes. In view of our results, we must thus disagree with the conclusion of Schäfer et al. about the grazing resistence of Alteromonas spp. That study nevertheless indicates that transient blooms of Alteromonas sp.-like bacteria as a response of food web manipulations may also occur in other coastal areas.
The most likely alternative explanation we are aware of is the selective elimination of the studied bacterial groups by viral infection (12). However, there are currently no methods available to study specific host-pathogen interactions in mixed assemblages. Moreover, aquatic viruses considered to be highly host specific (50) (e.g., many species of Pseudoalteromonas bacteriophages isolated from coastal North Sea waters were able to lyse only a small number of strains from this lineage [51]). To account for a decline of three bacterial groups in the dilution treatment (Fig. 3), we should therefore postulate the synchronized outbreak of at least three distinct viral populations. Such an assumption clearly violates the “Occam's Razor” principle, since the activity of one single size-selectively foraging predator species can explain the observed population dynamics equally well.
In the words of Tansley and Adamson (49), “The general conclusions are very much what might have been expected from the more or less vague knowledge we already possessed.” Our results indicate that some HNF species that are present in North Sea waters and that pass through filters with a nominal pore size of 0.8 μm can efficiently suppress planktonic blooms of Alteromonas, Vibrio, and Pseudoalteromonas spp. and thus potentially also suppress, for example, pathogenic Vibrio species (23). This process might in part be responsible for the typically low population densities of these readily culturable microbial genera in coastal marine picoplankton.
Acknowledgments
We thank Gunnar Gerdts and Antje Wichels for their continuous support on Helgoland, Martha Schattenhofer and Handana Hirhaj for help in the lab, and Barbara MacGregor for critical reading of the manuscript. The Biologische Anstalt Helgoland of the Alfred Wegener Institute for Polar and Marine Research, Bremerhaven, Germany, is acknowledged for providing lab facilities.
This work was supported by the European Union (EVK3-2001-00194 BASICS) and by the Max Planck Society.
REFERENCES
- 1.Button, D. K., B. R. Robertson, P. W. Lepp, and T. M. Schmidt. 1998. A small, dilute-cytoplasm, high-affinity, novel bacterium isolated by extinction culture and having kinetic constants compatible with growth at ambient concentrations of dissolved nutrients in seawater. Appl. Environ. Microbiol. 64:4467-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 4.Del Giorgio, P. A., J. M. Gasol, D. Vaque, P. Mura, S. Agusti, and C. M. Duarte. 1996. Bacterioplankton community structure: protists control net production and the propoportion of active bacteria in a coastal marine community. Limnol. Oceanogr. 41:1169-1179. [Google Scholar]
- 5.Eilers, H., J. Pernthaler, and R. Amann. 2000. Succession of pelagic marine bacteria during enrichment: a close look on cultivation-induced shifts. Appl. Environ. Microbiol. 66:4634-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eilers, H., J. Pernthaler, J. Peplies, F. O. Glöckner, G. Gerdts, and R. Amann. 2001. Isolation of novel pelagic bacteria from the German Bight and their seasonal contribution to surface picoplankton. Appl. Environ. Microbiol. 67:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson, R. L., E. N. Buckley, and A. V. Palumbo. 1984. Response of marine bacterioplankton to differential filtration and confinement. Appl. Environ. Microbiol. 47:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flärdh, K., P. S. Cohen, and S. Kjelleberg. 1992. Ribosomes exist in large excess over the apparent demand for protein synthesis during carbon starvation in marine Vibrio sp. strain CCUG 15956. J. Bacteriol. 174:6780-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs, B. M., M. V. Zubkov, K. Sahm, P. H. Burkill, and R. Amann. 2000. Changes in community composition during dilution cultures of marine bacterioplankton as assessed by flow cytometric and molecular biological techniques. Environ. Microbiol. 2:191-201. [DOI] [PubMed] [Google Scholar]
- 11.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1992. Novel major archaebacterial group from marine plankton. Nature 356:148-149. [DOI] [PubMed] [Google Scholar]
- 12.Fuhrman, J. A., and R. T. Noble. 1995. Viruses and protists cause similar bacterial mortality in coastal seawaters. Limnol. Oceanogr. 40:1236-1242. [Google Scholar]
- 13.Garcia-Martinez, J., S. G. Acinas, R. Massana, and F. Rodriguez-Valera. 2002. Prevalence and microdiversity of Alteromonas macleodii-like microorganisms in different oceanic regions. Environ. Microbiol. 4:42-50. [DOI] [PubMed] [Google Scholar]
- 14.Gasol, J. M., and X. A. G. Morán. 1999. Effects of filtration on bacterial activity and picoplankton community structure as assessed by flow cytometry. Aquat. Microb. Ecol. 16:251-264. [Google Scholar]
- 15.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 16.Giuliano, L., E. De Domenico, M. G. Höfle, and M. M. Yakimov. 1999. Identification of culturable oligotrophic bacteria within naturally occurring bacterioplankton communities of the Ligurian Sea by 16S rRNA sequencing and probing. Microb. Ecol. 37:77-85. [DOI] [PubMed] [Google Scholar]
- 17.Glöckner, F. O., R. Amann, A. Alfreider, J. Pernthaler, R. Psenner, K. Trebesius, and K.-H. Schleifer. 1996. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst. Appl. Microbiol. 19:403-406. [Google Scholar]
- 18.Goldman, J. C., and M. R. Dennett. 1985. Susceptibility of some marine-phytoplankton species to cell breakage during filtration and post-filtration rinsing. J. Exp. Mar. Biol. Ecol. 86:47-58. [Google Scholar]
- 19.Gonzalez, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez, J. M., E. B. Sherr, and B. F. Sherr. 1990. Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl. Environ. Microbiol. 56:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahn, M. W., and M. G. Hofle. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35:113-121. [DOI] [PubMed] [Google Scholar]
- 22.Hahn, M. W., and M. G. Höfle. 1999. Flagellate predation on a bacterial model community: interplay of size-selective grazing, specific bacterial cell size, and bacterial community composition. Appl. Environ. Microbiol. 65:4863-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heidelberg, J. F., K. B. Heidelberg, and R. R. Colwell. 2002. Seasonality of Chesapeake Bay bacterioplankton species. Appl. Environ. Microbiol. 68:5488-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jürgens, K., and C. Matz. 2002. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 81:413-434. [DOI] [PubMed] [Google Scholar]
- 25.Jürgens, K., J. Pernthaler, S. Schalla, and R. Amann. 1999. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl. Environ. Microbiol. 65:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 27.Massana, R., C. Pedros-Alio, E. O. Casamayor, and J. M. Gasol. 2001. Changes in marine bacterioplankton phylogenetic composition during incubations designed to measure biogeochemically significant parameters. Limnol. Oceanogr. 46:1181-1188. [Google Scholar]
- 28.Monger, B. C., and M. R. Landry. 1991. Prey-size dependency of grazing by free-living marine flagellates. Mar. Ecol. Prog. Ser. 74:239-248. [Google Scholar]
- 29.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pernthaler, A., J. Pernthaler, H. Eilers, and R. Amann. 2001. Growth patterns of two marine isolates: adaptations to substrate patchiness? Appl. Environ. Microbiol. 67:4077-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 32.Pernthaler, J., T. Posch, K. Simek, J. Vrba, R. Amann, and R. Psenner. 1997. Contrasting bacterial strategies to coexist with a flagellate predator in an experimental microbial assemblage. Appl. Environ. Microbiol. 63:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pernthaler, J., T. Posch, K. Simek, J. Vrba, A. Pernthaler, F. O. Glöckner, U. Nübel, R. Psenner, and R. Amann. 2001. Predator-specific enrichment of actinobacteria from a cosmopolitan freshwater clade in mixed continuous culture. Appl. Environ. Microbiol. 67:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinhassi, J., U. L. Zweifel, and A. Hagström. 1997. Dominant marine bacterioplankton species found among colony-forming bacteria. Appl. Environ. Microbiol. 63:3359-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Posch, T., M. Loferer-Krossbacher, G. Gao, A. Alfreider, J. Pernthaler, and R. Psenner. 2001. Precision of bacterioplankton biomass determination: a comparison of two fluorescent dyes and of allometric and linear volume-to-carbon conversion factors. Aquat. Microb. Ecol. 25:55-63. [Google Scholar]
- 36.Posch, T., J. Pernthaler, A. Alfreider, and R. Psenner. 1997. Cell-specific respiratory activity of aquatic bacteria studied with the tetrazolium reduction method, cyto-clear slides and image analysis. Appl. Environ. Microbiol. 63:867-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 38.Sanders, R. W., D. A. Caron, and U. G. Berninger. 1992. Relationship between bacteria and heterotrophic nanoplankton in marine and freshwaters: an inter-ecosystem comparison. Mar. Ecol. Prog. Ser. 86:1-14. [Google Scholar]
- 39.Schäfer, H., P. Servais, and G. Muyzer. 2000. Successional changes in the genetic diversity of a marine assemblage during confinement. Arch. Microbiol. 173:138-145. [DOI] [PubMed] [Google Scholar]
- 40.Sherr, B. F., E. B. Sherr, and C. Pedros Alio. 1989. Simultaneous measurement of bacterioplankton production and protozoan bacterivory in estuarine water. Mar. Ecol. Prog. Ser. 54:209-219. [Google Scholar]
- 41.Sherr, E. B., and B. F. Sherr. 1993. Protistan grazing rates via uptake of fluorescently labelled prey, p. 695-701. In P. Kemp, B. F. Sherr, E. B. Sherr, and J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis, Boca Raton, Fla.
- 42.Sherr, E. B., B. F. Sherr, and C. T. Sigmon. 1999. Activity of marine bacteria under incubated and in situ conditions. Aquat. Microb. Ecol. 20:213-223. [Google Scholar]
- 43.Simek, K., and T. H. Chrzanowski. 1992. Direct and indirect evidence of size-selective grazing on pelagic bacteria by freshwater nanoflagellates. Appl. Environ. Microbiol. 58:3715-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simek, K., J. Pernthaler, M. Weinbauer, K. Hornak, J. Dolan, J. Nedoma, M. Masin, and R. Amann. 2001. Changes in bacterial community composition, dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67:2723-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simek, K., J. Vrba, J. Pernthaler, T. Posch, P. Hartman, J. Nedoma, and R. Psenner. 1997. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl. Environ. Microbiol. 63:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon, M., H. P. Grossart, B. Schweitzer, and H. Ploug. 2002. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28:175-211. [Google Scholar]
- 47.Sonnemann, E. 1982. Allgemeine Lösungen multipler Testprobleme. EDV Med. Biol. 13:120-128. [Google Scholar]
- 48.Suzuki, M. T. 1999. Effect of protistan bacterivory on coastal bacterioplankton diversity. Aquat. Microb. Ecol. 20:261-272. [Google Scholar]
- 49.Tansley, A. G., and R. S. Adamson. 1925. Studies of the vegetation of the English chalk III: the chalk grasslands of Hampshire-Sussex border. J. Ecol. 13:177-223. [Google Scholar]
- 50.Thingstad, T. F. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 51.Wichels, A., S. S. Biel, H. R. Gelderblom, T. Brinkhoff, G. Muyzer, and C. Schütt. 1998. Bacteriophage diversity in the North Sea. Appl. Environ. Microbiol. 64:4128-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. A. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulfoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]
- 53.Zubkov, M. V., B. M. Fuchs, H. Eilers, P. H. Burkill, and R. Amann. 1999. Determination of total protein content of bacterial cells by SYPRO staining and flow cytometry. Appl. Environ. Microbiol. 65:3251-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]