Abstract
Pseudomonas strains with an atypical LOPAT profile (where LOPAT is a series of determinative tests: L, levan production; O, oxidase production; P, pectinolitic activity; A, arginine dihydrolase production; and T, tobacco hypersensibility) can be regarded as emergent pathogens in the Principality of Asturias (Spain), where they have been causing, since 1999, severe damage in at least three taxonomically unrelated orchard plants of agronomic importance: common bean (Phaseolus vulgaris), kiwifruit (Actinidia deliciosa), and lettuce (Lactuca sativa). These strains are mainly differentiated by production of yellowish mucoid material in hypersucrose medium, used for the levan test, and by a variable pectinolytic activity on different potato varieties. The atypical organisms were identified as Pseudomonas viridiflava based on their 16S rRNA sequences. Among them a certain intraspecies genetic heterogeneity was detected by randomly amplified polymorphic DNA (RAPD) typing. To differentiate between isolates of P. viridiflava and Pseudomonas syringae pathovars, a 16S ribosomal DNA restriction fragment length polymorphism method employing the restriction endonucleases SacI and HinfI was developed. This could be used as a means of reliable species determination after the usual phenotypical characterization, which includes the LOPAT tests.
The phytopathogenic oxidase-negative fluorescent Pseudomonas species have been traditionally grouped into two species, P. syringae and P. viridiflava (17), the former including more than 40 well-characterized pathovars. The term pathovar refers to strains grouped at the subspecies level on the basis of plant host range and symptoms and with the aid of biochemical tests (8, 25). Of these, the LOPAT determinative tests (L, levan production; O, oxidase production; P, pectinolytic activity; A, arginine dihydrolase production; and T, tobacco hypersensibility) (10) is widely applied to differentiate isolates. Although of practical interest, bacterial phenotypical characterization alone often fails to reveal genetic relationships within or between species. In fact, DNA hybridization analyses quite early revealed that P. syringae is a heterogeneous species (18), which is still subject to extensive revision (6, 21).
The Principality of Asturias (PA) is a region on the north coast of Spain in which agriculture is mainly centered in orchard plants and fruit trees. In the Phytopathology Laboratory of the Regional Service of Agrofood Research and Development of the PA (LPPA), P. syringae pv. syringae and P. syringae pv. phaseolicola (also named P. savastanoi pv. phaseolicola or P. amygdali) (16) have been identified as pathogens of plants with high agronomic value. Of these, P. syringae pv. syringae was found to be the pathovar displaying a wider host range, causing severe bacteriosis in practically all orchard plants under culture in Asturias as well as in fruit trees, whereas P. syringae pv. phaseolicola has been only collected from common bean plants with disease symptoms (7). P. viridiflava, rarely found in the PA, has been traditionally considered to be an epiphyte or opportunistic pathogen (11, 2). Since 1999, a new Pseudomonas type with an atypical LOPAT profile (convex colonies with uncharacteristic yellowish mucoid material in hypersucrose medium [L test]; O negative; P variable; A negative; and T positive) has been frequently isolated from and associated with disease in common bean plants. Later, it has also appeared in material from other plants with disease symptoms, including kiwifruits (from spring of 2000) and lettuce (from 2001). The aim of the present study was to identify the phytopathogenic Pseudomonas with the atypical LOPAT profile which can be regarded as emergent in the PA. The species was identified by sequencing of the DNA encoding 16S rRNA as P. viridiflava, the pathogenicity was verified according to Koch's postulates, and the genetic types causing disease were traced by randomly amplified polymorphic DNA (RAPD) segment analysis. In addition, a genetic procedure based on restriction fragment length polymorphism (RFLP) analysis of the 16S rRNA genes was developed to differentiate P. viridiflava from P. syringae isolates.
MATERIALS AND METHODS
Bacterial strains.
In this study 31 isolates belonging to the fluorescent Pseudomonas group and displaying an atypical LOPAT profile were analyzed. They were collected from common bean (19), kiwifruits (10), and lettuce (2) plants with disease symptoms from different orchard sites of the PA (Table 1). The PA is a coastal Cantabrian region with a wet and mild climate where the three mentioned species are important crops. In addition, other bacteria were included as reference or outgroup strains, including seven strains previously identified in the LPPA as P. viridiflava, P. syringae pv. syringae, or P. syringae pv. phaseolicola (one, three, and three strains, respectively), and three collection strains: P. viridiflava CECT 458 (ATCC 13223), P. syringae pv. syringae CECT 4429 (ATCC 19310), and P. syringae pv. phaseolicola CECT 321 (ATCC 19304).
TABLE 1.
Origin and relevant features of Pseudomonas strains analyzed in this work
| Strain/yr | Sample origin
|
Resultd
|
16S rDNA RFLP
|
RAPD pattern | Species | ||||
|---|---|---|---|---|---|---|---|---|---|
| Bost | Plant sample | Orchard locality | L test | P test | SacI | HinfI | |||
| LPPA 74/99 | Bean | Seed | Navia | + [y] | + | S-1 | H-2 | R-2 | P. viridiflava |
| LPPA 76/99 | Bean | Seed | Navia | + [y] | − | S-1 | H-2 | R-2 | P. viridiflava |
| LPPA 77/99 | Bean | Seed | Navia | + [y] | − | S-1 | H-2 | R-2 | P. viridiflava |
| LPPA 78/99 | Bean | Seed | Navia | + [y-g] | + | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 80/99 | Bean | Seed | Navia | + [y-g] | − | S-1 | H-2 | R-2 | P. viridiflava |
| LPPA 81/99 | Bean | Pod | Tapia | + [y] | v | S-1 | H-2 | R-2 | P. viridiflava |
| LPPA 83/00 | Bean | Pod | Villaviciosa | + [y-g] | v | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 84/00a,b,c | Bean | Pod | Villaviciosa | v | − | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 88/00 | Bean | Stem | Vegadeo | + [y] | + | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 89/01 | Bean | Seed | Valdés | + [y-g] | − | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 91/01 | Bean | Seed | Valdés | + [y] | − | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 94/01 | Bean | Seed | Vegadeo | + [y] | + | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 249/01 | Bean | Pod | Tineo | + [y] | − | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 252/01 | Bean | Pod | Tineo | + [y] | + | S-1 | H-2 | R-2 | P. viridiflava |
| LPPA 280/01 | Bean | Seed | Pravia | + [y-g] | − | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 287/02 | Bean | Seed | Valdés | + [y] | − | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 288/02a,b | Bean | Seed | Valdés | + [y] | + | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 290/02 | Bean | Seed | Valdés | + [y] | + | S-1 | H-2 | R-6 | P. viridiflava |
| LPPA 319/02 | Bean | Leaf | Villaviciosa | + [y] | − | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 102/00 | Kiwifruit | Leaf | Villaviciosa | + [y] | − | S-1 | H-2 | R-3 | P. viridiflava |
| LPPA 103/00 | Kiwifruit | Leaf | Villaviciosa | + [y] | + | S-1 | H-2 | R-3 | P. viridiflava |
| LPPA 118/00 | Kiwifruit | Leaf | Villaviciosa | + [y] | − | S-1 | H-2 | R-4 | P. viridiflava |
| LPPA 125/00 | Kiwifruit | Leaf | Villaviciosa | + [y] | + | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 126/00 | Kiwifruit | Leaf | Villaviciosa | + [y] | + | S-1 | H-2 | R-5 | P. viridiflava |
| LPPA 139/00a,b | Kiwifruit | Bud | Villaviciosa | + [y] | v | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 143/00a,b | Kiwifruit | Bud | Grado | + [y] | v | S-1 | H-2 | R-2 | P. viridiflava |
| LPPA 144/00a,b | Kiwifruit | Bud | Pravia | + [y] | + | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 147/00 | Kiwifruit | Bud | Llanes | + [y] | + | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 150/00 | Kiwifruit | Leaf | Llanes | + [y] | + | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 264/02a,b | Lettuce | Leaf | Villaviciosa | + [y] | + | S-1 | H-2 | R-1 | P. viridiflava |
| LPPA 265/02a,b | Lettuce | Leaf | Villaviciosa | + [y] | + | S-1 | H-2 | R-2 | P. viridiflava |
| Control strains | |||||||||
| CECT 458a,b | − | ++ | S-1 | H-2 | R-1 | P. viridiflava | |||
| CECT 4429a | + [w] | − | S-2 | H-1 | R-10 | P. syringae pv. syringae | |||
| CECT 321a | + [w] | − | S-2 | H-1 | R-20 | P. syringae pv. phaseolicola | |||
| LPPA 79/99 | Bean | Seed | Navia | − | + | S-1 | H-2 | R-2 | P. viridiflava |
| LPPA 86/00 | Bean | Pod | Vegadeo | + [w] | − | S-2 | H-1 | R-11 | P. syringae pv. syringae |
| LPPA 93/01 | Bean | Seed | Vegadeo | + [w] | − | S-2 | H-1 | R-12 | P. syringae pv. syringae |
| LPPA 136/00 | Kiwifruit | Leaf | Villaviciosa | + [w] | − | S-2 | H-1 | R-13 | P. syringae pv. syringae |
| LPPA 87/00 | Bean | Pod | Valdés | + [w] | − | S-2 | H-1 | R-20 | P. syringae pv. phaseolicola |
| LPPA 244/01 | Bean | Pod | Valdés | + [w] | − | S-2 | H-1 | R-20 | P. syringae pv. phaseolicola |
| LPPA 245/01 | Bean | Pod | Villayón | + [w] | − | S-2 | H-1 | R-20 | P. syringae pv. phaseolicola |
Strain tested for pathogenicity.
Strain whose 16S rDNA has been sequenced.
Polysaccharide-positive and-negative variants were tested for patogenicity.
Abbreviations and symbols: y, yellow; y-g, yellow-greenish; w, white; −, negative for the indicated feature; +, positive for the indicated feature; ++, strong pectinolytic activity; v, variable result.
Isolation procedure and phenotypic characterization of Pseudomonas strains.
Samples of seeds, stems, leaves, and pods from beans; floral buds from kiwifruits; and lettuce leaves with disease symptoms, collected at different times over the period 1999 to 2001, from different orchard sites (Table 1), were analyzed for bacteriosis. The bacterial isolation procedure on King's medium B was used (9). Fluorescent bacteria obtained on this medium were tested for biochemical traits, including Hugh-Leifson reaction; LOPAT profile; sculine and gelatin hydrolysis; and mannitol, erythritol, sorbitol, m-inositol, adonitol, betaine, homoserine, trigonelline, d-tartrate, and quinate assimilation in Ayers's minimal medium (16). Stability of the LOPAT profile was tested after successive subcultures in King's medium B. Isolates showing identical features, and collected from the same orchard site at the same period of the year, were considered to be a single strain.
Pathogenicity tests.
The virulence of different Pseudomonas strains, collected from different host plants and showing the atypical LOPAT pattern (Table 1), were tested for Koch's postulates, using the plant from which they were originally collected as the host. The inoculations were performed by spraying bacterial suspensions (106 and/or 109 CFU/ml) on sets of bean and lettuce plantules (10 plantules/set), or 6 to 10 buds of kiwifruit. P. syringae pv. syringae LPPA 86, P. syringae pv. phaseolicola LPPA 87, and P. viridiflava CECT 458 were tested in the same way. Beans and lettuce plant sets were maintained at 22°C with a light period of 16 h/day, covered with a transparent plastic bag during the initial 48 h. Kiwifruit buds in the field were also kept enclosed in plastic bags for 48 h. Assays were repeated at least twice. When disease symptoms appeared, part of the damaged tissues was processed and aliquots were plated onto King's medium B. After incubation (48 h at 25°C) fluorescent bacteria were recovered and subjected to biochemical and genetic tests (see below). All inoculated material was autoclaved before being discarded.
PCR amplification of 16S ribosomal DNA (rDNA).
Almost-full-length 16S rRNA genes were amplified from all the isolates with an atypical LOPAT pattern and control strains compiled in Table 1, using the pA and pH′ primers designed by Edwards et al. (5) for eubacteria. Amplifications were done in a MJ Research (Waltham, Mass.) PTC 100 PCR system, using 50-μl volumes that contained 1 μl of DNA extracted according to the method of Deener and Boychuck (3), 0.3 μM concentrations of each primer, a 0.2 mM concentration of each deoxynucleoside triphosphate, 2 U of DyNAZyme II DNA polymerase (Finnzymes Oy, Espoo, Finland), and 5 μl of the supplier's reaction buffer. Volume was made up to 50 μl with sterile double-distilled water. After a 3-min denaturation step at 94°C, the reaction mixture was run through 30 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 90 s, followed by a final step at 72°C during 10 min. The ca. 1,500-bp PCR products were purified with the Ultraclean PCR clean up DNA purification kit (MO-BIO, Inc.), as recommended by the manufacturer, and used directly for sequencing analysis and/or for RFLP profiling (see below).
16S rDNA sequencing and sequence analysis.
Selected PCR fragments, amplified from the isolates tested for pathogenicity and from the control strain P. viridiflava CECT 458, were sequenced in both strands, with the pA and pH′ primers by the Servicio de Secuenciación de DNA, Centro de Investigaciones Biológicas-Consejo Superior de Investigaciones Científicas, Madrid, Spain, using the BigDye Terminator cycle sequencing ready reaction FS kit and an ABI PRISM 3700 DNA sequencer (both from Applied Biosystems Div., Foster City, Calif.). To determine their phylogenetic affiliation, sequences were initially compared to the available databases by using BLAST (1). Phylogenetic trees were generated by neighbor joining, using Jukes-Cantor corrected distances, within the RDP-II PHYLIP interface (12). The accession numbers of the 16S rDNA sequences used for comparison were as follows: P. aeruginosa LMG 1242T, Z76651; P. amygdali LMG 2123T, Z76654; P. cichorii LMG 2162T, Z76658; P. fluorescens DSM 50090T, Z76662; P. marginalis LMG 2210T, Z76663; P. syringae pv. actinidiae, AB001439; P. syringae pv. maculicola, AB001444; P. syringae pv. morsprunorum, AB001445; P. syringae pv. phaseolicola, AB001448; P. syringae pv. theae, AB0001450; P. syringae pv. syringae LMG 1247 t1T, Z76669; and P. viridiflava LMG 2352T, Z76671 (14, 20).
RFLP and RAPD procedures.
Genetic types were traced by RFLP analysis of almost-entire 16S rRNAs genes and by RAPD typing of total genomic DNA of the bacteria under study. For RFLP analysis, 16S rDNA sequences generated in this work, together with relevant Pseudomonas sequences retrieved from databases, were examined by using the MAP-SORT program of the University of Wisconsin Genetics Computer Group. In this way, SacI and HinfI restriction enzymes were selected to distinguish between species. Restriction digestions were performed on 10 μl of PCR product, with enzymes supplied by Amersham Biosciences (Barcelona, Spain), in accordance with the manufacturer's instructions. RAPD typing of genomic DNA was carried out as described by Soto et al. (22), using random primer S (TCACGATGCA) (23).
Nucleotide sequence accession numbers.
Sequences generated in this work have been deposited in GenBank under accession numbers AY180967 to AY180972.
RESULTS
Phenotypic characterization of phytopathogenic Pseudomonas isolates with an atypical LOPAT profile.
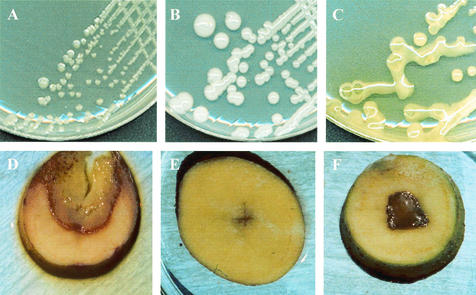
Since 1999, fluorescent bacteria with an atypical LOPAT profile (Table 1) have been isolated from plant parts of common beans (seeds, leaves, stems, and pods), lettuces (leaves), and kiwifruits (floral buds) with disease symptoms. Essentially, they were differentiated by the L test and their pectinolytic activity. They produced raised colonies with mucoid material of yellowish color (occasionally greenish) on hypersucrose medium, in contrast to the typical white mucoid material associated with levan producer strains, such as P. syringae pathovars (Fig. 1A to C). The mucoid material was not detected in sucrose-deficient medium. The strains showed a variable pectinolytic activity when assayed on potato slices, with rotting halos ranging from 0 up to about 10 mm surrounding two streaks inoculated in a cross pattern (Fig. 1D to F). This rotting capacity was remarkably lower than that of the control P. viridiflava strain. With respect to other LOPAT tests, the isolates were oxidase negative, arginine dihydrolase negative, and tobacco hypersensibility positive. However, the last of these characteristics, as well as production of the yellowish mucus, could be lost after subculture of some of the isolates in King's medium B but recovered after inoculation of the bacteria in the host plant. In contrast, the P. syringae pv. syringae strains used as controls remained levan positive after subculture. This LOPAT profile (which does not exactly fit that of any of the recognized phytopathogenic Pseudomonas spp.) is closest to those expected either for P. syringae or P. viridiflava. The newly isolated bacteria showed other biochemical properties in common (including oxidation of glucose in Hugh-Leifson medium, hydrolysis of esculin and gelatin, utilization of the same set of carbon compounds), which are all shared by the above-mentioned species.
FIG. 1.
Differential LOPAT tests of atypical Pseudomonas isolates. Growth on hypersucrose medium (L test) (A to C) and pectinolytic activity on potato slices (D to F) are shown. (A) P. viridiflava CECT 458 (negative control); (B) P. syringae pv. syringae CECT 4429 (positive control); (C) atypical P. viridiflava LPPA 144; (D) P. viridiflava CECT 458 (positive control); (E) P. syringae pv. syringae CECT 4429 (negative control); (F) atypical P. viridiflava LPPA 144.
Pathogenicity studies.
Symptoms observed in plants from which the atypical Pseudomonas strains were isolated displayed a considerable variation. In common beans, they ranged from the appearance of strikingly red spots, mainly in petioles and pods, to plant death due to systemic infection and associated to destruction of the medulla. In lettuce, soft rot of some of the leaves could also progress and result in plant death. In kiwifruits infection was characterized by the appearance of dark brown spots in floral buds that developed into extensive rot, leading to the dropping of most buds or to production of small and distorted fruits. To prove that disease symptoms were due to the atypical Pseudomonas, the virulence of seven isolates (two from common beans, two from lettuce, and three from kiwifruit) (Table 1) was tested through inoculation on plant sets of their original host as described in Materials and Methods. All the isolates were able to experimentally reproduce the symptoms of natural infection in their respective hosts, although those that have lost the ability to produce exopolysaccharide after subculture (LPPA 84/00) required longer. In general, the damage caused by the atypical isolates was significantly higher than that caused by the control P. syringae strains, whereas P. viridiflava CECT 458 only produced severe damage in kiwifruit floral buds, as expected.
Special attention was paid to the simultaneous presence of bacteria other than atypical Pseudomonas which could either contribute to the observed unspecific damage or even be the only responsible for disease development. P. syringae pv. syringae or P. syringae pv. phaseolicola were isolated from some of the bean's samples, P. syringae pv. syringae from some of the affected kiwifruits and Erwinia from some of the lettuces, although always in association with atypical Pseudomonas. In addition, the latter was the only bacteria collected from 28, 65, and 67% of bean, kiwifruit, and lettuce samples under investigation.
Genotypic identification of the Pseudomonas isolates.
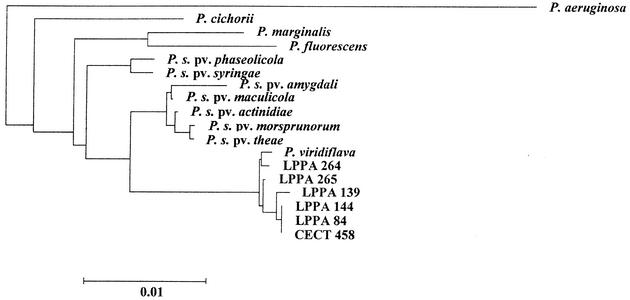
Nearly complete 16S rDNAs from the control strains P. syringae CECT 4429 and P. viridiflava CECT 458 and from six isolates with the atypical LOPAT pattern (Table 1), previously tested for pathogenicity, were sequenced after amplification. Analysis of the 16S rDNA sequences revealed that, independent of their origin, those from the atypical isolates were nearly identical (more than 99.9% identity over 1,439 nucleotides), and that they were most closely related to 16S rDNA sequences from several strains of P. viridiflava (including LMG2352 and CECT 458). Moreover, the sequence corresponding to hypervariable (hv) region 2, proposed by Moore et al. (14) as a signature for P. viridiflava, was found in the 16S rRNA genes of the new isolates. Also represented were the P. viridiflava sequences in hv regions 1 and 3, which distinguish this species from P. syringae or from P. fluorescens and some, but not all, of the P. syringae pathovars, respectively (14). In fact, with respect to P. syringae, higher identity values were observed with 16S rDNAs from strains of the P. actinidiae, P. maculicola, P. morsprunorum, and P. theae pathovars (more than 98% identity), all sharing hv region 3 with P. viridiflava. Finally, clustering of the atypical isolates 16S rDNA sequences with two control P. viridiflava sequences is revealed by the phylogenetic tree depicted in Fig. 2.
FIG. 2.
Inferred phylogenetic relationships among the atypical isolates and validly described members of the genus Pseudomonas (sensu stricto). See Materials and Methods for accession numbers and details on the construction of the tree.
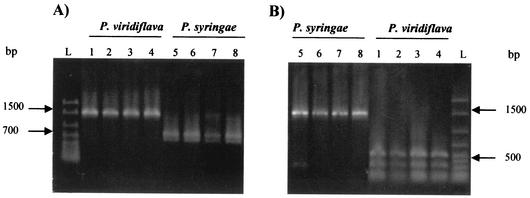
Comparisons of the sequences generated in this work with those of relevant Pseudomonas strains obtained from databases led to the identification of an additional hv region within the 16S rDNA of P. viridiflava strains, including the atypical isolates described in this work. It is located within the V5 region, helix 27 (corresponding to Escherichia coli 16S rRNA positions 829 to 857), described by Neefs et al. (15). This region contains a HinfI site which is absent from the rDNA of P. syringae pathovars, including those of P. actinidiae, P. maculicola, P. morsprunorum, and P. theae, as well as P. syringae and P. phaseolicola. Moreover, the equivalent region from these pathovars contains a single SacI site, which is absent from P. viridiflava. A differential HinfI site is also present in hv region 3 of P. viridiflava strains as well as in the P. syringae pathovars most closely related to P. viridiflava (see above). However it is absent from P. syringae pv. syringae and P. syringae pv. phaseolicola. Such differential restriction sites were used to develop an RFLP ribotyping method that allows a rapid identification of the atypical isolates when combined with phenotypical tests. In fact, SacI and HinfI digestions of 16S rRNA genes amplified with pA and pH′ from all bacteria depicted in Table 1 confirmed this approach. Examples of SacI and HinfI digestions of 16S rDNAs from relevant strains are shown in Fig. 3.
FIG. 3.
Restriction profiles of PCR-amplified 16S rDNA fragments from different phytopathogenic Pseudomonas. (A) Digestion with SacI; (B) digestion with HinfI. Lane L, molecular size standards; lanes 1 to 3, RFLP profiles corresponding to atypical isolates of P. viridiflava; lane 4, P. viridiflava CECT 458; lane 5, P. syringae pv. syringae LPPA 93/01; lane 6, P. syringae pv. syringae LPPA 136/00; lane 7, P. syringae pv. syringae CECT 4429; lane 8, P. syringae pv. phaseolicola CECT 321.
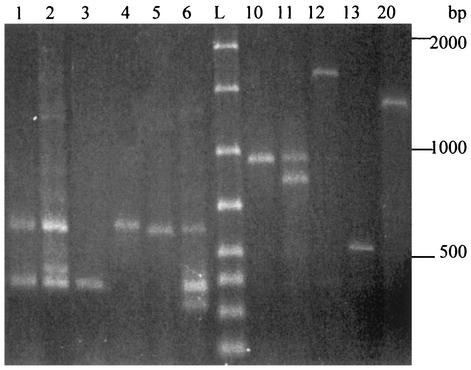
Finally, in order to gain some insight into a possible genetic heterogeneity of the pathogenic P. viridiflava atypical isolates, RAPD typing with primer S was performed. In this way P. viridiflava organisms could be discriminated into at least six profiles (R-1 to R-6). Most of the strains, including the control P. viridiflava CECT 548, belong to R-1 (Fig. 4). In contrast, each of the P. syringae control strains generated a distinctive RAPD profile. The distribution of all bacteria under study into RAPD profiles is compiled in Table 1.
FIG. 4.
RAPD typing of atypical Pseudomonas isolates performed with primer S. Lanes 1 to 6, profiles R-1 to R-6, respectively, generated by atypical P. viridiflava isolates; lanes 10 to 13, R profiles corresponding to P. syringae pv. syringae strains; lane 20, R profile of P. syringae pv. phaseolicola; lane L, molecular size standards. The distribution of the isolates within different profiles is shown in Table 1.
DISCUSSION
P. viridiflava strains are usually differentiated from P. syringae by their pathogenicity and by two of the biochemical tests represented in the LOPAT scheme. Whereas the first organism is considered as an epiphytic or opportunistic pathogen, the second, apart from being epiphytic, includes true pathogenic pathovars that cause different types of damage (such as necrosis, blights, rots, and spots) in a variety of plants. With respect to the LOPAT tests, P. viridiflava strains are L negative (i.e., they produce flat colonies on hypersucrose medium) and P positive (showing strong pectinolytic activity on potato slices). In contrast, P. syringae pathovars are L positive (forming convex colonies developing a strong white mucoid material on levan medium) and P negative. The emergent type described here shows a distinctive activity in both tests, with colonies characterized by a yellowish, rarely greenish, mucoid material in the L test, and a variable pectinolytic activity, ranging from negative or very weak to relatively strong (although it is generally weaker than that of the control P. viridiflava CECT 458 strain). It should be noted that the nature of the exopolysaccharide released by the atypical bacteria remains unknown, although its production in hypersucrose medium, but not in the same medium lacking sucrose, strongly suggests that it could be levan.
The species identification of the atypical phytopathogenic Pseudomonas isolates was achieved by analysis of the nucleotide sequences of 16S rRNA genes from representative isolates. Moore et al. (14) have previously demonstrated the potential of 16S rRNA sequence analyses to distinguish the species of the genus Pseudomonas, as well as for the establishment of phylogenetic lineages within the genus. In addition, sequence comparisons led to the identification of hypervariable regions that can be regarded as signatures for a certain species. In this work, similar experiments revealed the affiliation of the atypical isolates to P. viridiflava (Fig. 2). Accordingly, all signatures allowing discrimination of P. viridiflava from closely related species (mainly the P. syringae pathovars and the P. fluorescens lineage [14, 17]) were found in the 16S rRNA genes from the new isolates. On the other hand, RAPD typing proved to be a useful tool to differentiate P. syringae from P. viridiflava, and also for differentiation within P. viridiflava.
To the best of our knowledge, the new type of P. viridiflava reported in this investigation has not been previously described as a phytopathogen. However, remarkable damage on kiwifruit during the flowering phase caused by typical P. viridiflava isolates has been reported (13, 24). According to Young et al. (24) the population of the pathogen surviving epiphytically on the plant could represent an important source of inoculum and may cause damage under particular environmental conditions. Thus, mild temperatures, frequent rainfalls, and high relative humidity values would facilitate their spread. These weather conditions are frequently found in the PA, where the atypical P. viridiflava is causing severe damage, with the accompanying economical loss. With respect to the origin of this lineage, one can speculate that an L negative P. viridiflava strain could have recently gained the ability to produce exopolysaccharide on hypersucrose medium, which may contribute to the epiphytic fitness of the strains or even function as a virulence factor (19, 4). Alternatively, erroneous laboratory interpretations of the L test, which could have been considered to be positive based only on the appearance of convex mucous colonies, but ignoring pigmentation, cannot be ruled out. If so, pathogenic strains related to P. viridiflava could have been claimed to be P. syringae. In fact, this possibility has been already indicated by Lelliott (11). The RFLP method developed in the present investigation, using 16S rDNA digestions with selected endonucleases (SacI and HinfI) might constitute a rapid and accurate genetic procedure to aid phenotypical characterization in the identification of the atypical isolates.
Acknowledgments
This research was supported by project INIA-SC00-026 of the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (Madrid, Spain).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Andrés, M. F., F. García-Arenal, M. M. López, and P. Melgarejo. 2000. Manual de laboratorio, p. 526. Ministerio de Agricultura, Pesca y Alimentación, Madrid, Spain.
- 3.Deener, H. G., and I. Boychuck. 1991. Species-specific detection of Listeria monocytogenes by DNA amplification. Appl. Environ. Microbiol. 57:606-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denny, T. P. 1995. Involvement of bacterial polysaccharides in plant pathogenesis. Annu. Rev. Phytopathol. 33:173-197. [DOI] [PubMed] [Google Scholar]
- 5.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardan, L., H. Shafik, S. Belouin, R. Broch, F. Grimont, and P. A. D. Grimont. 1999. DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959). Int. J. Syst. Bacteriol. 49:469-478. [DOI] [PubMed] [Google Scholar]
- 7.González, A. J., E. Landeras, and M. C. Mendoza. 2000. Pathovars of Pseudomonas syringae causing bacterial brown spot and halo blight in Phaseolus vulgaris L. are distinguishable by ribotyping. Appl. Environ. Microbiol. 66:850-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hildebrand, D. C., H. N. Schroth, and D. C. Sands. 1988. Pseudomonas, p. 60-80. In N. W. Schaad (ed.), Laboratory guide for identification of plant pathogenic bacteria, 2nd ed. American Phytopathological Society, St. Paul, Minn.
- 9.King, E. D., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 10.Lelliott, R. A., E. Billing, and A. C. Hayward. 1966. A determinative scheme for fluorescent plant pathogenic bacteria. J. Appl. Bacteriol. 29:470-478. [DOI] [PubMed] [Google Scholar]
- 11.Lelliott, R. A. 1992. Pseudomonas viridiflava, p. 190. In I. M. Smith, J. Dunez, R. A. Lelliott, D. H. Phillips, and S. A. Archer (ed.), Manual de enfermedades de las plantas. Mundi-Prensa, Madrid, Spain.
- 12.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mansilla, J. P., and A. Abelleira. 1999. Presencia de Pseudomonas marginalis y Pseudomonas viridiflava sobre kiwi en Galicia. Bol. San. Veg. Plagas 25:175-180. [Google Scholar]
- 14.Moore, E. R. B., M. Mau, A. Arnscheidt, E. C. Böttger, R. A. Hutson, M. D. Collins, Y. Van de Peer, R. de Wachter, and K. N. Timmis. 1996. The determination and comparison of the 16s rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of the natural intrageneric relationships. Syst. Appl. Microbiol. 19:478-492. [Google Scholar]
- 15.Neefs, J.-M., Y. Van de Peer, L. Hendriks, and R. de Wachter. 1990. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 18:2237-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noval, C. 1991. Manual de laboratorio, p. 379-410. Ministerio de Agricultura, Pesca y Alimentación, Madrid, Spain.
- 17.Palleroni, N. J. 1984. Genus I. Pseudomonas Migula 1894, 237, p. 141-199. In N. J. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 18.Pecknold, P. C., and R. G. Grogan. 1973. Deoxyribonucleic acid homology groups among phytopathogenic Pseudomonas species. Int. J. Syst. Bacteriol. 23:111-121. [Google Scholar]
- 19.Rudolf, K., M. Gross, F. Ebrahim-Nesbat, M. Noellenburg, and A. Zomorodian. 1994. The role of extracellular polysaccharides as virulence factors for phytopathogenic pseudomonads and xanthomonads, p. 357-378. In C. I. Kado and J. H. Corsa (ed.), Molecular mechanisms of bacterial virulence. Kluwer Academic Press, Dordrecht, The Netherlands.
- 20.Sawada, H., S. Kanaya, M. Tsuda, F. Suzuki, K. Azegami, and N. Saitou. 2002. A phylogenetic study of the OCTase genes in Pseudomonas syringae pathovars: the horizontal transfer of the argK-tox cluster and the evolutionary history of OTCase genes on their genomes. J. Mol. Evol. 54:437-457. [DOI] [PubMed] [Google Scholar]
- 21.Schaad, N. W., A. K. Vidaver, G. H. Lacy, K. Rudolph, and J. B. Jones. 2000. Evaluation of proposed amended names of several pseudomonads and xanthomonads and recommendations. Phytopathology 90:208-213. [DOI] [PubMed] [Google Scholar]
- 22.Soto, S. M., B. Guerra, M. A. Gónzález-Hevia, and M. C. Mendoza. 1999. Potential of a three-way randomly amplified polymorphic DNA analysis as a typing method within twelve Salmonella serotypes. Appl. Environ. Microbiol. 65:4830-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams, J. G. K., A. R. Kubelin, K. J. Livak, J. A. Rafalski, and S. V. Tingey. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young, J. M., G. J. Cheesmur, F. V. Welham, and W. R. Henshall. 1988. Bacterial blight of kiwifruit. Ann. Appl. Biol. 112:91-105. [Google Scholar]
- 25.Young, J. M., Y. Takikawa, L. Gardan, and D. E. Stead. 1992. Changing concepts in the taxonomy of plant pathogenic bacteria. Annu. Rev. Phytopathol. 30:67-105. [Google Scholar]