Abstract
In plants, chalcones are precursors for a large number of flavonoid-derived plant natural products and are converted to flavanones by chalcone isomerase or nonenzymatically. Chalcones are synthesized from tyrosine and phenylalanine via the phenylpropanoid pathway involving phenylalanine ammonia lyase (PAL), cinnamate-4-hydroxylase (C4H), 4-coumarate:coenzyme A ligase (4CL), and chalcone synthase (CHS). For the purpose of production of flavanones in Escherichia coli, three sets of an artificial gene cluster which contained three genes of heterologous origins—PAL from the yeast Rhodotorula rubra, 4CL from the actinomycete Streptomyces coelicolor A3(2), and CHS from the licorice plant Glycyrrhiza echinata—were constructed. The constructions of the three sets were done as follows: (i) PAL, 4CL, and CHS were placed in that order under the control of the T7 promoter (PT7) and the ribosome-binding sequence (RBS) in the pET vector, where the initiation codons of 4CL and CHS were overlapped with the termination codons of the preceding genes; (ii) the three genes were transcribed by a single PT7 in front of PAL, and each of the three contained the RBS at appropriate positions; and (iii) all three genes contained both PT7 and the RBS. These pathways bypassed C4H, a cytochrome P-450 hydroxylase, because the bacterial 4CL enzyme ligated coenzyme A to both cinnamic acid and 4-coumaric acid. E. coli cells containing the gene clusters produced two flavanones, pinocembrin from phenylalanine and naringenin from tyrosine, in addition to their precursors, cinnamic acid and 4-coumaric acid. Of the three sets, the third gene cluster conferred on the host the highest ability to produce the flavanones. This is a new metabolic engineering technique for the production in bacteria of a variety of compounds of plant and animal origin.
In plants, the phenylpropanoid pathway is responsible for the synthesis of a wide variety of secondary metabolic compounds, including lignins, salicylates, coumarins, hydroxycinnamic amides, pigments, and flavonoids. In addition to their roles in the structure and protection of the plant, phenylpropanoid compounds have an important effect on plant qualities, such as texture, flavor, color, and other characteristics (29). Recently, the flavonoid-derived plant natural products have drawn much attention because of their benefits to human health, such as antimicrobial, cancer chemopreventive, antioxidant, and antiasthmatic activities (6, 14). These phenylpropanoid and flavonoid biosynthetic enzymes are therefore attractive targets for metabolic engineering to provide enhancement or initiation of the production of economically desirable traits or compounds. The phenylpropanoid and flavonoid biosynthetic pathways and their regulation have been the subject of many studies (4, 5, 25, 29). Recent advances in the regulation of the pathways and the biochemistry of specific enzymes and enzyme complexes have opened up strategies for the enhancement of flavonoid compounds by modifying the pathways (4).
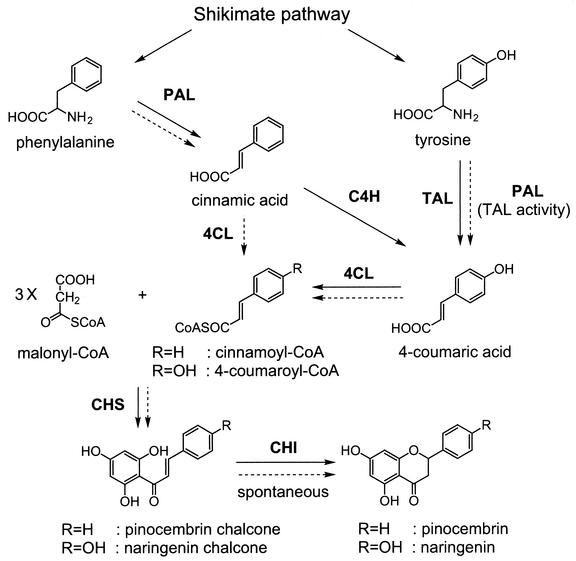
The phenylpropanoid pathway in plants converts phenylalanine into naringenin chalcone (Fig. 1). As the first step, phenylalanine is deaminated to yield cinnamic acid by the action of phenylalanine ammonia lyase (PAL). Cinnamic acid is hydroxylated by cinnamate-4-hydroxylase (C4H) to 4-coumaric acid, which is then activated to 4-coumaroyl-coenzyme A (CoA) by the action of 4-coumarate:CoA ligase (4CL). Chalcone synthase (CHS) catalyzes the stepwise condensation of three acetate units from malonyl-CoA with 4-coumaroyl-CoA to yield naringenin chalcone, the precursor for a large number of flavonoids (29). Naringenin chalcone is converted to naringenin by chalcone isomerase or nonenzymatically in vitro (5, 19, 24, 25). Because some of the PALs show tyrosine ammonia lyase activity, tyrosine is also used as the precursor (2, 15, 22, 26).
FIG. 1.
Flavanone biosynthetic pathway in plants. The dashed arrows represent the expected flavanone biosynthetic pathway in E. coli containing the artificial gene cluster including PAL, 4CL, and CHS. TAL, tyrosine ammonia-lyase; CHI, chalcone isomerase.
Production of flavanones or flavonoids by genetically engineered bacteria has not been reported before, although the heterologous expression of phenylpropanoid biosynthetic enzymes in bacteria has been reported (1, 8, 16, 30). One of the barriers for the production of these compounds is the difficulty in expression of active C4H, which would not be efficiently expressed due to its instability and the lack of its specific cytochrome P-450 reductase in bacteria (8, 20). We have recently discovered a 4CL in the gram-positive filamentous bacterium Streptomyces coelicolor A3(2) that can activate cinnamic acid to cinnamoyl-CoA, as well as 4-coumaric acid to 4-coumaroyl-CoA (12). The use of the bacterial 4CL enzyme would bypass the C4H step for the production of pinocembrin chalcone from phenylalanine via the phenylpropanoid pathway (Fig. 1). On the basis of this idea, we constructed an artificial gene cluster containing PAL from a yeast, Rhodotorula rubra; 4CL from an actinomycete, S. coelicolor A3(2); and CHS from a licorice plant, Glycyrrhiza echinata, to produce a plant-specific flavanone, pinocembrin. As expected, the Escherichia coli cells containing the gene cluster produced pinocembrin. In addition, the E. coli cells produced naringenin, because the yeast PAL enzyme also used tyrosine as a substrate and yielded 4-coumaric acid, as is found for many PALs (2, 22, 26), and the 4CL enzyme used cinnamic acid and 4-coumaric acid to yield the corresponding CoA thioester compounds. This paper describes successful production of plant-specific flavanones, pinocembrin and naringenin, in E. coli. Furthermore, we compared the yields of the flavanones by E. coli cells that contained the gene clusters in different organizations.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
E. coli JM109 and pUC18, purchased from Takara Biochemicals, were used for DNA manipulation (23). E. coli BL21(DE3) and pET26b, purchased from Novagen, were used for the production of flavanones.
DNA manipulation.
Restriction enzymes, T4 DNA ligase, and Taq DNA polymerase were purchased from Takara Biochemicals. Recombinant DNA techniques were described previously (23). PAL cDNA from R. rubra (4a) was obtained from S. Kawai, Tokyo University of Agriculture and Technology. CHS cDNA from G. echinata L. was from T. Akashi and S. Ayabe, Nihon University (T. Akashi, unpublished data). 4CL from S. coelicolor A3(2) was cloned and characterized in our laboratory (12). After the DNA manipulation, the absence of undesired alterations during PCR was checked by nucleotide sequencing on an automated nucleotide sequencer.
Construction of pET26b-3GS.
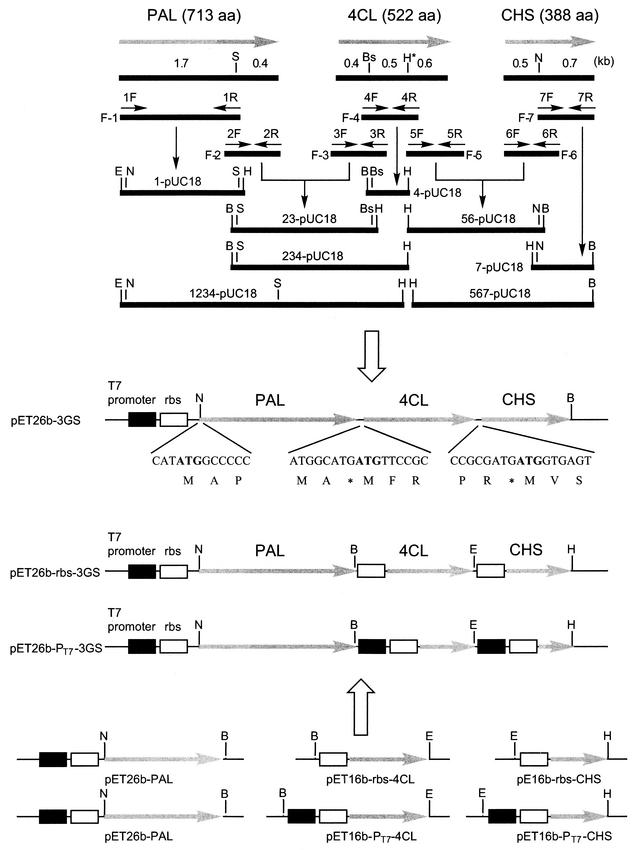
Plasmid pET26b-3GS was constructed by standard DNA manipulation, including several cycles of fragment-primed PCR as follows (Fig. 2). Using PAL, 4CL, or CHS cDNA as a template, seven DNA fragments were amplified by PCR with appropriate pairs of primers (Table 1). Fragments 1, 4, and 7 were cloned into pUC18 using EcoRI plus HindIII, BamHI plus HindIII, and HindIII plus BamHI, resulting in 1-pUC18, 4-pUC18, and 7-pUC18, respectively. Fragments 2 and 3 were connected by a fragment-primed PCR procedure and cloned between the BamHI and HindIII sites of pUC18, resulting in 23-pUC18. Fragments 5 and 6 were also connected by a fragment-primed PCR procedure and cloned between the HindIII and BamHI sites of pUC18, resulting in 56-pUC18. The absence of undesired alterations in PCR was confirmed by nucleotide sequencing. A 0.5-kb BstPI-HindIII fragment from 4-pUC18 was cloned between the corresponding sites of 23-pUC18, resulting in 234-pUC18. A 1.8-kb EcoRI-SplI fragment from 1-pUC18 was cloned between the corresponding sites of 234-pUC18, resulting in 1234-pUC18. A 0.7-kb NdeI-BamHI fragment from 7-pUC18 was cloned between the corresponding sites of 56-pUC18, resulting in 567-pUC18. Finally, both a 3.1-kb NdeI-HindIII fragment from 1234-pUC18 and a 1.8-kb HindIII-BamHI fragment from 567-pUC18 were cloned between the NdeI and BamHI sites of pET26b by three-fragment ligation, resulting in pET26b-3GS. In this construction, the start codons of 4CL and CHS overlapped the termination codons of the preceding genes.
FIG.2.
Schematic representation of the strategies used for construction of pET26b-3GS, pET26b-rbs-3GS, and pET26b-PT7-3GS. The following abbreviations are used for restriction enzymes: S, SplI; Bs, BstPI; H, HindIII; N, NdeI; E, EcoRI; and B, BamHI. A HindIII site (H∗) was created within the 4CL coding sequence without changing the amino acid sequence. By DNA manipulation, including several cycles of fragment-primed PCR, PAL encoding a 713-amino-acid (aa) protein, 4CL encoding a 522-aa protein, and CHS encoding a 388-aa protein are placed under the control of the T7 promoter and an RBS in a high-copy-number vector, pET26b. In pET26b-3GS, the start codons of 4CL and CHS are overlapped with the stop codons of the preceding genes. In pET26b-rbs-3GS, the RBS is placed in front of all three genes. In pET26b-PT7-3GS, both the RBS and the T7 promoter are placed in front of the three genes.
TABLE 1.
Primers used in this study

Refer to Fig. 2 for the position of each primer.
Underlining indicates restriction enzyme cleavage sites: GAATTC, EcoRI; CATATG, NdeI; AAGCTT, HindIII; CGTAGC, SplI; GGATCC, BamHI; and CCATGG, NcoI. Capital and lowercase letters indicate nucleotides matching the template sequence and nucleotides designed for mutagenesis, respectively. Boxes and shaded boxes indicate parts of the RBS and the T7 promoter (PT7), respectively. Boldface, start codon; italics, stop codon.
Construction of pET26b-rbs-3GS and pET26b-PT7-3GS.
An ∼0.4-kb 3′ fragment (nucleotide positions 587 to 713, taking the first nucleotide of the translational start codon of PAL as 1) of PAL was amplified by PCR using two primers, PAL-F-EcoRI and PAL-R-BamHI (Table 1), and pET-3GS as a template. The amplified fragment was cloned into pUC18 using the EcoRI and BamHI sites, resulting in pUC18-3′-fragment-PAL. An EcoRI-SplI fragment of 1234-pUC18 and an SplI-BamHI fragment of pUC18-3′-fragment-PAL was cloned between the EcoRI and BamHI sites of pUC18 by three-fragment ligation to create pUC18-PAL. PAL was excised from pUC18-PAL by using NdeI plus BamHI and cloned into pET26b, resulting in pET26b-PAL (Fig. 2). For construction of pET16b-4CL, 4CL was amplified by PCR from pUC18-4CL as a template with the two primers 4CL-F-NcoI and 4CL-R-BamHI. The amplified fragment was cloned into previously prepared pUC18N, which had an NcoI site instead of the original SmaI site of pUC18. 4CL was cloned into pET16b by using NcoI and BamHI, resulting in pET16b-4CL. For construction of pET16b-CHS, CHS was amplified by PCR from pUC18-CHS as a template with the two primers CHS-F-NcoI and CHS-R-BamHI. The amplified fragment was cloned into pUC18N and recombined into pET16b, resulting in pET16b-CHS. 4CL with the T7 promoter (PT7) and/or the ribosome-binding sequence (RBS) was then amplified by PCR from pET16b-4CL as a template with two primer sets, rbs-4CL-F-BamHI plus 4CL-R-EcoRI and PT7-4CL-F-BamHI plus 4CL-R-EcoRI, respectively. Both amplified fragments were cloned into pUC18. CHS with PT7 and/or RBS was similarly amplified (primer sets rbs-CHS-F-EcoRI plus CHS-R-HindIII and PT7-CHS-F-EcoRI plus CHS-R-HindIII, respectively) and cloned into pUC18. Finally, rbs-4CL and rbs-CHS or PT7-4CL and PT7-CHS were introduced into pET26b-PAL stepwise to construct pET26b-rbs-3GS and pET26b-PT7-3GS, respectively.
Expression and fermentation.
E. coli BL21(DE3) harboring pET26b-3GS, pET26b-rbs-3GS, or pET26b-PT7-3GS was precultured in Luria-Bertani liquid medium containing 5 μg of kanamycin/ml at 30°C for 16 h with reciprocal shaking. Isopropyl β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 1 mM. After an incubation period of 2 h at 26°C, the cells were harvested by centrifugation and disrupted by sonication to analyze the total proteins by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Soluble fractions were obtained by ultracentrifugation of the cell lysates.
For identification of compounds produced by E. coli harboring the expression plasmids or pET26b, a portion (20 ml) of the preculture was inoculated into 200 ml of M9 medium with 5 μg of kanamycin/ml and cultured for 5 h at 26°C in the presence of 1 mM IPTG. The cells were then harvested and washed once with M9 medium. A portion of the cells (500 mg [wet weight]) was transferred to 200 ml of fresh M9 medium with 5 μg of kanamycin/ml and 1 mM IPTG and cultivated at 26°C for 65 h with reciprocal shaking.
Extraction and analysis of flavanones.
The culture broth was prepared, and the pH was adjusted to pH 9.0 with NaOH. After the broth had stood at room temperature for 1 h, the materials in the broth were extracted with the same volume of ethyl acetate. The organic layer was evaporated to dryness, and the residue was dissolved in 100 ml of acetonitrile containing 0.1% acetic acid for high-performance liquid chromatography (HPLC) analysis on a Waters 600E chromatograph. The compounds produced were separated on a reversed-phase DOCOSIL-B column (C22; Senshu Scientific Co.), maintained at 40°C, by elution with an acetonitrile-water gradient, both containing 0.1% acetic acid, at a flow rate of 1.0 ml/min. The HPLC conditions were as follows: for detection of 4-coumaric acid, cinnamic acid, and naringenin, 20 to 30% CH3CN for 55 min and 30 to 100% CH3CN for 5 min; for detection of pinocembrin, 30 to 40% CH3CN for 10 min, 40% CH3CN for 30 min, and 40 to 100% CH3CN for 5 min. Absorbances at 309, 277, and 290 nm were monitored for 4-coumaric acid, cinnamic acid, and flavanones, respectively. The retention times of the authentic samples under these HPLC conditions were 8.67 (4-coumaric acid), 34.29 (cinnamic acid), 45.56 (naringenin), and 31.89 (pinocembrin) min. These authentic compounds, except for pinocembrin, were purchased from Sigma-Aldrich. Pinocembrin was a gift from H. Kuroda, Kyoto University.
LC-APCIMS.
Liquid chromatography-atmospheric pressure chemical ionization mass spectrometry (LC-APCIMS) was performed on a Thermo Quest LCQ apparatus equipped with a reversed-phase DOCOSIL-B column with detection at 254 and 290 nm under the same conditions as for HPLC. The negative ion values by LC-APCIMS and the retention times of the authentic compounds were as follows: 4-coumaric acid, m/z 163.3[M − H]−, 9.22 min; cinnamic acid, m/z 147.5[M − H]−, 33.76 min; naringenin, m/z 271.3[M − H]−, 44.67 min; pinocembrin, m/z 255.4[M − H]−, 31.01 min. The m/z values and retention times of the four compounds produced by E. coli harboring pET26b-3GS were identical to those of the respective authentic samples.
RESULTS
Construction and expression of an artificial gene cluster in E. coli.
We first designed a gene cluster in pET26b-3GS so that phenylalanine, a primary metabolite, was converted into pinocembrin chalcone by the successive actions of PAL, 4CL, and CHS in E. coli (Fig. 2). Pinocembrin chalcone was thought to be rapidly converted into pinocembrin by nonenzymatic isomerization under alkaline conditions (19). It was also expected that naringenin was produced at the same time, in addition to pinocembrin, because some PALs use tyrosine as the substrate to yield 4-coumaric acid (2, 22, 26) and 4CL from S. coelicolor A3(2) uses both 4-coumaric acid and cinnamic acid as substrates (12). Three genes of heterologous origins—PAL from a yeast, R. rubra; 4CL from an actinomycete, S. coelicolor A3(2); and CHS from a licorice plant, G. echinata—were placed in that order under the control of the T7 promoter in an E. coli vector, pET26b, resulting in pET26b-3GS (Fig. 2). An RBS, AAGGAGA, was present 7 nucleotides upstream of the start codon of PAL. The termination codons of PAL and 4CL were overlapped with the initiation codons of the following genes, on the assumption that these three genes were translationally coupled because of the absence of intervening sequences between the adjacent genes. In bacteria, translational coupling seems to be common in gene clusters, since the overlapping of a start codon and a stop codon between the two adjacent genes is often observed (18). We therefore expected efficient transcription and translation of the three genes in E. coli.
The cells were cultured at 26°C to avoid the formation of inclusion bodies of the proteins, since SDS-polyacrylamide gel electrophoretic analysis of the total proteins showed that, when grown at 37°C, a large amount of 78-kDa PAL was found in both the insoluble and soluble fractions (data not shown). Even by culturing at 26°C, a considerable amount of the 78-kDa PAL was still recovered in the insoluble fraction. On the other hand, no apparent increased production of proteins with molecular masses of 57 and 42 kDa, representing 4CL and CHS, respectively, was detectable by SDS-polyacrylamide gel electrophoresis. Because of the very low production of 4CL and CHS, we attached the RBS and/or the T7 promoter in front of each of the three genes, as described below.
Analysis of fermentation products.
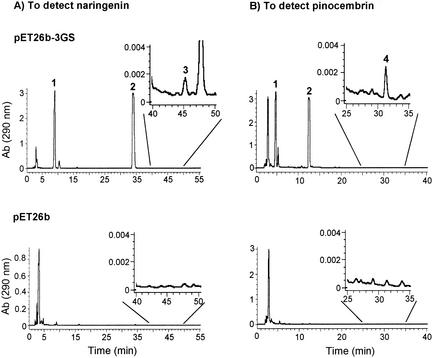
Chalcones are converted into flavanones by the action of chalcone isomerase (11) or nonenzymatically under alkaline conditions (19). After the fermentation, the culture broth was collected and the pH was adjusted to 9.0 to convert chalcones into the corresponding flavanones. Comparison by HPLC analysis of the fermentation products of the E. coli cells harboring pET26b-3GS and the vector pET26b revealed that four new compounds were reproducibly detected in the engineered strain (Fig. 3). The retention times of peaks 1, 2, 3, and 4 were identical to those of the authentic compounds 4-coumaric acid, cinnamic acid, naringenin, and pinocembrin, respectively.
FIG. 3.
HPLC analysis of the culture broth of E. coli harboring pET26b-3GS or pET26b. For detection of peaks 1, 2, and 3 (4-coumaric acid, cinnamic acid, and naringenin, respectively) and of peaks 1, 2, and 4 (4-coumaric acid, cinnamic acid, and pinocembrin, respectively), different elutions were used. See Materials and Methods for the details of the HPLC conditions. Ab, absorbance.
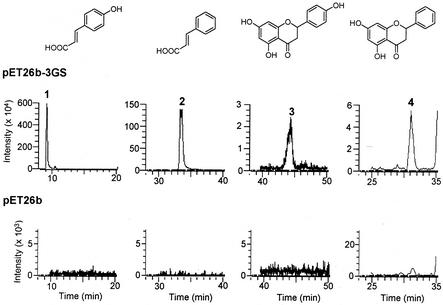
We further analyzed these compounds by HPLC coupled to LC-APCIMS in the negative-ion mode (Fig. 4). The major compounds (peaks 1 and 2) appeared at m/z 163.3[M − H]− and m/z 147.5[M − H]−, respectively, indicating that they were 4-coumaric acid and cinnamic acid, respectively. The minor compounds (peaks 3 and 4) appeared at m/z 271.3[M − H]− and m/z 255.4[M − H]−, respectively, indicating that they were naringenin and pinocembrin, respectively. No CoA compounds as the precursors of CHS were detected in the cell extracts of this strain.
FIG. 4.
Selected ion chromatograms by LC-APCIMS of the compounds produced by E. coli BL21(DE3) harboring pET26b-3GS (top) and pET26b (bottom). Peaks 1 to 4 were measured on the basis of m/z 162.5-163.5, 146.5-147.5, 270.5-271.5, and 254.5-255.5[M − H]−, respectively, obtained from the authentic samples. Peak identities: 1, 4-coumaric acid; 2, cinnamic acid; 3, naringenin; 4, pinocembrin. See Materials and Methods for the details of HPLC and LC-APCIMS protocols.
Feeding of amino acid precursors to the engineered E. coli cells.
When cultured in M9 medium with glucose as a carbon source, E. coli containing the artificial gene cluster accumulated large amounts of 4-coumaric acid and cinnamic acid (0.47 and 1.23 mg/liter) but only small amounts of the flavanones naringenin and pinocembrin (0.27 and 0.17 μg/liter, respectively) (Table 2). To increase the yields of these compounds, we added phenylalanine and tyrosine as the precursors. As expected, exogenous supply of phenylalanine to the culture at a final concentration of 2 mM (0.33 g/liter) enhanced the yields of cinnamic acid ∼10-fold and of pinocembrin 1.2-fold. The 10-fold decrease in the yields of 4-coumaric acid and naringenin can be explained by means of dominant use of phenylalanine as the starting material of the artificial pathway, as a result of which the yields of 4-coumaric acid and 4-coumaroyl-CoA, the substrates of 4CL and CHS, respectively, are decreased. A similar supply of 2 mM (0.36 g/liter) tyrosine enhanced the yields of coumaric acid 19-fold and of naringenin 2-fold. In this case, the yield of pinocembrin was decreased 3.5-fold, although that of cinnamic acid was not decreased. This is probably due to much higher activity of PAL toward phenylalanine than toward tyrosine.
TABLE 2.
Production of plant-specific flavanones and related compounds by E. coli BL21(DE3) harboring pET26b-3GS, pET26b-rbs-3GS, or pET26b-PT7-3GSa
| Plasmid and carbon source | Production (μg/liter) |
|||
|---|---|---|---|---|
| 4-Coumaric acid | Cinnamic acid | Naringenin | Pinocembrin | |
| pET26b-3GS | ||||
| Glc | 472 | 1,234 | 0.27 | 0.17 |
| Glc + Phe | 43 | 11,800 | 0.03 | 0.20 |
| Glc + Tyr | 8,990 | 1,960 | 0.57 | 0.05 |
| pET26b-rbs-3GS | ||||
| Glc | 334 | 905 | 12.1 | 10.9 |
| Glc + Phe | 68 | 5,550 | 2.5 | 8.7 |
| Glc + Tyr | 3,030 | 1,060 | 48.5 | 6.1 |
| pET26b-PT7-3GS | ||||
| Glc | 78 | 220 | 83.3 | 156.6 |
| Glc + Phe | 36 | 3,640 | 9.6 | 751.9 |
| Glc + Tyr | 2,130 | 492 | 452.6 | 38.9 |
The recombinant E. coli cells were cultured in M9-glucose (Glc) medium supplemented with 2 mM phenylalanine (Phe) or tyrosine (Tyr). The amounts of the compounds were determined by integration of the peak areas in the HPLC profiles (Fig. 3) and comparison of the areas with those obtained from the definite amounts of the authentic samples. The values are the averages obtained from two independent experiments. The wavelengths measured for 4-coumaric acid, cinnamic acid, naringenin, and pinocembrin were 309, 277, 290, and 290 nm, respectively.
Effects of transcription and translation of the genes on the yields of flavanones.
The gene organization of PAL, 4CL, and CHS in pET26b-3GS mimicked those of the functionally related genes, such as for antibiotic biosynthesis and xenobiotics. Contrary to our expectation, however, the yields of the flavanones produced by the E. coli cells harboring pET26b-3GS were very low, perhaps due to apparently low production of 4CL and CHS, as suggested by SDS-polyacrylamide gel electrophoresis of the cell lysate. We therefore constructed pET26b-rbs-3GS and pET26b-PT7-3GS (Fig. 2) to assess the possible contribution of the RBS and the T7 promoter in front of each of the three genes to the production of 4CL and CHS and to the yields of flavanones. The three genes in pET26b-rbs-3GS were organized so that they contained their own RBS at appropriate positions and were transcribed by a single T7 promoter upstream of PAL. Those in pET26b-PT7-3GS were organized so that all three genes were separately under the control of both the T7 promoter and the RBS.
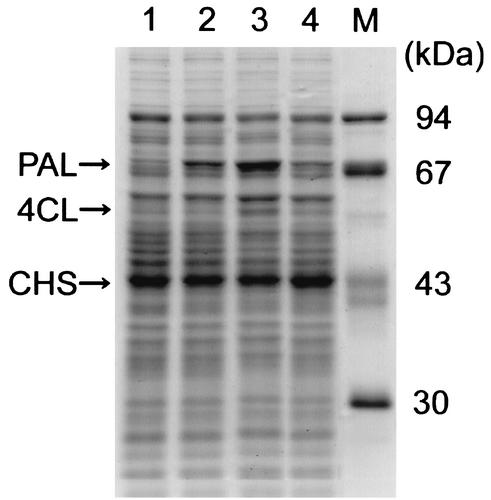
The soluble fractions of the E. coli cells harboring pET26b, pET26b-3GS, pET26b-rbs-3GS, or pET26b-PT7-3GS were analyzed by SDS-polyacrylamide gel electrophoresis (Fig. 5). As mentioned above, apparently increased production of a 78-kDa protein, probably representing PAL, was observed in E. coli harboring pET26b-3GS, but no increased production of 4CL (57 kDa) or CHS (42 kDa) was detectable. On the other hand, E. coli harboring pET26b-rbs-3GS or pET26b-PT7-3GS produced increased amounts of 78-, 57-, and 42-kDa proteins, although the degrees of increase varied depending on the culture. In the particular cultures shown in Fig. 5, increased production of the 42-kDa protein in lane 3 and the 78-kDa protein in lane 4 was not very clear. Our repeated experiments showed that E. coli harboring pET26b-PT7-3GS produced larger amounts of 4CL and CHS than E. coli harboring pET26b-3GS or pET26b-rbs-3GS. This observation is consistent with the yields of flavanones produced by E. coli harboring these plasmids, as described below.
FIG. 5.
SDS-polyacrylamide gel electrophoresis of the soluble fractions of E. coli harboring the expression plasmids. The soluble fractions of E. coli BL21(DE3) harboring pET26b as a control (lane 1), pET26b-3GS (lane 2), pET26b-rbs-3GS (lane 3), and pET26b-PT7-3GS (lane 4) were run, together with molecular mass markers (lane M).
We cultured the E. coli cells harboring pET26b-rbs-3GS or pET26b-PT7 -3GS and measured the amounts of the flavanones (Table 2). As expected, the recombinant E. coli cells produced much greater amounts of pinocembrin and naringenin than those harboring pET26b-3GS. Exogenous supply of the amino acid precursors phenylalanine and tyrosine further increased the yields. Addition of the RBS to the three genes (plasmid pET26b-rbs-3GS) enhanced the yields ∼45-fold for pinocembrin and ∼85-fold for naringenin, showing the importance of the presence of the RBS in front of each gene. This means that the anticipated translational coupling by means of overlap of the termination and initiation codons does not work in this case. Furthermore, addition of both the RBS and the T7 promoter in front of all three genes (plasmid pET26b-PT7-3GS) enhanced the yields of pinocembrin and naringenin 810- and 3,760-fold, respectively, compared to those produced by E. coli harboring pET26b-3GS. The amounts of 4-coumaric acid and cinnamic acid that remained in these culture broths were smaller than those in the culture broth of E. coli harboring pET26b-3GS, which showed that 4CL and CHS in increased amounts converted these precursors more efficiently in E. coli harboring pET26b-3GS or pET26b-rbs-3GS.
DISCUSSION
The present study clearly shows that the artificial gene cluster containing PAL, 4CL, and CHS directed the synthesis of the functionally active enzymes that convert phenylalanine to pinocembrin chalcone and tyrosine to naringenin chalcone. Production of the two chalcones can be ascribed to the substrate specificity of the enzymes (Fig. 1); the PAL from a yeast uses both phenylalanine and tyrosine as the substrates, the 4CL from an actinomycete uses both cinnamic acid and 4-coumaric acid, and CHS uses cinnamoyl-CoA and 4-coumaroyl-CoA as the starter. Our success in flavanone production in E. coli is due to the use of the actinomycete 4CL that can ligate CoA thioester to both cinnamic acid and 4-coumaric acid at almost the same efficiency (12). Without this 4CL, we had to overcome the difficulty in expressing functional C4H. C4H is a membrane-bound cytochrome P-450 hydroxylase that requires molecular oxygen and a reducing equivalent from NADPH delivered via cytochrome P-450 reductase (8, 20).
The organization of PAL, 4CL, and CHS in pET26b-3GS mimics those of the clusters of functionally related genes often found in microorganisms. However, the overlap of the termination and initiation codons appears not to lead to efficient translational coupling, because 4CL and CHS were produced in apparently larger amounts by E. coli harboring pET26b-rbs-3GS than by E. coli harboring pET26b-3GS. The finding showing the importance of the RBS for each gene in the cluster is useful for efficient gene expression of artificially organized gene clusters. The importance of the amount of mRNA is also evident, since the addition of the T7 promoter in front of each gene enhanced the amounts of the proteins and the yields of the flavanones.
We may increase the yields of the flavanones in several ways. An increase in the amount of malonyl-CoA by overexpressing the acetyl-CoA carboxylase gene (3, 17) is useful, because its concentration is only 4 to 90 μM (0.01 to 0.23 nmol/mg [dry weight]) (27). Control of the culture conditions, for example, by continuous feeding of glucose and the amino acid precursors, is also promising. Concerning the continuous feeding of the amino acids, genetically engineered hosts, in which the carbon flux is mainly toward the aromatic acid biosynthetic pathway (28), are useful. A mutant of Corynebacterium glutamicum, an amino acid fermenter that produces aromatic amino acids in large amounts, is a candidate for the host (10, 13).
Incorporation into the recombinant host of the genes that are required for the biosynthesis of flavonoids and isoflavonoids perhaps leads to production of these compounds that have been believed to be specific to plants. Once flavonoid and isoflavonoid skeletons are produced in bacteria, they can be further modified by so-called combinatorial biosynthesis (7, 9, 21), i.e., by introducing the genes whose products modify the skeletons, for example, by reduction, hydroxylation, and methylation at specific positions and by altering the substrate specificities of the gene products. Furthermore, assembly of genes of various origins can theoretically be applied to production of all the biologically produced compounds. Metabolic engineering techniques of this type can be combined with combinatorial biosynthesis for the production of a variety of compounds, including “unnatural” natural compounds.
Acknowledgments
E. I. Hwang was supported by the Japan Society for the Promotion of Science. This work was supported by a research grant from the Noda Institute for Scientific Research, by the Bio Design Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan (BDP-03-VI-2-1), and by a grant from the Industrial Technology Research Grant Program 2000 of the New Energy and Industrial Technology Development Organization of Japan (00A03004).
REFERENCES
- 1.Baedeker, M., and G. E. Schulz. 1999. Overexpression of a designed 2.2 kb gene of eukaryotic phenylalanine ammonia-lyase in E. coli. FEBS Lett. 457:57-60. [DOI] [PubMed] [Google Scholar]
- 2.Beaudoin-Eagan, L. D., and T. A. Thorpe. 1985. Tyrosine and phenylalanine ammonia lyase activities during shoot initiation in tobacco callus cultures. Plant Physiol. 78:438-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis, M. S., J. Solbiati, and J. E. Cronan, Jr. 2000. Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J. Biol. Chem. 275:28593-28598. [DOI] [PubMed] [Google Scholar]
- 4.Dixon, R. A., and C. L. Steele. 1999. Flavonoids and isoflavonoids—a gold mine for metabolic engineering. Trends Plant Sci. 4:394-400. [DOI] [PubMed] [Google Scholar]
- 4a.Filpula, D., C. A. Vaslet, A. Levy, A. Sykes, and R. L. Strausberg. 1988. Nucleotide sequence of gene for phenylalanine ammonia-lyase from Rhodotorula rubra. Nucleic Acids Res. 16:11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hahlbrock, K., and H. Grisebach. 1979. Enzymatic controls in the biosynthesis of lignin and flavonoids. Annu. Rev. Plant Physiol. 30:105-130. [Google Scholar]
- 6.Hollman, P. C., and M. B. Katan. 1998. Bioavailability and health effects of dietary flavonols in man. Arch. Toxicol. 20(Suppl.):237-248. [DOI] [PubMed] [Google Scholar]
- 7.Hopwood, D. A. 1997. Genetic contributions to understanding polyketide synthases. Chem. Rev. 97:2465-2497. [DOI] [PubMed] [Google Scholar]
- 8.Hotze, M., G. Schröder, and J. Schröder. 1995. Cinnamate 4-hydroxylase from Catharanthus roseus, and a strategy for the functional expression of plant cytochrome P450 proteins as translational fusions with P450 reductase in E. coli. FEBS Lett. 374:345-350. [DOI] [PubMed] [Google Scholar]
- 9.Hutchinson, C. R., and I. Fujii. 1995. Polyketide synthase gene manipulation: a structure-function approach in engineering novel antibiotics. Annu. Rev. Microbiol. 49:201-238. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda, M., K. Okamoto, and R. Katsumata. 1999. Cloning of the transketolase gene and the effect of its dosage on aromatic amino acid production in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 51:201-206. [DOI] [PubMed] [Google Scholar]
- 11.Jez, J. M., M. E. Bowman, R. A. Dixon, and J. P. Noel. 2000. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat. Struct. Biol. 7:786-791. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko, M., Y. Ohnishi, and S. Horinouchi. 2003. Cinnamate:coenzyme A ligase from the filamentous bacterium Streptomyces coelicolor A3(2). J. Bacteriol. 185:20-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katsumata, R., and M. Ikeda. 1993. Hyperproduction of tryptophan in Corynebacterium glutamicum by pathway engineering. Bio/Technology 11:921-925. [Google Scholar]
- 14.Knekt, P., R. Jarvinen, A. Reunanen, and J. Maatela. 1996. Flavonoid intake and coronary mortality in Finland: a cohort study. Br. Med. J. 312:478-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyndt, J. A., T. E. Meyer, M. A. Cusanovich, and J. J. Van Beeumen. 2002. Characterization of a bacterial tyrosine ammonia lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS Lett. 512:240-244. [DOI] [PubMed] [Google Scholar]
- 16.Lee, D., and C. J. Douglas. 1996. Two divergent members of a tobacco 4-coumarate:coenzyme A ligase (4CL) gene family. Plant Physiol. 112:193-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, S. J., and J. E. Cronan, Jr. 1992. The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J. Biol. Chem. 267:855-863. [PubMed] [Google Scholar]
- 18.Martin, J. F., and P. Liras. 1989. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 43:173-206. [DOI] [PubMed] [Google Scholar]
- 19.Mol, J. N. M., M. P. Robbins, R. A. Dixon, and E. Veltkamp. 1985. Spontaneous and enzymic rearrangement of naringenin chalcone to flavanone. Phytochemistry 24:2267-2269. [Google Scholar]
- 20.Pompon, D., B. Louerat, A. Bronine, and P. Urban. 1996. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 272:51-64. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez, E., and R. McDaniel. 2001. Combinatorial biosynthesis of antimicrobials and other natural products. Curr. Opin. Microbiol. 4:526-534. [DOI] [PubMed] [Google Scholar]
- 22.Rösler, J., F. Krekel, N. Amrhein, and J. Schmid. 1997. Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol. 113:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Sankawa, U., and T. Hakamatsuka. 1997. Biosynthesis of isoflavone and related compounds in tissue cultures of Pueraria lobata, p. 25-48. In K. Ogura and U. Sankawa (ed.), Dynamic aspects of natural products chemistry. Kodansha, Tokyo, Japan.
- 25.Schröder, J. 1997. A family of plant-specific polyketide synthases: facts and predictions. Trends Plant Sci. 2:373-378. [Google Scholar]
- 26.Scott, D. A., P. M. Hammond, G. M. Brearley, and C. P. Price. 1992. Identification by high-performance liquid chromatography of tyrosine ammonia-lyase activity in purified fractions of Phaseolus vulgaris phenylalanine ammonia-lyase. J. Chromatogr. B 573:309-312. [DOI] [PubMed] [Google Scholar]
- 27.Takamura, Y., and G. Nomura. 1988. Changes in the intracellular concentration of acetyl-CoA and malonyl-CoA in relation to the carbon and energy metabolism of Escherichia coli K12. J. Gen. Microbiol. 134:2249-2253. [DOI] [PubMed] [Google Scholar]
- 28.Tatarko, M., and T. Romeo. 2001. Disruption of a global regulatory gene to enhance central carbon flux into phenylalanine biosynthesis in Escherichia coli. Curr. Microbiol. 43:26-32. [DOI] [PubMed] [Google Scholar]
- 29.Weisshaar, B., and G. I. Jenkins. 1998. Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1:251-257. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi, T., F. Kurosaki, D. Y. Suh, U. Sankawa, M. Nishioka, T. Akiyama, M. Shibuya, and Y. Ebizuka. 1999. Cross-reaction of chalcone synthase and stilbene synthase overexpressed in E. coli. FEBS Lett. 460:457-461. [DOI] [PubMed] [Google Scholar]