Abstract
Eukaryotic repair of double-strand DNA breaks can occur either by homologous recombination or by nonhomologous DNA end joining (NHEJ). NHEJ relies on Ku70/86, XRCC4, DNA ligase IV, and DNA-dependent protein kinase. NHEJ involves a synapsis step in which the two ends are maintained in proximity, processing steps in which nucleases and polymerases act on the ends, an alignment step in which a few nucleotides of terminal homology guide the ends into preferred alignments, and a ligation step. Some of the steps, such as ligation, rely on a single enzymatic component. However, the processing steps begin and end with a wide array of alternative substrates and products, respectively, and likely involve multiple nucleases and polymerases. Given the alternative pathways that can be catalyzed by the remaining nucleases and polymerases, no one of these processing enzymes is likely to be essential. The only requirement for the processing enzymes, as a collective, is to generate a ligatable configuration, namely a ligatable nick on each strand. Here, we have tested the two major known 5′-specific nucleases of Saccharomyces cerevisiae for involvement in NHEJ. Whereas EXO1 does not appear to be involved to any detectable level, deleting RAD27 (FEN-1 of yeast) leads to a 4.4-fold reduction specifically of those NHEJ events predicted to proceed by means of 5′ flap intermediates. Because Rad27/FEN-1 acts specifically at 5′ flap structures, these results suggest that the NHEJ alignment step precedes nucleolytic processing steps in a significant fraction of NHEJ events.
Keywords: recombination, repair, 5′ DNA flaps, structure-specific nucleases, double-strand DNA breaks
Double-strand DNA breaks are repaired by either homologous recombination or nonhomologous DNA end joining (NHEJ). NHEJ is the major pathway in multicellular eukaryotes, particularly during G0, G1, and early S phase of the cell cycle (1). NHEJ may be among the most complex forms of DNA repair because the two free DNA ends must be constrained to prevent diffusion away from each other, and yet the ends must be accessible to nucleases and polymerases before ligation typically can proceed.
DNA ligase IV is the enzyme responsible for the final step of NHEJ in yeast (2–4) and human cells (5), and it exists in a complex with XRCC4 (6, 7). The substrate for the XRCC4/DNA ligase IV complex appears to be two ligatable nicks, either in a blunt or a staggered configuration (2, 6), and DNA ligase I is unable to substitute for it (5, 8).
Though the enzyme for the final step in NHEJ has now been determined, the proteins and nucleic acid steps for the earlier phases of NHEJ are not as fully defined. The initial steps of NHEJ appear to involve the Ku70/86 heterodimer (Ku70 is HDF1 in Saccharomyces cerevisiae). In multicellular eukaryotes, DNA-dependent protein kinase can bind to DNA ends (9) and is the only protein kinase thus far identified that requires free DNA ends as an essential cofactor for phosphorylation of protein targets (10, 11). The physiologic protein phosphorylation targets of DNA–PK are not yet identified (12). The precise mechanics of Ku and DNA–PK at the DNA ends are still under study, but possibilities include bringing the two DNA ends together, recruiting other repair components, remodeling the local chromatin, or halting the cell cycle until the double-strand breaks are repaired. DNA–PK has no known enzymatic action directed at the DNA itself. Though one laboratory reports weak helicase activity by Ku when acting on substrates with overhangs longer than 84 nt (13), the overhangs in NHEJ appear to be only a few nucleotides (14).
Between the first steps and the final ligation step, the two broken DNA ends must be altered to achieve the ligatable form. These alterations require nucleases to remove excess DNA and polymerases to fill any gaps. The nucleases and polymerases for this processing phase have not yet been identified. Genetic and biochemical methods must be used to identify these components. Genetic identification of components has the advantage of occurring under relatively physiologic conditions.
In considering genetic strategies for identification of such nucleases and polymerases, it is critical to note the fundamental difference between the processing step and the synapsis or ligation steps. First, for steps like ligation, there appears to be only one ligase. This may not be surprising given that the substrate is singular in its structure. Hence, genetic knockout of DNA ligase IV essentially ablates DNA end joining. In marked contrast, processing of the DNA ends at double-strand breaks is intrinsically diverse. For the simplest double-strand DNA break, both DNA strands are broken to create two blunt ends with 5′ phosphate configurations. This blunt-end configuration would require no nucleases or polymerases before ligation. However, breaks caused by physiologic pathways, like variable (diversity) joining segment recombination and class switch recombination, or pathologic breaks caused by ionizing radiation or oxidative free radicals generate a diverse set of biochemical DNA end configurations. For pathologic damage, the array of events is not simply determined by which nucleotide and overhang is generated for each end, but also by whether there is a 5′ phosphate or hydroxyl group, whether there is damage to the deoxyribose sugar, whether there is damage to the base, and whether there are intrastrand crosslinks (15). This array of outcomes can proceed to a ligatable form through many different alternatives. Many of the joining events proceed in a manner that indicates that one to four nucleotide stretches of adventitious homology align the two ends (14). This may occur as the two ends undergo breathing (transient melting) while they are held in proximity by proteins in the synapsis step. Some pairs of DNA ends align at these short stretches of adventitious homology close to the DNA terminus and others align farther in, but typically no more than 5 to 10 bp internally. Once such regions of microhomology anneal, there typically would be excess DNA beyond the point of annealing (either 5′ or 3′ overhangs), which must be removed either endonucleolytically as flaps or, perhaps, exonucleolytically. In addition, gaps exist that must be filled in.
The nature and number of different pathways is determined not only by the nucleic acid chemistry, but also by the enzymes able to act at the DNA ends. It is not yet clear how many nucleases and polymerases participate in this process. However, one point is critical to note. The genetic knockout of any one of the processing components has no possibility of blocking all NHEJ, because the ends can be joined in many different ways. As mentioned, blunt-end configurations conceivably would not require any nucleases or polymerases. However, for the full range of other combinations of two DNA ends to be joined together, at a minimum one can expect at least two nucleases to be used, one to cleave 5′ overhangs or flaps and one for 3′ ones. Regarding polymerases, we have preliminary indications that polymerase β can clearly account for some, but not all, gap fill-in synthesis (T.E.W. and M.R.L., unpublished work). Hence, there appear to be at least two polymerases. Given the minimal involvement of two polymerases and two nucleases, theoretically there are already a very large number of possible combinations for the order and number of the four enzymes that could contribute to the joining of two DNA ends. Knockout of any one of these four enzymes eliminates at most only one-fourth of these numerous pathways. Therefore, the genetic effect expected from the knockout of processing components like polymerases or nucleases is certain to be small and much less dramatic than that seen by us and others for the knockout of single enzymatic steps in NHEJ, such as the DNA ligase IV knockout in S. cerevisiae (2–4) or in mammalian cells (5, 8).
Concerning nucleases, as mentioned, one can anticipate roles for nucleases that deal with excess 5′ as well as 3′ DNA. The 5′–3′ exonuclease activities that have been identified in nuclear extracts of S. cerevisiae are exonuclease I (EXOI), SEP1 and RAD27 (FEN-1). EXOI has been shown to be biochemically nonprocessive and acts preferentially on double-stranded DNA in a 5′ to 3′ direction on each strand (17). EXOI is the major 5′ nuclease in S. cerevisiae. SEPI, also called XRN1, catalyzes DNA strand exchange in vitro and has a potent 5′,3′-exoribonuclease. Though it has a weak exonuclease activity (19), it appears to have a major role in RNA metabolism (20).
FEN-1 (RAD27) acts as an endonuclease on branched DNA structures (also called flap structures) of any length, even those as short as one nucleotide (21). These types of DNA intermediates exist during DNA replication (22, 23), but they also may exist during DNA end-joining reactions. Interestingly, the double mutant for rad27 and exoI is nonviable (18). FEN-1 and EXOI are both members of the FEN-1 family of nucleases, based on homology alignment (17). [Two ORFs (DIN7 and YER041W) that encode proteins with some homology to FEN-1 have not yet been demonstrated to have enzymatic activity, and these have not been examined here (17).]
In this study, we do a targeted deletion of RAD27 and EXOI independently and examine the effects on processing excess 5′ DNA at the junction of two incompatible DNA ends. In the rad27 mutant, there is a 4.4-fold reduction in the use of pathways that would require removal of two nucleotide 5′ flaps. In the exoI mutant, there is no effect. Complementation of the rad27 mutant with an expression vector for RAD27 restores usage of the flap pathway back to normal. These studies suggest a role for FEN-1 (Rad27) in NHEJ, and such a role indicates the order of annealing and processing steps in these cases.
MATERIALS AND METHODS
Reagents.
Components for yeast media were from United States Biochemical. Chemical reagents were from Sigma. Restriction endonucleases and DNA modifying enzymes were from NEB (Beverly, MA). Oligonucleotides were made by Operon Technologies (Alameda, CA). Reagents for Pwo-PCR were purchased from Boehringer Mannheim.
Yeast Strains and Genetic Methods.
Complete deletions of RAD27 and EXOI were performed as follows. rad27 and exoI strains were made in YW112 by replacement of the chromosomal RAD27 sequence or EXOI with the HIS3 gene, followed by PCR verification (24). For the RAD27 deletion, the primers Rad27–1 and Rad27-a were used to generate the HIS3 gene products tailed with 45 nucleotides that are homologous to either the 5′ or 3′ end of RAD27. Primers Rad27–2 and His-376 were used to verify that the RAD27 genomic sequence was deleted by the HIS3 gene. To make the EXOI deletion mutant, the primers ExoI-1, ExoI-a, ExoI-2, and His-376 were used. The sequences for all these primers are as follows. Rad27–1 GCATACA TTGGAAAGAAATAGGAAACGGACACCGGAAGAAAAAATGGCCTCCTCTAGTACACTC. Rad27-a TGCCAAGGTGAAGGACCAAAAGAAGAAAGTGGAAAAAGAACCCCCGCGCGCCTCGTTCAGAATG. Rad27–2 ATAGGAAACGCGACGCGTAAC. Exo1–1 ATTAAAATAAAAGGAGCTCGTAAAAACTGAAAGGCGTAGAAAGGAGGCCTCCTCTAGTACACTC. Exo1-a ATTTGAAAAATATACCTCCGATATGAAACGTGCAGTACTTAA CTTGCGCGCCTCGTTCAGAATG. Exo1–2 TTGCGTGATTGATAGAAGGCA. His-376 GCCTCATCCAAAGGCGC. All yeast strains are haploid and derived from the parent strain YW112, itself a derivative of YW104 (also known as YPH499). YW112: genotype MATa, ura3–52, trp1Δ −63, his3Δ −200, leu2Δ −1, lys2–801, ade2Δ∷TRP1. Yeast medium was either YPAD (1% yeast extract/2% peptone/2% glucose/40 μg/ml adenine) or minimal medium (1.7 g/l yeast nitrogen base/5 g/l ammonium sulfate) supplemented with the appropriate “CSM” nutrient dropout mix (BIO101, adenine concentration is 10 μg/ml) and carbon source (2% glucose or 2% raffinose plus 2% galactose). The adenine concentration was increased to 40 μg/ml for liquid culture in minimal medium. Yeasts were grown at 30°C for all experiments.
The disruption of the RAD27 gene results in several phenotypes that are well established, including a growth defect (25). Growth is very slow at 37°C and is reduced at 30°C. The temperature-sensitive lethality was almost fully complemented by transformation of the mutant with a plasmid carrying Myc-tagged RAD27, but not by the plasmid vector alone (data not shown).
Construction of Plasmids.
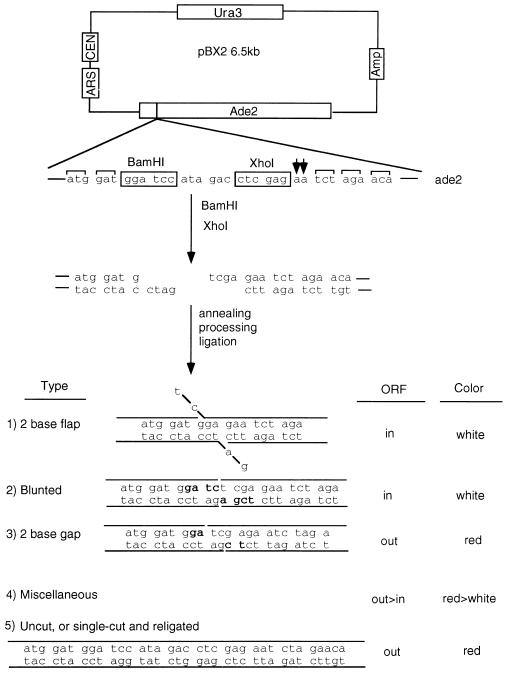
pBX2 substrate is a derivative of pES16, which was described previously (2). Two PCR products were simultaneously ligated into pES16 digested with NotI and BglII. The upstream PCR products were made with a forward primer that annealed to the beginning of the ADE2 promoter and was tailed with a NotI site (A2NF: GAGGCGGCCGCGGATTCATGCTTATGGGTTAG) and with reverse primers that spanned the start codon of the ADE2 gene and were tailed with a BamHI site (BX2-a TACGGATCCATCCATACTTGATTGTTTTGTCCG). Downstream PCR products were made by using the forward primer BX2–1 (CTAGGATCCATAGACCTCGAGAATCTAGAACAGTTGGTATATTAGG) and the reverse primer A2–1800-R (CCGTTAACAGATCTCACAATC). The primer BX2–1 anneals just after the start codon of the ADE2 gene and contains both BamHI and XhoI sites with a two-nucleotide addition to shift the ORF of ADE2 gene. The primer A2–1880-R spans the BglII site in the ADE2 gene.
The Rad27 expression plasmid was derived from the LEU2/2 μARS plasmid, pCWX200, which contains the tetracycline repressor coding sequence under the control of the ADH1 promoter (26). To generate the His-Myc-tagged Rad27 fusion protein, first the tetracycline repressor coding sequence was replaced by a HindIII–BamHI PCR fragment encoding a His9–Myc3 epitope tag. The Rad27 PCR fragment was made by using BglII-tailed primer Rad27–5 and SalI-tailed primer Rad27-c. This fragment was then ligated into the above construct after digestion with BamHI–SalI enzymes, which are immediately downstream of His9–Myc3 sequence. The sequences for both Rad27–5 and Rad27-c are as follows. Rad27 −5: GGAAGATCTTGGGTATTAAAGGTTTGAATGCA and Rad27-c: CCGCTCGAGTCATCTTCTTCCCTTTGTGACTT.
Western Blot Analysis.
The rad27 strain was transformed with either a control plasmid or the plasmid expressing a Myc fusion protein from the strong constitutive ADH1 promoter. The log-phase culture was harvested, and the cells were lysed in lysis buffer by using glass beads in the presence of 1 mM phenylmethylsulfonyl fluoride/2 μg/ml aprotinin/1 μg/ml pepstatin A/0.5 μg/ml of leupeptin. Whole-cell extracts were made according to procedures described previously (27). Anti-Myc Western blot analysis of N-terminal-tagged RAD27 was performed by using chemiluminescence.
Plasmid Transformation Assay.
The substrate pBX2 was prepared by overdigestion with either BamHI or BamHI–XhoI, followed by phenol/chloroform extraction and ethanol precipitation. To determine whether the plasmids were completely digested, the digested substrates were examined by ethidium bromide staining after agarose gel electrophoresis. The YW112, rad27, exoI, hdf1, and dnl4 yeast strains or the complemented rad27 strain were transformed by using the lithium acetate procedure described previously (28). In brief, the yeasts were grown from multiple colonies to saturation overnight in the appropriate medium, diluted 10-fold into a total volume of 10 ml YPAD, and allowed to grow to log phase (OD600 0.6–0.9). Two milliliters of culture were harvested, washed with water or 0.1 M lithium acetate, and then resuspended in a mixture of lithium acetate, polyethylene glycol 3350, substrate DNA, and 250 μg of single-stranded salmon sperm carrier DNA. After incubation at 30°C for 30 minutes, the cells were heat shocked at 42°C for 15 minutes, followed by washing, resuspending in H2O, and plating on the appropriate selective medium at 30°C. Replicate experiments used different mutant isolates. The substrate DNA, pBX2 was transformed in either supercoiled or linearized form (after being cut with the designated restriction enzymes).
RESULTS AND DISCUSSION
Experimental Strategy.
To study 5′ nucleases in NHEJ (29, 30), we developed an assay in which restriction enzyme-linearized plasmids are introduced into yeast and rejoined intracellularly, specifically by generating breaks in an ADE2 gene carried on a URA3-selectable centromeric plasmid named pBX2. The plasmid was constructed such that BamHI and XhoI sites are inserted immediately downstream of the ATG of the ADE2 gene (Fig. 1). To prevent plasmid-borne ADE2 from being repaired by homologous recombination with the chromosome, the chromosomal ADE2 gene has been deleted in the parent yeast strain, YW112. Both rad27 and exo1 mutant strains were made by the complete deletion of RAD27 or EXO1 ORFs by using the HIS3 gene by standard gene inactivation procedures in the YW112 background. Supercoiled, BamHI single-cut, or BamHI–XhoI double-cut pBX2 plasmid were transformed into the wild-type yeast (strain YW112), the rad27 mutant (18), the exo1 mutant (17), or a strain in which the rad27 mutant is complemented with overexpression of an N-terminal Myc-tagged Rad27 protein.
Figure 1.
Substrate structure and possible recircularization products. pBX2 is a centromeric plasmid with a URA3 selectable marker and carries the ADE2 gene as a reporter. It is derived from pES16. To destroy the XhoI site in the polylinker, pES16 was digested with SalI–XhoI and recircularized. Two PCR fragments (NotI–XhoI and XhoI–BglII) were ligated between the BglII site of the ADE2 gene and the NotI site in the polylinker, resulting in the junction sequence as shown. The addition of two AA nucleotides shifts the ADE2 gene ORF (shown by two arrows). BamHI–XhoI-linearized pBX2 with the indicated 5′-overhanging ends was transformed into yeast. The two ends are joined intracellularly to produce different types of end joints (lower portion of figure). A flap structure is formed by first annealing CT to GA, followed by flap cleavage. A blunted structure is the result of four-nucleotide fill-in synthesis on each end (fill-in nucleotides shown in bold). A gap structure is produced by first annealing AG to TC, then fill-in synthesis of the gap on each side (bold nucleotides). Miscellaneous products are made by trimming each end to varying extents, followed by apparent blunt ligation. The flap, blunted, and some of the miscellaneous products yield an ORF of the ADE2 gene to yield white colonies (ADE2 yeast). The gap and most miscellaneous products, as well as the starting plasmid, do not generate an ORF of the ADE2 gene; hence, they generate red colonies (ade2 yeast). A small fraction of the plasmid was uncut or was cut by only one of the two restriction enzymes (BamHI or XhoI), and these yield red colonies with the sequence shown at the bottom of the figure.
To examine flap end joining formation specifically, the strategy of reading frame shifting of the ADE2 gene was used. The two incompatible ends generated by double-digestion with BamHI and XhoI are recircularized by S. cerevisiae to recreate either an ADE2 or ade2 genotype, which corresponds to white or red phenotypes of yeast, respectively. Specifically, a symmetrical two-base flap (i.e., two-base deletion on each 5′ end) regenerates ADE2 (T.E.W. and M.R.L., unpublished work) and produces white colonies, in contrast to uncut plasmids and those joined by means of the dominant two-base gap intermediate (Fig. 1).
Table 1 shows the transformation efficiency and ratio of white vs. total colonies using supercoiled, BamHI single-digested, and BamHI–XhoI double-digested pBX2 substrates among the wild-type, rad27 mutant, complemented rad27, exo1, hdf1, and dnl4 strains. There was a reduction in overall transformability of rad27 yeast relative to the YW112 strain for both cut and uncut plasmids (approximately 10-fold). Reduced transformability is not uncommon among nucleic acid metabolism mutants and is of uncertain cause. This reduced transformability was complemented to the level of wild type by overexpression of Myc epitope-tagged Rad27 from an episomal plasmid. It is important to note that when results were normalized to the supercoiled efficiency, all strains had the same relative rate of transformation by the BamHI- and BamHI–XhoI-cut substrates. This indicates that the rad27 and exo1 mutant strains have no general quantitative defect in joining either compatible or incompatible substrates. Further, the ratio of white to total colonies was not significantly different among the four strains transformed with each substrate, but, as expected, was the greatest for the BamHI–XhoI-cut plasmid because of the increased potential for flap and blunted joints. To verify that these joints are indeed genuine events in the NHEJ pathway, dnl4 (DNA ligase IV) and hdf1 (Ku70) mutants were tested with BamHI–XhoI-cut pBX2. As expected, the ratio of white vs. total colonies dropped to zero in both strains, which indicates that the flap and blunted joints are completely abolished in the presence of a generalized NHEJ deficiency (Table 1 and ref. 2).
Table 1.
Transformation efficiency and number of white and total colonies in yeast strains
| Strains | Transformants/microgram of
DNA
|
White/total, %
|
||||
|---|---|---|---|---|---|---|
| Supercoiled × 104 | BamHI × 103 | BamHI–XhoI | Supercoiled | BamHI | BamHI–XhoI | |
| YW112 | 2.4 ± 0.31 | 4.6 ± 0.59 (19%) | 676 ± 88 (2.8%) | 0 | 1.8 | 23.4 |
| rad27 | 0.22 ± 0.35 | 0.38 ± 0.05 (17%) | 61 ± 14 (27%) | 0 | 2.1 | 14.8 |
| rad27 complemented | 1.7 ± 0.27 | 3.3 ± 0.31 (19%) | 391 ± 43 (2.3%) | 0 | 2.8 | 21.4 |
| exo1 | 3.5 ± 0.51 | 3.3 ± 0.48 (9.4%) | 505 ± 39 (1.4%) | 0 | 0.6 | 27.7 |
| hdf1 | 1.75 ± 0.15 | 0.31 ± 0.015 (1.8%) | 60 ± 7 (0.34%) | 0 | 0 | 0 |
| dnl4 | 2.53 ± 0.05 | 0.31 ± 0.02 (1.2%) | 62.5 ± 2.5 (0.25%) | 0 | 0 | 0 |
Yeast cells were grown to log phase and transformed with supercoiled BamHI or BamHI–XhoI double-cut linearized pBX2 by lithium acetate transformation, with addition of single-strand salmon sperm DNA as a carrier. The transformation efficiency numbers in parentheses are simply relative to the supercoiled efficiency. Wild-type (YW112), rad27 (mutant), exol (mutant), hdf1 (mutant), and dnl4 (mutant) strains were plated to minimal medium lacking uracil. The overexpression strain harboring expressing plasmid and the substrate was plated onto minimal medium lacking both uracil and leucine. For the purpose of normalization, a parallel transformation was done with supercoiled pBX2. Results are averaged from three independent experiments; each replicate experiment uses different mutant isolates (see Methods). The total transformants after BamHI or BamHI–XhoI in the hdf1 and dnl4 mutants are derived from uncut plasmids.
Marked Reduction Specifically in NHEJ Events Proceeding by the Flap Pathway in the rad27 Mutant.
To characterize the spectrum of end joining in these strains, plasmids were recovered from white colonies transformed with BamHI–XhoI-cut pBX2, sequenced, and analyzed. We found that in the wild-type strain, joining by the flap pathway (two-base deletion on each 5′ end) was responsible for 60% of the white colonies, which is comparable to that in the exo1 mutant strain (64.5%) (Table 2). This clearly shows that inactivation of EXO1 does not affect the rate of the flap pathway even though, biochemically, Exo1 has been demonstrated to be the major 5′–3′ exonucleolytic activity in yeast extracts (17). In contrast, we find that contribution by the flap pathway is significantly decreased in a strain bearing a deletion of RAD27 (reduced from 60% to 22.5%). This represents a 4.4-fold reduction in the flap joint rate when corrected for the transformation efficiency (see calculation in Table 2 legend). However, RAD27 expression from a plasmid restores the protein (by Western blot, not shown) and, importantly, the normal joint pattern (Table 2).
Table 2.
Distribution of end joining types in yeast strains
| Strains | No. sequenced | End joining
|
P value* | ||
|---|---|---|---|---|---|
| Flap (%) | Blunt (%) | Miscellaneous (%) | |||
| YW112 | 40 | 24 (60) | 5 (12.5) | 11 (27.5) | NA |
| rad27 | 40 | 9 (22.5) | 13 (32.5) | 18 (45) | 0.0024 |
| rad 27 complemented | 34 | 18 (52.9) | 5 (14.7) | 11 (32.4) | 0.830 |
| exo1 | 31 | 20 (64.5) | 3 (9.7) | 8 (25.8) | 0.905 |
The strains from YW112, rad27, rad27 complemented with RAD27 plasmid, and exol strains were transformed with BamHI–XhoI double-digested pBX2 substrate. For YW112, exol, and rad27, plated to minimal medium lacking uracil and containing 10 μg/ml adenine. The overexpression strain pretransformed with the expression plasmid was plated on minimal medium lacking both uracil and leucine and containing 10 μg/ml adenine after being retransformed with substrate. To efficiently recover the low-copy ura3 plasmid from the overexpression strain, the high-copy LEU2 plasmid was first segregated out by consecutively streaking yeast on minimal medium lacking uracil. All ura3-plasmids were recovered from white colonies for each strain and sequenced. The percentage of events proceeding by the flap configuration defined in Fig. 1 is shown in bold.
The appropriate comparison between the wild-type and mutant strains is for the formation rate of a particular type of joint in the different strains. This formation rate can be calculated by multiplying the normalized rate of formation of the appropriate colony color by the percentage of those colonies that contain the joint of interest. Thus, the flap joint rate for YW112 is 0.028 × 0.234 × 0.60 = 0.0039, which is the product of the BamHI–XhoI transformation efficiency from Table 1 times the percentage of white to total colonies in the last column of Table 1 times the percentage of flap joints (bold). The flap joint rate for rad27 is 0.027 × 0.148 × 0.225 = 0.00090. The ratio of these two rates is 4.4; hence, the rad27 strain has a >4-fold reduction in the rate of flap joining.
P values are the χ2 probability obtained from the 2 × 3 table of strain vs. joint type, each in comparison to YW112. The P value of the 4 × 3 table of strain vs. joint type is 0.007, increasing to 0.913 for the 3 × 3 table that excludes the rad27 strain.
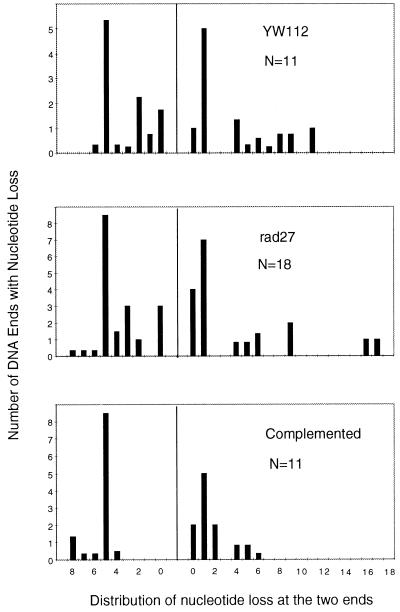
Mutation of RAD27 does not generally affect other types of NHEJ events that do not rely on the flap pathway. First, mutation of RAD27 does not reduce the rate of joining of compatible 5′ overhangs (Table 1). Second, mutation of RAD27 does not affect the rate of BamHI–XhoI joints characterized either by four-base fill-in synthesis (blunted joints) or by varying extents of exonucleolytic degradation followed by blunt-end ligation (so-called “miscellaneous” joints, Table 2). The preservation of blunted and miscellaneous joints is evident as an increase in their relative contribution to the white colonies obtained in the rad27 mutant. Further, we find that mutation of RAD27 does not affect the quality of the miscellaneous joints (Fig. 2). Specifically, the distribution of exonucleolytic chewback was very similar for both DNA ends, regardless of whether RAD27 was wild type, mutant, or complemented. Thus, the rad27 effect is specific to the deletion pathway (flap), indicating that Rad27 protein acts at a subset of NHEJ events that require processing of 5′ flaps, in a manner independent of the exonucleolytic processing that can also occur during NHEJ (Fig. 2). As shown in Table 2, there is some level of potential flap joints (22.5%) in the complete absence of Rad27 activity. These are presumably contributed by other unidentified 5′ nucleases in yeast, and it is uncertain whether these residual joints proceed by means of a flap or nonflap intermediate.
Figure 2.
Comparison of nucleotide loss from the miscellaneous category of products in YW112, rad27, and the complemented strain. A histogram is shown for double-strand break junctional sequences, showing the number of ends along the y axis and the number of nucleotides lost on the x axis. N is the total number of ends scored for each histogram. When the precise number of nucleotides lost from an end was ambiguous because of microhomology use at the junction, the nucleotide loss from each end was calculated as half of the nucleotides lost from the junction; then, the loss was evenly distributed among all potential positions within each end.
Order of Alignment and Processing Steps in NHEJ.
The order of events in NHEJ will vary depending on the configuration of the DNA ends. Compatible ends can be, and usually are, also joined without any need for nucleases or polymerases. However, pathologic breaks will typically not generate compatible or blunt configurations. In these cases, synapsis, processing, terminal homology alignment, and ligation must occur. The specifics of each end combination will dictate the number of possible enzymatic activities that may participate, and this will dictate the number of pathways. The greater the number of enzymes involved, the greater the number of possible pathways, and the smaller the effect of removing any one of those pathways or removing any one component needed for that pathway. For this reason, we were fortunate to find a combination of DNA ends that allowed us to demonstrate a quantitative reliance on the 5′ flap endonuclease Rad27, indicating its involvement in NHEJ. This analysis is specific for 5′ flap configurations. The other 5′ nuclease, Exo1, was not involved. Further, the absence of Rad27 protein in no way affects nucleic acid metabolism in a way that is manifest in the nucleotide loss from the miscellaneous subset of NHEJ events (Fig. 2).
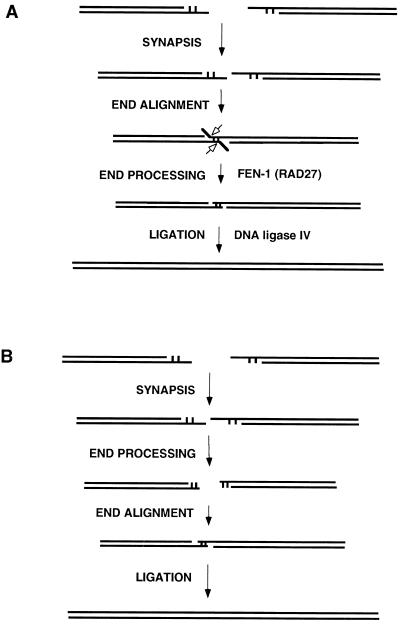
A limitation of studying certain processing events, such as end filling, is that one cannot know whether processing has occurred before or after end alignment. In contrast, Rad27 and its mammalian counterpart, FEN-1, do not act on free 5′ overhangs or blunt ends (21, 31–33). Hence, for those NHEJ events using Rad27, the order of events must be such that DNA end alignment occurs as a discrete step before the action of the 5′ flap nuclease.
Involvement of RAD27 (FEN-1) permits us to know the order of events throughout the process (Fig. 3). The predilection of the two-base flap pathway (Table 2) suggests that this particular terminal homology alignment may be more stable, thereby establishing a transient flap configuration (Fig. 3A). Once the flaps are clipped, other possible pathway options are limited, thus contributing to the predilection for products derived from this pathway. While it is clear that our results regarding two-base flap joints are not necessarily applicable to all joint types, we think it is likely that NHEJ does generally proceed according to a model in which end annealing precedes end processing. Thus, the sequence of events during NHEJ would first entail end recognition and stabilization, presumably involving Ku and, perhaps, the SIR proteins (24, 34). Next, the joining machinery would search for end complementarity to provide a metastable alignment for the subsequent processing steps; this alignment would presumably have to be stabilized by proteins, probably the same ones as in the earlier steps. The fact that blunt ends join inefficiently is consistent with the notion that this is a critical step in the joining process. Once the ends are synapsed and annealed, end processing would proceed by recruitment of the relevant enzymes as needed to the active complex. The basis of this recruitment is unknown but of great interest. The data presented here indicate that Rad27 shares features that promote recruitment. The last processing event is strand ligation by XRCC4 and DNA ligase IV, which results in the final resolution of the break (2).
Figure 3.
Models for nonhomologous DNA end joining involving FEN-1 (RAD27). (A) Double-strand DNA breaks can have 5′ or 3′ overhangs or can be blunt. The two ends must be brought together into synapsis. This provides an opportunity for microhomology search of one DNA end by the other, resulting in end alignment. If 5′ flaps are generated by this alignment, then FEN-1 (called RAD27 in S. cerevisiae) is the predominant nuclease to remove the flaps (open arrows), as part of the end processing in preparation for ligation by DNA ligase IV. (B) Same as in A, except as follows. The nucleolytic removal of nucleotides from overhangs occurs by exonucleases in a nonprocessive (one at a time) fashion. A subset of these end-processing events happen to anneal, resulting in alignment, followed by ligation. A nonprocessive removal of one nucleotide at a time from each end would generate a diversity of outcomes, each with a small percentage contribution to the diverse array of possible products. This is not consistent with the 60% flap usage seen in Table 2. Moreover, such a pathway would not be affected by deletion of RAD27 because the FEN-1 nuclease, including RAD27 in S. cerevisiae, is not active on single-stranded 5′ ends or blunt ends (21, 31, 32).
Concluding Remarks.
Though our experiments are directed at 5′ flaps, they do not address how 3′ flap structures are resolved in cells. Recent genetic work indicates that the Rad1/Rad10 endonuclease, DNA polymerase δ, the helicase Srs2, and the mismatch repair proteins Msh2 and Msh3 are involved in dealing with such structures when encountered during homologous repair of double-strand DNA breaks (29, 35, 36). Recent biochemical work indicates that Mre11 can function as a 3′ to 5′ exonuclease on double-stranded DNA (37).
FEN-1 (RAD27 in S. cerevisiae) appears to function in excision repair (38) and in DNA replication (22). The studies here indicate that FEN-1 also functions in double-strand break repair. Because of its role in Okazaki fragment processing, mutation of FEN-1 results in genetic instability at regions of short and long repetitive DNA (18, 39). Recently, it has become clear that such regions are detectable as breaks at the chromosomal level (40). Our data raise the possibility that increased genetic instability in the absence of FEN-1 is caused not only by increased generation of double-strand breaks, but also by compromised repair of these double-strand breaks.
Even when NHEJ proceeds optimally, the nature of the process is such that nucleotides are commonly lost at the junction (Fig. 3). This means that such sites of double-strand DNA breakage always leave a residual loss of information or “scar” in the genome. It will be interesting to determine whether the accumulation of such scars over time can account for pathologic states in multicellular organisms. Early indications from NHEJ mutants in mice raise this possibility (16).
Acknowledgments
T.E.W. was supported by a Howard Hughes Medical Institute Fellowship. This work was supported by National Institutes of Health grants to M.R.L. M.R.L. is a Leukemia Society of America Scholar and is the Rita and Edward Polusky Basic Cancer Research Professor.
ABBREVIATIONS
- NHEJ
nonhomologous DNA end joining
- EXOI
exonuclease I
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Takata M, Sasaki M S, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson T E, Grawunder U, Lieber M R. Nature (London) 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 3.Schar P, Herrmann G, Daly G, Lindahl T. Genes Dev. 1997;11:1912–1924. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teo S H, Jackson S P. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber M R. Mol Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 6.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson T E, Mann M, Lieber M R. Nature (London) 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 7.Critchlow S, Bowater R, Jackson S P. Curr Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 8.Frank K M, Sekiguchi J M, Seidl K J, Swat W, Rathbun G A, Cheng H-L, Davidson L, Kangaloo L, Alt F W. Nature (London) 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 9.Yaneva M, Kowaleski T, Lieber M R. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter T H, Vancurova I, Sun I, Lou W, DeLeon S. Mol Cell Biol. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lees-Miller S P, Chen Y-R, Anderson C W. Mol Cell Biol. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson C W, Carter T H. In: The DNA-Activated Protein Kinase-DNA-PK, in Molecular Analysis of DNA Rearrangements in the Immune System. Jessberger R, Lieber M R, editors. Heidelberg: Springer; 1996. pp. 91–112. [Google Scholar]
- 13.Tuteja N, Tutega R, Ochem A, Taneja P, Huang N, Simoncsits A, Susic S, Rahman K, Marusic L, Chen J, Zhang J, Wang S, Ponger S, Falaschi A. EMBO J. 1994;13:4991–5001. doi: 10.1002/j.1460-2075.1994.tb06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roth D, Wilson J. In: Illegitimate Recombination in Mammalian Cells, in Genetic Recombination. Kucherlapapti R, Smith G R, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 621–653. [Google Scholar]
- 15.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 16.Gao Y, Sun Y, Frank K, Dikkes P, Fujiwara Y, Seidl K, Sekiguchi J, Rathbun G, Swat W, Wang J, et al. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 17.Fiorentini P, Huang K N, Tishkoff D X, Kolodner R D, Symington L S. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tishkoff D X, Filosi N, Gaida G, Kolodner R D. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 19.Johnson A W, Kolodner R D. J Biol Chem. 1991;266:14046–14054. [PubMed] [Google Scholar]
- 20.Schwer B, Mao X, Shuman S. Nucleic Acids Res. 1998;26:2050–2057. doi: 10.1093/nar/26.9.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrington J J, Lieber M R. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bambara R A, Murante R S, Henricksen L A. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 23.Lieber M R. BioEssays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 24.Ramsden D A, Gellert M. EMBO J. 1998;17:609–614. doi: 10.1093/emboj/17.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reagan M S, Pittenberger C, Siede W, Friedberg E C. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu C W, Mendelsohn A R, Brent R. Proc Natl Acad Sci USA. 1997;94:12473–12478. doi: 10.1073/pnas.94.23.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1996. [Google Scholar]
- 28.Schiestl R H, Gietz R D. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 29.Haber J E. BioEssays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 30.Kanaar R, Hoeijmakers J H J. Genes Funct. 1997;1:165–174. doi: 10.1046/j.1365-4624.1997.00016.x. [DOI] [PubMed] [Google Scholar]
- 31.Harrington J J, Lieber M R. Genes Dev. 1994;8:1344–1355. doi: 10.1101/gad.8.11.1344. [DOI] [PubMed] [Google Scholar]
- 32.Murante R, Huang L, Turchi J, Bambara R. J Biol Chem. 1994;269:1191–1196. [PubMed] [Google Scholar]
- 33.Murante R S, Rust L, Bambara R A. J Biol Chem. 1995;270:30377–30383. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]
- 34.Tsukamoto Y, Kato J, Ikeda H. Nature (London) 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- 35.Sugawara N, Paques F, Colaiacovo M, Haber J E. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paques F, Haber J E. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paull T, Gellert M. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 38.Klungland A, Lindahl T. EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson R, Kovvali G, Prakash L, Prakash S. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 40.Freudenreich C H, Kantrow S M, Zakian V A. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]