Abstract
Human Lyme borreliosis (LB) is the most prevalent arthropod-borne infection in temperate climate zones around the world and is caused by Borrelia spirochetes. We have identified 10 variable-number tandem repeat (VNTR) loci present within the genome of Borrelia burgdorferi and subsequently developed a multiple-locus VNTR analysis (MLVA) typing system for this disease agent. We report here the successful application of MLVA for strain discrimination among a group of 41 globally diverse Borrelia isolates including B. burgdorferi, B. afzelii, and B. garinii. PCR assays displayed diversity at these loci, with total allele numbers ranging from two to nine and Nei's diversity (D) values ranging from 0.10 to 0.87. The average D value was 0.53 across all VNTR loci. A clear correlation exists between the repeat copy number and the D value (r = 0.62) or the number of alleles (r = 0.93) observed across diverse strains. Cluster analysis by the unweighted pair-group method with arithmetic means resolved the 30 observed unique Borrelia genotypes into five distinct groups. B. burgdorferi, B. afzelii, and B. garinii clustered into distinct affiliations, consistent with current 16S rRNA phylogeny studies. Genetic similarity and diversity suggest that B. afzelii and B. garinii are close relatives and were perhaps recently derived from B. burgdorferi. MLVA provides both phylogenetic relationships and additional resolution to discriminate among strains of Borrelia species. This new level of strain identification and discrimination will allow more detailed epidemiological and phylogenetic analysis in future studies.
Human Lyme borreliosis (LB) is the most prevalent arthropod-borne infection in temperate climate zones around the world and is caused by Borrelia spirochetes (3, 19, 30, 32). In 1996, more than 16,000 cases of LB were reported in North America, totaling 100,000 cases in a 14-year period (9, 10). Borrelia spirochetes are 5 to 25 μm long and 0.2 to 0.5 μm wide (24). These organisms are highly motile, microaerophilic, slow growing, and fastidious (24). Lyme disease is an inflammatory disorder characterized by the skin lesion erythema migrans (25) and the potential development of neurologic, cardiac, and joint abnormalities (24). Three Borrelia species frequently cause Lyme disease in humans: Borrelia burgdorferi sensu stricto, Borrelia garinii, and Borrelia afzelii (6, 19). Specific Borrelia species can cause distinct clinical manifestations of Lyme disease. B. burgdorferi can cause arthritis (2, 28). B. garinii is known to cause serious neurological manifestations (2, 28). B. afzelii causes a distinctive skin condition known as acrodermatitis chronica atrophicans (27). Each of the three Borrelia species causes a characteristic erythema migrans (2, 28).
The taxonomy of B. burgdorferi has undergone extensive revision. Ten species of B. burgdorferi sensu lato have been characterized and subsequently placed within the B. burgdorferi complex. B. burgdorferi sensu stricto is found primarily in North America and Europe (6, 15, 19, 33). B. garinii, B. afzelii, B. valaisiana, and B. lusitaniae have been isolated throughout Eurasia (33). B. japonica, B. tanukii, and B. turdi are found primarily in Japan (17, 20). B. andersonii and B. bissettii are distributed predominantly in North America (22, 31). Ixodes scapularis, I. pacificus, and I. ricinus are the three primary tick reservoirs for B. burgdorferi sensu lato (5). The tick reservoir hosts include numerous small mammal species and birds (1, 18, 26).
Strains of B. burgdorferi sensu lato are genetically diverse. The bacterium possesses the largest number of extrachromosomal elements, plasmids, of any known bacterial species: nine circular plasmids and 12 linear plasmids (7, 16). Borrelia spp. also have some of the smallest bacterial genomes (910 kb). The combined chromosome-plasmid nucleotide content is approximately 1.5 Mb. Although the Borrelia genome mostly evolves in a clonal way (12), ospC gene studies suggest that lateral transfer does exist (11, 13, 23). These genetic exchanges could be due to whole-plasmid lateral transfer or, more likely, to a gene transfer agent (11). The molecular mechanisms by which this genetic exchange occurs are unknown. The Borrelia genome exhibits significant genetic redundancy and carries 161 to 175 paralogous gene families (7). Such families may serve as foci for interplasmid homologous recombination. At least one linear plasmid gene is found within each of 107 gene families, creating a significant amount of redundancy and an unusually large number of pseudogenes (7). Approximately 90% of Borrelia plasmid genes show little similarity to genes of other bacteria (7). It is possible that these linear plasmids are in a phase of rapid evolution and undergo antigenic variation because of immune selection.
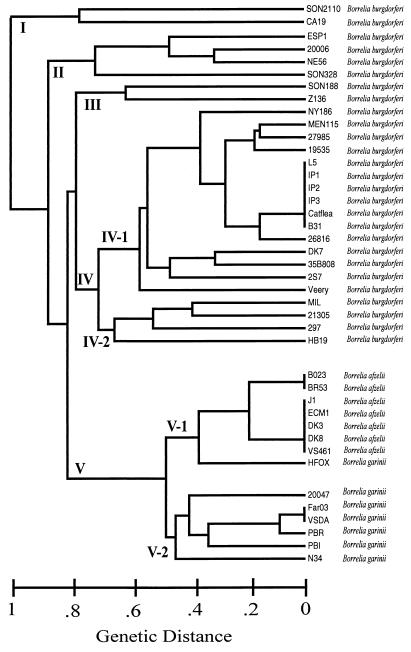
Numerous molecular techniques have recently been used to characterize Borrelia species, including 16S rRNA gene sequence analysis, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blot analysis, pulsed-field gel electrophoresis, plasmid fingerprinting, randomly amplified polymorphic DNA analysis, restriction fragment length polymorphism analysis, fatty acid profile analysis, and serotyping (4, 8, 15, 33). For a more thorough review of the molecular typing methods used for Borrelia characterization, see reference 36. Although a previous study suggested that randomly amplified polymorphic DNA analysis is effective for strain discrimination within and among Borrelia species (35), its routine application to determine robust evolutionary relationships remains questionable because of the method's reduced capacity to provide reproducible data crucial for cladistic character analysis (33, 34). Simple sequence repeats or variable-number tandem repeats (VNTRs) have been shown to provide a high level of discriminatory power (21). This stems from the significant mutability of the repeat copy number. Many of the genomes examined contain numerous VNTRs, and in combination, these can be used to develop a robust PCR-based marker typing system. Multiple-locus VNTR analysis (MLVA) has previously shown great discriminatory capacity and accurate estimation of genetic relationships within bacterial pathogens such as Francisella tularensis and Bacillus anthracis (14, 21). We report here the successful application of MLVA for strain discrimination among a group of 41 globally diverse Borrelia isolates, including B. burgdorferi, B. afzelii, and B. garinii. Ten VNTR loci were identified from genomic and plasmid sequences in this study. Polymorphisms at these loci were then used to resolve 30 unique genotypes into five to seven distinct groups (Fig. 1).
FIG. 1.
Genetic relationships among Borrelia isolates. UPGMA cluster analysis based upon allelic differences from 10 VNTR markers across 41 B. burgdorferi, B. afzelii, and B. garinii isolates was used to construct this dendrogram. The designation to the right of each branch corresponds to the individual sample identity (Table 2), followed by the Borrelia species name. The horizontal lines indicate genetic distances as fractions of the allelic differences. Roman numerals indicate arbitrary groupings.
MATERIALS AND METHODS
Genomic analysis.
The B. burgdorferi sensu stricto strain B31 genomic sequence was downloaded from the National Center for Biotechnology Information web page (http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/framik?gi=132&db=Genome) and used to identify potential VNTR loci. We screened sequences from the 946-kb genome, the 12 linear plasmids, and the 9 circular plasmids of B. burgdorferi. Each sequence contig was screened for the presence of tandem repeats with the DNASTAR software program Genequest (Lasergene, Inc., Madison, Wis.). This program locates and displays tandemly and nontandemly repeated arrays. Confirmation of the repeated sequence structure was performed by dot plot similarity analysis with the software program Megalign (Lasergene, Inc.).
PCR amplification of VNTR loci.
MLVA primers were developed around 46 potential VNTR loci with the DNASTAR program PrimerSelect. A total of 10 primer sets amplified polymorphic VNTR loci (Table 1), while 36 loci proved monomorphic. The reagents used in the PCRs were obtained from Life Technologies. Primers were designed with annealing temperatures of 65 to 61°C. Individual primer pair annealing temperatures were designed within 2°C of each other.
TABLE 1.
Sequences of Borrelia primers used in this study
| Marker | Primer sequence |
|
|---|---|---|
| Forward | Reverse | |
| BR-V1 | GTTCAAGATATGGTTAAGGGCAATTTAGATAAAGATC | GAAGACTTACATGCCAGTTCATCAAGAGTC |
| BR-V2 | GTATAATGAAGTTAGTGGGCGTTACTCTTGGGTAC | GAAACCATAAAACCATCTAAAGATACAAATCATTC |
| BR-V3 | GTTTGTCGTTGCCAAAACTGCTTTCATAATTC | GGGATTAAATATGAAAATATATTTAGTTTGTGTGCATTATATCTGC |
| BR-V4 | GTTTCTGCGACTAGGTATGGAACAACTAATAGCTC | GCAGTGGGCACAACTACTACTGCAATAATAACTAC |
| BR-V5 | GCAATCCAAAATATTCAAGATCGTATAAAAATGTC | GATGATAAAATTTTCAAATGTATATCTTTTTTTAAGAAAGGC |
| BR-V6 | GGATCGATCGTACTGTGCAGCCACAAACGTGCTGCGC | GTAGCGTACGTAGCTGCGCGTAGTATTTTTATCGTAGCGCGAGC |
| BR-V7 | GCTTCAAAATGCTGCTTCAATTGCTGGAC | GCAAAAACACAAGCTTGCCGGTGAAAC |
| BR-V8 | GATCTAATTCATTAAAAAATTTTGTGAAAGGGGCTTC | GATAAATAACTTGCAATATTTCCGCTTAAGGTAGTTTTC |
| BR-V9 | GTCATCTTTAGTGTCTAATTTTAGAATTTTATTAACTTTTTCTTTGC | GTCATGCTTATATCAATGCCCTATGCCTCAAC |
| BR-V10 | GCTTTTAACGCTAAATTATAAAGAAAAATTATTTCATTTCGGC | GTCAAAATTATGCTTCCAAAAGCATTACAATTAAAAAAATC |
PCR amplification of the 10 variable loci from 41 Borrelia isolates was carried out with a mixture containing 2 mM MgCl2; 1× PCR buffer; 0.1 mM deoxynucleoside triphosphates; 1 μM R110, R6G, or Tamra phosphoramide fluorescently labeled dUTP (Perkin-Elmer Biosystems); 0.5 U of Taq polymerase; 1.0 μl of template DNA; 0.5 μM forward primer; 0.5 μM reverse primer; and filtered sterile water to a volume of 12.5 μl. The reaction mixtures were incubated at 94°C for 5 min and then cycled at 94°C for 30 s, 61 or 56°C for 30 s, 72°C for 30 s, and 94°C for 30 s for 35 cycles, with a final incubation of 72°C for 5 min.
Bacterial thermolysates.
Borrelia strains were grown in BSK medium (Sigma) until they reached 107 bacteria/ml. One milliliter was harvested by centrifugation, washed in phosphate-buffered saline, resuspended in 100 μl of water, and then heated at 100°C for 20 min.
Automated genotyping.
Fluorescently labeled amplicons were sized by polyacrylamide gel electrophoresis in an ABI 377 DNA sequencer. Analysis was accomplished with the Genescan and Genotyper software (14). The PCR product was diluted threefold and mixed 1:1 with equal parts of a 5:1 formamide-dextran blue dye and size standard prior to electrophoresis. The Bioventures Rox 1000 size standard was used with filter set D.
Statistical analysis.
Pairwise genetic differences among isolates were estimated with a simple matching coefficient. The clustering method used to evaluate genetic relationships was the unweighted pair-group method with arithmetic means (UPGMA) with the PAUP4a software (D. Swofford, Sinauer Associates, Inc., Publishers, Sunderland, Mass.). The diversity index (D) for each marker was calculated as 1 − Σ(allele frequency)2 (37).
RESULTS AND DISCUSSION
VNTR marker identification and diversity.
Our analysis of the genomic sequence of B. burgdorferi type strain B31 revealed 225 genomic sequence motifs that potentially represent VNTR loci. An additional 167 potential VNTR loci were identified among the plasmid sequences of B. burgdorferi type strain B31. We arbitrarily chose 46 repeated sequence motifs from these for MLVA. MLVA revealed that 36 were monomorphic and only 10 proved to be polymorphic loci (see Table 3) among 41 globally diverse B. burgdorferi, B. afzelii, and B. garinii strains (Table 2). However, all loci did not support PCR amplification. A total of 19 isolates failed to yield PCR products across markers BR-V4, -V6, -V8, and -V10 (see Table 4). Sixteen of these 19 failures occurred within plasmid-based loci (see Table 4).
TABLE 3.
VNTR marker attributes
| Marker locus | Repeat motif | Genome or plasmid coordinatea | Repeat size (nucleotides) | Repeat number |
No. of allelesb | D valuec | ||
|---|---|---|---|---|---|---|---|---|
| Borrelia (B31) array | Smallest array | Largest array | ||||||
| BR-V1 | Complex array | CH-844,650 | CXd | 6 | 0.74 | |||
| BR-V2 | TAAAT | CH-590,955 | 5 | 5 | 8 | 11 | 4 | 0.67 |
| BR-V3 | TA | LP17-10,530 | 2 | 5 | 22 | 29 | 4 | 0.14 |
| BR-V4 | Complex array | LP28-2-28,142 | CXe | 8 | 0.55 | |||
| BR-V5 | AAG | CH-456,964 | 3 | 4 | 2 | 4 | 3 | 0.63 |
| BR-V6 | TGA | CH-720,032 | 3 | 4 | 1 | 3 | 3 | 0.51 |
| BR-V7 | TGC | CH-690,090 | 3 | 4 | 13 | 14 | 2 | 0.1 |
| BR-V8 | —f | LP17-13,155 | 21 | 8.3 | 6 | 14 | 9 | 0.89 |
| BR-V9 | TTC | LP28-3-4,235 | 3 | 4 | 3 | 4 | 3 | 0.1 |
| BR-V10 | AATATTAAATA | LP54-20,145 | 11 | 5.5 | 1 | 9 | 7 | 0.75 |
CH indicates chromosome locus; LP indicates linear plasmid locus.
The average number of alleles was 4.9.
D = 1 − ∑(allele frequency)2. The average D value was 0.51.
CX indicates the complex nature of the repeat motif, which makes accurate array size calculation difficult. The B31 sequence at this locus consists of four tandem repeats. For example, a 32-bp motif repeated 2.2 times is listed here in the form (32 × 2.2). Other arrays that contribute to the complexity observed at this locus include the following: (32 × 3.2) + (32 × 2.0) + (41 × 2.0).
(86 × 2.2) + (32 × 4.0) + (32 × 2.6).
The 21-bp repeat TAATTAATATGTGATATAAAA was found.
TABLE 2.
Borrelia strains used in this study
| Strain | Species | Location | Source | Provider or strain |
|---|---|---|---|---|
| ESP1 | B. burgdorferi | Spain | I. ricinus | R. C. Johnson |
| SON328 | B. burgdorferi | California | I. pacificus | M. Janda |
| IP2 | B. burgdorferi | Tours, France | Human CSFa | G. Baranton |
| SON2110 | B. burgdorferi | California | I. pacificus | M. Janda |
| HB19 | B. burgdorferi | Connecticut | Human blood | A. Barbour |
| IP1 | B. burgdorferi | Poitiers, France | Human CSF | G. Baranton |
| B31 | B. burgdorferi | New York | I. scapularis | ATCC 35210 |
| ZS7 | B. burgdorferi | Germany | I. ricinus | L. Gem |
| 20006 | B. burgdorferi | France | I. ricinus | J. F. Anderson |
| VEERY | B. burgdorferi | Connecticut | Veery bird | R. T. Marconi |
| MEN115 | B. burgdorferi | California | I. pacificus | M. Janda |
| CA19 | B. burgdorferi | California | I. pacificus | T. Schwan |
| 19535 | B. burgdorferi | New York | Peromyscuc leucopus | J. F. Anderson |
| MIL | B. burgdorferi | Slovakia | I. ricinus | A. Livesley |
| Cat flea | B. burgdorferi | Texas | Ctenocephalides felis | D. Ralph |
| 21305 | B. burgdorferi | Connecticut | Peromyscuc leucopus | J. F. Anderson |
| NY186 | B. burgdorferi | New York | Human skin | R. T. Marconi |
| DK7 | B. burgdorferi | Denmark | Human skin | M. Theisen |
| 297 | B. burgdorferi | Connecticut | Human CSF | R. C. Johnson |
| 26816 | B. burgdorferi | Rhode Island | Microtus pennsylvanicus | J. F. Anderson |
| SON188 | B. burgdorferi | California | I. pacificus | M. Janda |
| IP3 | B. burgdorferi | Pau, France | Human CSF | G. Baranton |
| Z136 | B. burgdorferi | Germany | I. ricinus | A. Vogt |
| 35B808 | B. burgdorferi | Germany | I. ricinus | A. Schönberg |
| NE56 | B. burgdorferi | Switzerland | I. ricinus | L. Gem |
| 27985 | B. burgdorferi | Shelter Island, N.Y. | I. scapularis | J. F. Anderson |
| L5 | B. burgdorferi | Austria | Human skin | G. Stanek |
| DK3 | B. afzelii | Denmark | Human skin | R. C. Johnson |
| BR53 | B. afzelii | Czech Republic | Aedes vexans | Z. Hubalek |
| ECM1 | B. afzelii | Sweden | Human skin (EM)b | S. Bergstrom |
| J1 | B. afzelii | Japan | I. persulcatus | R. T. Marconi |
| B023 | B. afzelii | Germany | Human skin (EM)c | A. Vogt |
| VS461 | B. afzelii | Switzerland | I. ricinus | O. Peter |
| DK8 | B. afzelii | Denmark | Human skin | R. C. Johnson |
| PBI | B. garinii | Germany | Human CSF | C. Kodner |
| VSDA | B. garinii | Switzerland | Human CSF | O. Peter |
| N34 | B. garinii | Germany | I. ricinus | J. Ackerman |
| 20047 | B. garinii | France | I. ricinus | J. F. Anderson |
| HFOX | B. garinii | Japan | Fox heart | E. Isogai |
| PBR | B. garinii | Germany | Human CSF | B. Wilske |
| FAR03 | B. garinii | Sweden | Seabird | S. Bergström |
CSF, cerebrospinal fluid.
EM, erythema migrans.
TABLE 4.
Borrelia allele sizes
| Strain | Allele size (bp) at marker locus: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BR-V1 | BR-V2 | BR-V3 | BR-V4 | BR-V5 | BR-V6 | BR-V7 | BR-V8 | BR-V9 | BR-V10 | |
| SON188 | 706 | 173 | 144 | 522 | 116 | 89 | 206 | 300 | 201 | 476 |
| NY186 | 800 | 178 | 144 | 522 | 116 | 89 | 206 | 321 | 204 | 476 |
| MIL | 750 | 173 | 144 | 697 | 116 | 89 | 206 | 404 | 204 | 465 |
| MEN115 | 800 | 178 | 144 | 697 | 116 | 89 | 206 | 384 | 204 | 465 |
| L5 | 800 | 178 | 144 | 697 | 116 | 89 | 206 | 321 | 204 | 509 |
| IP1 | 800 | 178 | 144 | 697 | 116 | 89 | 206 | 321 | 204 | 509 |
| IP2 | 800 | 178 | 144 | 697 | 116 | 89 | 206 | 321 | 204 | 509 |
| ESP1 | 750 | 173 | 150 | 697 | 119 | 89 | 206 | 342 | 204 | 454 |
| IP3 | 800 | 178 | 144 | 697 | 116 | 89 | 206 | 321 | 204 | 509 |
| DK7 | 800 | 178 | 144 | 697 | 119 | 89 | 206 | 279 | 204 | 465 |
| Cat flea | 800 | 178 | 144 | 697 | 116 | 89 | 206 | 321 | 204 | 509 |
| 19535 | 800 | 178 | 144 | 697 | 116 | —a | 206 | 321 | 204 | 465 |
| 20006 | 750 | 178 | 144 | 697 | 119 | 89 | 206 | 363 | 204 | 454 |
| B31 | 800 | 178 | 144 | 697 | 116 | 89 | 206 | 321 | 204 | 509 |
| SON2110 | 750 | 183 | 144 | 638 | 116 | 89 | 204 | 300 | 204 | 476 |
| SON328 | 750 | 173 | 144 | 638 | 119 | 86 | 206 | 363 | 204 | 542 |
| CA19 | 750 | 173 | 144 | 522 | 116 | 86 | 204 | 285 | 204 | 454 |
| 297 | 750 | 173 | 144 | 802 | 116 | 86 | 206 | 454 | 204 | 465 |
| 21305 | 750 | 178 | 144 | 835 | 116 | — | 206 | 404 | 204 | — |
| VEERY | 750 | 178 | 144 | — | 119 | 89 | 206 | 321 | 204 | 520 |
| 27985 | 800 | 178 | 144 | 697 | 116 | 89 | 206 | 300 | 204 | 465 |
| ZS7 | 800 | 178 | 144 | 642 | 119 | 89 | 206 | 300 | 204 | 454 |
| HB19 | 750 | 178 | 144 | 608 | 116 | 89 | 206 | 300 | 207 | 465 |
| 35B808 | 800 | 173 | 144 | 697 | 119 | 89 | 206 | 300 | 204 | 465 |
| Z136 | 706 | 173 | 144 | 697 | 116 | 89 | 206 | 384 | 204 | 608 |
| 26816 | 800 | 178 | 144 | 697 | 116 | 89 | 206 | 342 | 204 | 509 |
| NE56 | 750 | 183 | 154 | 697 | 119 | 89 | 206 | 363 | 204 | 454 |
| B023 | 750 | 168 | 144 | 697 | 113 | 68 | 206 | — | 204 | 465 |
| J1 | 706 | 168 | 144 | 697 | 113 | 68 | 206 | — | 204 | 465 |
| ECM1 | 706 | 168 | 144 | 697 | 113 | — | 206 | — | 204 | — |
| DK3 | 706 | 168 | 144 | 697 | 113 | 68 | 206 | 300 | 204 | 465 |
| DK8 | 706 | 168 | 144 | 697 | 113 | 68 | 206 | — | 204 | 465 |
| BR53 | 750 | 168 | 144 | 697 | 113 | 68 | 206 | — | 204 | — |
| VS461 | 706 | 168 | 144 | 697 | 113 | 68 | 206 | — | 204 | 465 |
| 20047 | 655 | 168 | 144 | 697 | 113 | 89 | 206 | 321 | 204 | 509 |
| N34 | 655 | 168 | 142 | 697 | 113 | 89 | 206 | 342 | 204 | 465 |
| FAR03 | 692 | 168 | 144 | 731 | 113 | 89 | 206 | — | 204 | 465 |
| VSDA | 692 | 168 | 144 | — | 113 | 89 | 206 | — | 204 | 465 |
| PBI | 655 | 168 | 144 | 638 | 113 | 89 | 206 | 363 | 204 | — |
| PBR | 692 | 168 | 144 | 697 | 113 | 89 | 206 | — | 204 | 465 |
| HFOX | 467 | 168 | 144 | 802 | 113 | 68 | 206 | — | 204 | 465 |
—, missing data due to lack of PCR amplification.
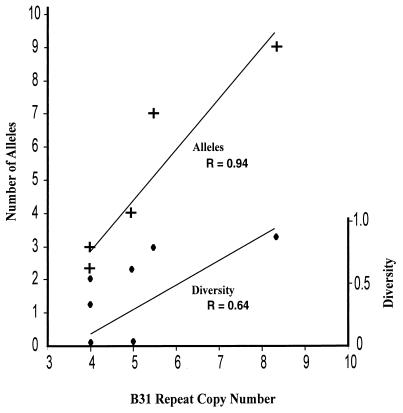
Because the ultimate utility of VNTR loci lies in their diversity, we examined marker diversity by using both allele number and frequency. The allele numbers observed ranged from two (BR-V7) to nine (BR-V8) (Table 3). We observed that the larger the repeat array in strain B31, the greater the VNTR diversity (r = 0.62) and number of alleles (r = 0.94) among globally diverse strains (Fig. 2). This has also been observed previously in F. tularensis and B. anthracis (14, 21). For example, marker BR-V8 has a repeat copy number of 8.3 in type strain B31 and exhibited nine alleles (Table 3). In contrast, marker BR-V9, with a copy number of only three, exhibited only three alleles in our study (Table 3). We observed repeat motifs ranging from 2 bp for BR-V3 to 21 bp for BR-V8 (Table 3). The minimum array sizes observed across all alleles ranged from 1 (BR-V10) to 29 (BR-V3) (Table 3). D values ranged from 0.1 to 0.89, with an overall average D value of 0.51 (Table 3). VNTR markers that exhibit high D values, such as BR-V8 (D = 0.89), possess great discriminatory capacity for identification of genetically similar strains. Less-diverse markers, such as BR-V9 (D = 0.10) (Table 3), may be applied with greater utility for species identification and analysis of evolutionary relationships. The ability to predict VNTR diversity on the basis of array size will allow guided selection of marker loci in future studies.
FIG. 2.
Correlation between repeat copy number and D values. The B. burgdorferi strain B31 repeat copy number (Table 1) was compared with the D value (Pearson coefficient r = 0.62) and the total observed allele number (Pearson coefficient r = 0.94) at each marker locus. A plus sign indicates the marker's total observed allele number versus the repeat copy number at an individual marker locus. A diamond indicates the marker's calculated D value versus the repeat copy number of an individual marker. Analysis was performed with only data from the eight Borrelia markers with noncomplex repeat motifs.
Genetic relationships among isolates.
Ten VNTR marker loci were used to calculate genetic distances among Borrelia strains. UPGMA analysis then revealed 30 distinct genotypes among the 41 Borrelia isolates, with five unique subdivisions evident within these affiliations (Fig. 1). No fixed allelic differences between these clusters were present (Table 4); therefore, cluster formation is due to overall allelic frequency. Clusters I, II, III, and IV include only B. burgdorferi sensu stricto isolates (Fig. 1). All B. burgdorferi strains revealed unique marker allele size combinations, with the exception of B. burgdorferi strains L5, IP1, IP2, IP3, Cat flea, and B31, which were identical at all marker loci (Fig. 1). Isolates B31 and Cat flea were isolated in North America, while strains IP1, IP2, and IP3 are human cerebrospinal fluid isolates from France (Table 2). Nineteen of the 27 B. burgdorferi sensu stricto strains grouped within cluster IV (Fig. 1). MLVA revealed substantial discrimination between B. afzelii and B. garinii in cluster V (Fig. 1). This cluster included seven B. afzelii strains and seven B. garinii strains (Fig. 1). All seven B. afzelii strains assembled within the single subgroup of cluster V-1 (Fig. 1). B. afzelii isolates B023 and BR53 showed 100% marker identity, as did isolates J1, ECM1, DK3, DK8, and VS461 (Fig. 1). Six unique genotypes are evident among the B. garinii isolates, with strains FAR03 and VSDA showing 100% marker identity (Fig. 1). Although the Japanese B. garinii strain (HFOX) loosely clustered within the B. afzelii subgroup (Fig. 1), this strain exhibited only a single B. afzelii-specific chromosomal allelic state (Table 4). The HFOX isolate also exhibited a B. burgdorferi-specific plasmidic allele and a unique allele specific to this isolate alone (Table 4). The loose affiliation of HFOX with B. afzelii (cluster V-1, Fig. 1) does not appear robust. This apparent affiliation is not contradictory to the identity of HFOX in this unrooted tree, as HFOX is actually more closely related to the B. garinii isolates than to the B. afzelii isolates. Overall, the phylogenetic relationships observed in this study are in general agreement with the previous 16S rRNA sequence analysis (31), with the Borrelia MLVA system developed here having a greater ability to discriminate individual strains.
The diversity within the three species is dramatically different and suggests phylogenetic relationships and evolutionary history. For example, we observed that four out of the five clusters contain only B. burgdorferi sensu stricto members. These four groups have great diversity, especially when contrasted with the B. afzelii (group V-1, Fig. 1) and B. garinii (V-2, Fig. 1) clusters. The cohesiveness of the two latter species in one group argues for a more recent common evolutionary derivation, perhaps from a B. burgdorferi sensu stricto ancestor. Certainly, their lack of diversity is due to either a recent origin or a common and pronounced genetic bottleneck.
A more subtle diversity trend was observed within B. burgdorferi sensu stricto when North America and European strains were compared (Fig. 2). We observed greater genetic diversity among the 15 North American samples (mean genetic distance = 0.46) than among 12 European samples (mean genetic distance = 0.41). Perhaps because of a relatively small sample size, this trend is not statistically significant (t = 0.009) but it is consistent with previous evolutionary models postulating a founder effect as North American B. burgdorferi sensu stricto moved to the Old World (15, 23). However, diversity within B. burgdorferi sensu stricto could likewise be affected by lateral transfer of genetic material from other species. In previous studies, four diverse isolates (NE56, 20006, Z136, and ESP1) were shown to have obtained the ospC gene from other species (23). Hence, genetic mixing via lateral transfer may provide an additional mechanism of evolutionary change.
MLVA provides a convenient and rapid method for discrimination of Borrelia strains. Previously, most Borrelia analyses have been performed either phenotypically with monoclonal antibodies or by using DNA sequencing or small-fragment restriction fragment length polymorphisms. These analyses involved single genes or limited genomic loci, which do not effectively reflect the characteristics of the whole organism. In addition, previous studies either were restricted to one species (29) or used a small number of strains (30). In contrast, we examined multiple strains from three species at 10 genetically diverse and informative loci. The characterization of molecular diversity we achieved by applying MLVA to the strain typing of B. burgdorferi, B. afzelii, and B. garinii suggests that this method can be harnessed for the rapid discrimination and identification of the remaining major Borrelia species and will allow further phylogenetic and epidemiological analyses of these genetically diverse organisms.
Acknowledgments
This work was funded by the Cowden Endowment in Microbiology.
REFERENCES
- 1.Anderson, J. F. 1991. Epizoology of Lyme borreliosis. Scand. J. Infect. Dis. Suppl. 77:23-34. [PubMed] [Google Scholar]
- 2.Balmelli, T., and J. C. Piffaretti. 1995. Association between different clinical manifestations of Lyme disease and different species of Borrelia burgdorferi sensu lato. Res. Microbiol. 146:329-340. [DOI] [PubMed] [Google Scholar]
- 3.Barbour, A. G., and D. Fish. 1993. The biological and social phenomenon of Lyme disease. Science 260:1610-1616. [DOI] [PubMed] [Google Scholar]
- 4.Belfaiza, D., D. Postic, E. Bellenger, G. Baranton, and I. S. Girons. 1993. Genomic fingerprinting of Borrelia burgdorferi sensu lato by pulsed-field gel electrophoresis. J. Clin. Microbiol. 31:2873-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgdorfer, W., J. F. Anderson, L. Gern, R. S. Lane, J. Piesman, and A. Spielman. 1991. Relationship of Borrelia burgdorferi to its arthropod vectors. Scand. J. Infect. Dis. Suppl. 77:35-40. [PubMed] [Google Scholar]
- 6.Busch, U., C. Hizo-Teufel, R. Boehmer, V. Fingerle, H. Nitschko, B. Wilske, and V. Preac-Mursic. 1996. Three species of Borrelia burgdorferi sensu lato (B. burgdorferi sensu stricto, B. afzelii, and B. garinii) identified from cerebrospinal fluid isolates by pulsed-field gel electrophoresis and PCR. J. Clin. Microbiol. 34:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casjens, S., N. Palmer R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 8.Casjens, S., M. Delange, H. L. Ley III, P. Rosa, and W. M. Huang. 1995. Linear chromosomes of Lyme disease agent spirochetes: genetic diversity and conservation of gene order. J. Bacteriol. 177:2769-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 1997. Lyme disease—United States, 1996. Morb. Mortal. Wkly. Rep. 46:531-535. [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 1999. Availability of Lyme disease vaccine. Morb. Mortal. Wkly. Rep. 48:35-36. [PubMed]
- 11.Dykhuizen, D. E., and G. Baranton. 2001. The implications of a low rate of horizontal transfer in Borrelia. Trends Microbiol. 9:344-350. [DOI] [PubMed] [Google Scholar]
- 12.Dykhuizen, D. E., D. S. Polin, J. J. Dunn, B. Wilske, V. Preac-Mursic, R. J. Dattwyler, and B. J. Luft. 1993. Borrelia burgdorferi is clonal: implications for taxonomy and vaccine development. Proc. Natl. Acad. Sci. USA 90:10163-10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eggers, C. H., B. J. Kimmel J. L. Bono, A. F. Elias, P. Rosa, and D. S. Samuels. 2001. Transduction by phiBB-1, a bacteriophage of Borrelia burgdorferi. J. Bacteriol. 183:4771-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farlow, J., K. L. Smith, J. Wong, M. Abrams, M. Lytle, and P. Keim. 2001. Francisella tularensis strain typing with multiple-locus, variable-number tandem repeat analysis. J. Clin. Microbiol. 39:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foretz, M., D. Postic, and G. Baranton. 1997. Phylogenetic analysis of Borrelia burgdorferi sensu stricto by arbitrarily primed PCR and pulsed-field gel electrophoresis. Int. J. Syst. Bacteriol. 47:11-18. [DOI] [PubMed] [Google Scholar]
- 16.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 17.Fukunaga, M., A. Hamase, K. Okada, and M. Nakao. 1996. Borrelia tanukii sp. nov. and Borrelia turdae sp. nov. found from ixodid ticks in Japan: rapid species identification by 16S rRNA gene-targeted PCR analysis. Microbiol. Immunol. 40:877-881. [DOI] [PubMed] [Google Scholar]
- 18.Gern, L., A. Estrada-Pena, F. Frandsen, J. S. Gray, T. G. Jaenson, F. Jongejan, O. Kahl, E. Korenberg, R. Mehl, and P. A. Nuttall. 1998. European reservoir hosts of Borrelia burgdorferi sensu lato. Zentbl. Bakteriol. 287:196-204. [DOI] [PubMed] [Google Scholar]
- 19.Hubalek, Z., and J. Halouzka. 1997. Distribution of Borrelia burgdorferi sensu lato genomic groups in Europe, a review. Eur. J. Epidemiol. 13:951-957. [DOI] [PubMed] [Google Scholar]
- 20.Kawabata, H., T. Masuzawa, and Y. Yanagihara. 1993. Genomic analysis of Borrelia japonica sp. nov. isolated from Ixodes ovatus in Japan. Microbiol. Immunol. 37:841-848. [DOI] [PubMed] [Google Scholar]
- 21.Keim, P., L. B. Price A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marconi, R. T., D. Liveris, and I. Schwartz. 1995. Identification of novel insertion elements, restriction fragment length polymorphism patterns, and discontinuous 23S rRNA in Lyme disease spirochetes: phylogenetic analyses of rRNA genes and their intergenic spacers in Borrelia japonica sp. nov. and genomic group 21038 (Borrelia andersonii sp. nov.) isolates. J. Clin. Microbiol. 33:2427-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marti Ras, N., D. Postic, M. Foretz, and G. Baranton. 1997. Borrelia burgdorferi sensu stricto, a bacterial species “made in the USA”? Int. J. Syst. Bacteriol. 47:1112-1117. [DOI] [PubMed] [Google Scholar]
- 24.Murray, P. R., E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th edition. ASM Press, Washington, D.C.
- 25.Nadelman, R. B., and G. P. Wormser. 1998. Lyme borreliosis. Lancet 352:557-565. [DOI] [PubMed] [Google Scholar]
- 26.Nicholls, T. H., and S. M. Callister. 1996. Lyme disease spirochetes in ticks collected from birds in mid-western United States. J. Med. Entomol. 33:379-384. [DOI] [PubMed] [Google Scholar]
- 27.Ohlenbusch, A., F. R. Matuschka, D. Richter, H. J. Christen, R. Thomssen, A. Spielman, and H. Eiffert. 1996. Etiology of the acrodermatitis chronica atrophicans lesion in Lyme disease. J. Infect. Dis. 174:421-423. [DOI] [PubMed] [Google Scholar]
- 28.Peter, O., A. G. Bretz, D. Postic, and E. Dayer. 1997. Association of distinct species of Borrelia burgdorferi sensu lato with neuroborreliosis in Switzerland. Clin. Microbiol. Infect. 3:423-431. [DOI] [PubMed] [Google Scholar]
- 29.Picken, R. N., F. Strle, M. M. Picken, E. Ruzic-Sabljic, V. Maraspin, S. Lotric-Furlan, and J. Cimperman. 1998. Identification of three species of Borrelia burgdorferi sensu lato (B. burgdorferi sensu stricto, B. garinii, and B. afzelii) among isolates from acrodermatitis chronica atrophicans lesions. J. Investig. Dermatol. 110:211-214. [DOI] [PubMed] [Google Scholar]
- 30.Postic, D., N. M. Ras, R. S. Lane, P. Humair, M. M. Wittenbrink, and G. Baranton. 1999. Common ancestry of Borrelia burgdorferi sensu lato strains from North America and Europe. J. Clin. Microbiol. 37:3010-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Postic, D., N. M. Ras, R. S. Lane, M. Hendson, and G. Baranton. 1998. Expanded diversity among California Borrelia isolates and description of Borrelia bissettii sp. nov. (formerly Borrelia group DN127). J. Clin. Microbiol. 36:3497-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmid, G. P. 1985. The global distribution of Lyme disease. Rev. Infect. Dis. 7:41-50. [DOI] [PubMed] [Google Scholar]
- 33.Smith, J. J., J. S. Scott-Craig, J. R. Leadbetter, G. L. Bush, D. L. Roberts, and D. W. Fulbright. 1994. Characterization of random amplified polymorphic DNA (RAPD) products from Xanthomonas campestris and some comments on the use of RAPD products in phylogenetic analysis. Mol. Phylogenet. Evol. 3:135-145. [DOI] [PubMed] [Google Scholar]
- 34.Tyler, K. D., G. Wang, S. D. Tyler, and W. M. Johnson. 1997. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J. Clin. Microbiol. 35:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, G., A. van Dam, L. Spanjaard, and J. Dankert. 1998. Molecular typing of Borrelia burgdorferi sensu lato by randomly amplified polymorphic DNA fingerprinting analysis. J. Clin. Microbiol. 36:768-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, G., A. P. van Dam, I. Schwartz, and J. Dankert. 1999. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin. Microbiol. Rev. 12:633-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weir, B. S. 1990. Genetic data analysis. Sinauer Associates, Inc., Sunderland, Mass.