Abstract
A study of 59 isolates of Bartonella henselae reveals relatively limited diversity among those of human origin (n = 28). Either of two distinct alleles of both gltA and 16S ribosomal DNA (rDNA) was found in all isolates, with a high level of congruity between 16S and gltA inheritance among proven human pathogens. Human isolates from all over Eastern Australia were most commonly 16S rDNA (Bergmans) type I, with the same gltA allele as the type strain (Houston-1). Comparable feline isolates were more commonly 16S type II, with less congruity of inheritance between 16S and gltA alleles. Previously described arbitrarily primed PCR and EagI-HhaI infrequent restriction site PCR fingerprinting techniques separated Bartonella species effectively but lacked discriminating power within B. henselae. Examination of the 16-23S intergenic spacer region revealed for several strains several point mutations as well as a repeat sequence of unknown significance which is readily detected by HaeIII restriction fragment length polymorphism analysis. The bacteriophage-associated papA gene was present in all isolates. Enterobacterial repetitive intergenic consensus PCR proved to be a useful and robust typing tool and clearly separated human isolates (including imported strains) from the majority of feline isolates. Our data are consistent with published evidence and with previous suggestions of intragenomic rearrangements in the type strain and suggest that human isolates come from a limited subset of B. henselae strains. They strengthen arguments for careful exploration of genotype-phenotype relationships and for the development of a multilocus enzyme electrophoresis and multilocus sequence typing-based approach to the phylogeny of B. henselae.
Bartonella henselae is a ubiquitous flea-borne feline pathogen which may infect other animals, including humans, in which it is the most common cause of cat scratch disease (CSD) and bacillary angiomatosis (BA). Cats are infected in proportion to their exposure to the pathogen and the principal vector, Ctenocephalides felis (12). Bacteremic infection in Australian cats may exceed 30% (8), and approximately 5% of Australian blood donors are seropositive (11). However, human infections may be underrecognized, as they are commonly subclinical and it is difficult to culture directly from tissue specimens (17). Phase variation in vitro (3) and a lack of well-established animal models further complicate the assessment of pathogenicity.
The first suggestion that human pathogens may arise from within a limited subset of B. henselae isolates came from a Dutch study of lymph nodes obtained from patients with CSD (5). 16S ribosomal DNA (rDNA) sequences were used to distinguish two genotypes, into which representative strains of the two serogroups recognized among known human pathogenic isolates (Houston, 16S type I; Marseille, 16S type II) (9) could be placed.
DNA fingerprinting methods such as repetitive extragenic palindromic PCR and enterobacterial repetitive intergenic consensus (ERIC)-PCR have also been used to distinguish subgroups within human B. henselae isolates (23), and a variety of such methods applied to 17 German feline isolates suggested four distinct subgroupings (26), all of which were clearly different from the Houston-1 type strain. A subsequent study suggested that 16S type I was more common than type II B. henselae in lymph nodes from CSD patients (25). Similar findings were seen in a more recent European study, which demonstrated at least seven different clonal types among 19 feline isolates from Berlin, most of which were 16S type II, while a human isolate (Berlin-1, obtained from a BA lesion) was indistinguishable by SmaI pulsed-field gel electrophoresis from the Houston-1 (16S type I) strain (2).
There have been a number of attempts to infer phylogenetic relationships from less highly conserved sequences. Sequence variation within the Hsp60 chaperonin gene groEL (18) and the RNA polymerase B subunit gene rpoB (22) separates B. henselae clearly from other species in the same genus and separates Marseille strains from Houston-1 strains (18, 22). groEL sequence variation within six human isolates of disparate origin separated one group (in which B. henselae Houston-1, SA-2, and 90-615 were all identical) from another, in which latter cluster one isolate (Fizz) could be distinguished from the other two (Marseille and CAL-1) by minor sequence variation (29). The deduced sequence of the cell division protein FtsZ in 15 human and feline strains also fell into three groups in a recent study, with all three types represented only among cat isolates (10). There were no contradictions between FtsZ and groEL-derived groupings, although few common strains were tested, and the reported differences were minor.
The citrate synthase (gltA) gene sequence can be used to distinguish between Bartonella species (6, 20) and between the Marseille and Houston-1 strains of B. henselae (6). Sequence variation in the riboflavin synthase gene ribC (4) was described for the (Sander) variant II group of 17 previously described feline isolates, all of which were quite distinct from Houston-1, and for the Bergmans 16S rDNA type II isolates (26).
We are therefore faced with a confusing array of typing methods and systems which have been developed in different strain sets, often with small sample numbers (particularly of proven human pathogenic strains). There appear to be two distinct serotypes which infect humans (9), and sequence divergence in conserved regions points to at least two lineages. The Houston-1 (type) and Marseille strains may thus represent distinct phylogenetic groupings, but our understanding of geographical distribution is incomplete and clear genotype-pathogenicity correlations remain undefined. We therefore compared a group of human and feline isolates in an Australian setting, using a number of these methods.
(This study was presented in part at the ASR-Bartonella joint conference, Big Sky, Mont., August 2001.)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Thirty-one Bartonella isolates from cats and 28 B. henselae isolates from humans were obtained from the sources listed in Table 1. Human isolates were obtained by telephone calls and letters to every large medical microbiology laboratory in Australia to identify existing isolates (most were freezer-stored isolates) and comprise all of the isolates in Australia available to us at the commencement of the study. Fresh blood was collected by aseptic venipuncture of cats anesthetized after injury or for castration and was added as soon as possible after collection to an equal volume of presterilized lysing solution (0.6 g of saponin and 0.6 g of liquoid in 100 ml of 0.9% saline solution) in a sterile centrifuge tube. After mixing and standing for 5 min, sediment obtained at 5,000 × g was plated onto chocolate blood agar (Oxoid blood agar base no. 2 with 5% defibrinated sheep blood and vitamin K-hemin) and incubated for 10 days. All strains were subcultured in the laboratory on chocolate blood agar (Oxoid blood agar base no. 2) containing 5% horse blood (CBA) at 37°C in 5% CO2 for 5 to 7 days or were grown in liquid medium (27) at 37°C in 5% CO2 with intermittent shaking (100 rpm) in cotton wool-plugged Pyrex Erlenmeyer flasks (as specified in text). Bartonella quintana (J. Robson) and Bartonella vinsonii (ATCC 51672) were cultured under similar conditions. Bartonella bacilliformis (ATCC 35686) was grown on a CBA slope at 26°C for 10 to 14 days. The passage number and agar-pitting phenotype for B. henselae strains is listed in Table 1.
TABLE 1.
B. henselae isolates studieda
| Isolate(s) | Source | 16S type | gltA type | AP type | ERIC type | IRS type |
|---|---|---|---|---|---|---|
| Feline isolates (n = 31) | ||||||
| NU4695 | Blood | I | H | H | E4H | 1 |
| RMC10† | Blood | I | H | H | E4H | 1 |
| HC54 | Blood | I | H | H | E4H | 2 |
| HC69 | Blood | I | H | M | E4H | 2 |
| HC62† | Blood | I | H | H | E1 | 2 |
| RMC3* | Blood | I | H | H | E5 | 2 |
| NU4428† | Blood | I | H | M | E2 | 2 |
| HC71 | Blood | I | M | H | E2 | 2 |
| NU4714*† | Blood | II | H | H | E4H | 1 |
| HC35*, HC55 | Blood | II | H | H | E3a | 1 |
| RMC8* | Blood | II | H | Ha | E3a | 2 |
| NU4681* | Blood | II | H | H | E3a | 1 |
| HC60† | Blood | II | H | H | E3b | 1 |
| RMC1* | Blood | II | H | H | E1 | 2 |
| RMC12 | Blood | II | H | M | E3a | 1 |
| NU4423† | Blood | II | M | H | E4H | 2 |
| RMC11 | Blood | II | M | M | E7 | 1 |
| RMC4 | Blood | II | M | M | E7 | 2 |
| HC41, HC48†, HC61, HC63†, HC77 | Blood | II | M | M | E2 | 2 |
| RMC2† | Blood | II | M | M | E6 | 2 |
| RMC5 | Blood | II | M | M | E1 | 2 |
| RMC6†, RMC9† | Blood | II | M | M | E5 | 2 |
| RMC7, Berlin-2 | Blood | II | M | M | E5 | 2 |
| NU4713 | Blood | II | M | H | E2 | 2 |
| Human isolates (n = 28) | ||||||
| BH2 | CSD | I | H | H | E4H | 1 |
| ATCC 49793/NZRm3492 (s4) | BA | I | H | H | E4H | 1 |
| ATCC 49882 (Houston-1)†, Berlin-1 | BA | I | H | H | E4H | 2 |
| BH3(s4), BH5s2*, R987, JR1, JR2, JR3, JR5, JR6, JR7, JR8, JR9, JR12, JR13, JR14, JR15, JR17, JR18, JR19, JR20, JR22, JR23 | CSD | I | H | H | E4H | 2 |
| R1073† | CSD | II | H | H | E4H | 2 |
| BH4 | CSD | II | M | M | E4M | 2 |
| URLLY-8 (Marseille)† | BA | II | M | M | E4M | 2 |
Symbols: *, Isolate embeds into solid agar and displays ratchet motility at this passage number; †, 16S type was determined by direct sequencing. s2, second passage after initial isolation; (s4), third to fifth passage after initial isolation. If not specified, RMC-prefix isolates (this work) were studied at passage numbers 2 to 3; HC-prefix isolates were a gift of Tom Gottlieb (8) and were studied at passage numbers 4 to 6; NU-prefix isolates (15) were studied at passage numbers 3 to 5; R- and JR-prefix strains (this work and Fournier et al. [12a]) were all studied at passage numbers 4 to 7. Other isolates were isolated in our laboratory or obtained from the following: Mardjan Arvand, Hygiene-Institute, Heidelberg, Germany (2) (Berlin-1 and -2); R. Bunter, Goulburn Valley Base Hospital, Goulburn, NSW, Australia (BH2); the New Zealand quality control strain NZRm3492 is thought to be a lab-passaged derivative of the widely used strain ATCC 49793, and they are inseparable by all typing methods; Jock Harkness, Royal North Shore Hospital, Sydney, Australia [BH3(s4)]; T.H.E. Pathology, Fairfield Heights, NSW, Sydney, Australia [BH5(S2)]; and D. Raoult, Unite des Rickettsies, Marseille, France (9) (URLLY-8 [Marseille]). Strains with the ATCC prefix were obtained from the American Type Culture Collection, Manassas, Va. 16S-23S regions were sequenced in all feline strains, in all Sydney and imported human strains, and in JR2.
Preparation of DNA and lysates.
Bacteria grown on CBA plates were washed and resuspended in 1 ml of 0.01 M phosphate-buffered saline, pH 7.2, to a final optical density at 550 nm of 0.5, incubated for 10 min at 95°C, and centrifuged at 13,000 × g for 30 s. Supernatants (lysates) were used as templates in PCRs. Genomic DNA was extracted from 2 ml of early-stationary-phase (7-day) broth culture with a commercially available kit (Promega Corporation, Madison, Wis.) according to the manufacturer's instructions.
PCR, biological reagents, gel electrophoresis, and sequencing reactions.
Unless otherwise stated, the following concentrations of reagents were used for PCR: deoxynucleoside triphosphates, 200 μM each (Astral Scientific, Sydney, Australia); MgCl2, 1.5 mM (Astral Scientific); 2 U of Taq polymerase (Promega); 1× PCR buffer (Promega); and 2 μl of bacterial lysate in 50-μl reaction volumes. Control strain lysates and distilled water served as controls. DNA amplification was performed in a PC-960C thermal cycler (Corbett Research, Sydney, Australia). PCR products were purified with the GeneClean kit (Bio 101 Inc., Carlsbad, Calif.), and both strands were sequenced by use of an automated sequencer (ABI 373 Genetic Analyzer; Perkin-Elmer Biosystems). Alignments were performed with EclustalW (ANGIS), and finalized sequences were submitted to the GenBank database.
Electrophoresis of DNA was performed in 1 to 2% (wt/vol) agarose (Promega) and visualized using ethidium bromide on an UV transilluminator using 667 Polaroid film. Images were scanned with an HP ScanJet 5300C (Hewlett Packard) and Adobe PhotoShop 4.0 (Microsoft).
16S rRNA type-specific PCR and sequencing.
Primers BH1 and BH2 were used in conjunction with the broad-host-range primer 16SF according to the method of Bergmans et al. (5) at 100 nM and at annealing temperatures of 54 to 56°C, as specified in the text. 16S sequencing was performed using primers 16SF [5′-AGAGTTTGATCCTGG(CT)TCAG-3′] and 16SR [5′-CTTTACGCCCA(AG)TAA(AT)TCCG-3′] (19).
Amplification of gltA and 16S-23S intergenic region and ITS restriction fragment length polymorphism.
16S-23S intergenic spacer region (ITS)-PCR was performed as previously described (19), with minor modifications. External primers RPC5 (5′-AAGTCGTAACAAGGTA-3′) and R23S2693 (5′-TACTGGTTCACTATCGGTCA-3′) (200 nM) were used in a 100-μl PCR mixture (19). Internal primers BHITSF (5′-AAGAGGATGCCCGGGAAGGT-3′) and BHITSR (5′-GCGTTCTCTGCCTTGTGCAA-3′) (200 nM) were designed (this study) and used to amplify a partial ITS region for sequence analysis as follows: 5-min denaturation at 95°C; 35 cycles of 95°C for 30 s, 50°C for 60 s, and 72°C for 90 s; and final extension at 72°C for 10 min. Restriction enzymes (HaeIII and AluI) from New England Biolabs were used according to the manufacturer's instructions to digest 16S-23S amplicons. Previously described primers (BhCS.1137 [5′-AATGCAAAAAGAACAGTAAACA-3′] and CS140f [5′-TTACTTATGATCCKGGYTTTA-3′] [200 nM]) were used to generate and sequence gltA amplicons (6).
AP-PCR and ERIC-PCR.
Arbitrarily primed (AP)-PCR was performed with M13 bacteriophage core sequence primer (5′-GAGGGTGGCGGTTCT-3′ [200 nM]) as described previously (26). Primers targeting the ERIC sequence, ERIC-1R (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC-2 (5′-AAGTAAGTGACTGGGGTGAGAGCG-3′) both at 200 nM), were used with 4 μl of bacterial lysate in the ERIC-PCR (28) under the following conditions: initial denaturing at 95°C for 5 min; 35 cycles of denaturing at 95°C for 20 s, annealing at 50°C for 60 s, and extension at 72°C for 2 min; and a final extension at 72°C for 10 min. ERIC-PCR fingerprints were analyzed by applying the Dice coefficient to peaks, using GelCompar software, version 4 (Applied Maths, Kortijk, Belgium), according to the manufacturer's instructions. The unweighted pair group method with arithmetic means was used with a 1.2% tolerance in band positions to analyze clustering of fingerprint patterns.
IRS-PCR with HhaI and EagI.
PCR reagent concentrations and amplification conditions for infrequent restriction site (IRS)-PCR have been described by Handley and Regnery (13). PCR products were separated on a 6.5% polyacrylamide gel in 1× Tris-borate-EDTA buffer for 3 h at 150 V.
pap PCR.
Amplification reactions were carried out in a 50-μl final volume containing 200 nM (each) PAP-1 and PAP-2 primers (1). Amplifications were performed as follows: initial denaturation at 95°C for 5 min; 30 cycles of 95°C for 30 s, 50°C for 1 min, and 72°C for 90 s; and final extension at 72°C for 10 min.
Statistical analysis.
Chi-square analysis was performed where indicated, using Minitab for Windows release 12 (Minitab Inc., State College, Pa.).
Nucleotide sequence accession numbers.
The 16S-23S rRNA intergenic spacer region sequences of the following isolates from this study have been deposited in the GenBank and EMBL nucleotide sequence databases under the following accession numbers: HC54, AJ439687; HC69, AJ439688; RMC3, AJ441257; RMC10, AJ441256; BH2, AJ457178; and JR2, AJ457177. The gltA sequence of isolate BH4 has been deposited under accession number AJ439406.
RESULTS
16S rDNA type I is more common in human isolates.
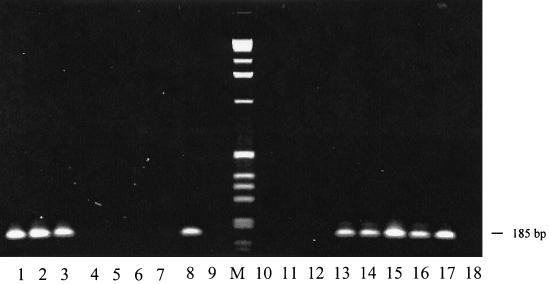
We studied 59 isolates, half of human origin and half of feline origin (Table 1). Human isolates were mostly from eastern Australia (New South Wales [NSW] and Queensland) and the feline isolates were from two Sydney (NSW) collections and one New Zealand collection. The majority of feline isolates from Sydney (and New Zealand) were 16S rDNA type II (Table 1). In contrast, almost all (22 of 24) Australian human isolates yielded positive results in the type I-specific PCR; one Brisbane strain (R1073) and one Sydney strain (BH4) were clearly type II. Clear-cut results were obtained from boiled lysates in the majority (48 of 59) of cases; 11 samples (HC48, HC60, HC62, HC63, NU4423, NU4428, NU4714, RMC2, RMC6, RMC9, and RMC10) reacted in both assays at low stringency (Tm, 54°C) (Fig. 1). For five of these samples, accurate and unambiguous results were also obtained by PCR at a slightly higher stringency (Tm, 56°C). Typing of these 11 and certain additional strains was confirmed by direct sequencing as indicated in Table 1.
FIG. 1.
Representative gel electrophoresis of 16S rDNA type-specific PCR. Lanes 1 to 9, BH1 type-specific PCR; lanes 10 to 18, BH2 type-specific PCR. Lanes 1 and 10, JR1; lanes 2 and 11, ATCC 49793; lanes 3 and 12, ATCC 49882 (Houston-1); lanes 4 and 13, URLLY-8 (Marseille); lanes 5 and 14, R1073; lanes 6 and 15, NU4423; lanes 7 and 16, HC35; lanes 8 and 17, NU4428; lanes 9 and 18, negative (water). M, 0.07- to 12.2-kbp marker (Roche Diagnostics, Mannheim, Germany).
A common variable repeat region in the 16S-23S ITS.
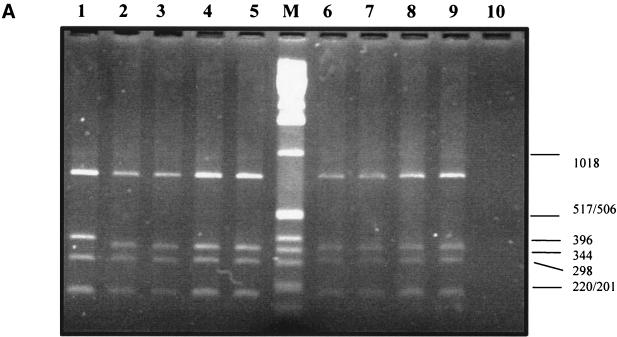
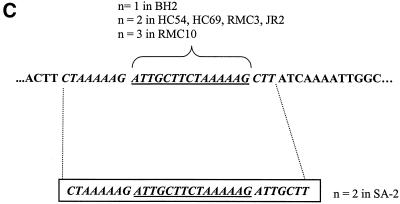
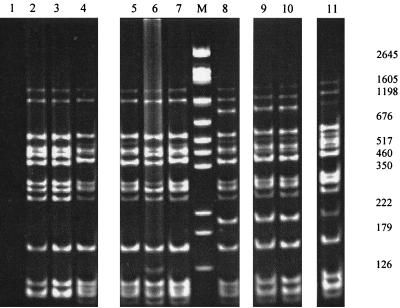
For B. bacilliformis and B. vinsonii, the expected 700-bp and 1.6-kbp fragments, respectively, were generated (data not shown). HaeIII digests generated four fragments for each of the 59 isolates which ranged in size from 190 to 700 bp, with some (feline isolates HC54, HC69, RMC3, and RMC10 and human isolates JR2 and BH2) having one slightly larger fragment (Fig. 2A). Forty isolates, including R1073, R987, JR2, and all those from Sydney and overseas, were sequenced. Additional repeat sequences were evident only in the six isolates with HaeIII polymorphism (Fig. 2B and C), and all these repeats differed slightly from that previously described for a U.S. (human) isolate, SA-2 (14) (Fig. 2C). There were several point mutations (Fig. 2B), which did not usefully separate groups. The majority of fully sequenced isolates (23 of 40) were identical to the Houston-1 and Marseille 16S-23S ITS strains.
FIG. 2.
(A) Representative HaeIII restriction fragment length polymorphism patterns of PCR-amplified 16S-23S ITS region. Lane 1, JR2; lane 2, JR3; lane 3, URLLY-8 (Marseille); lane 4, ATCC 49882 (Houston-1); lane 5, ATCC 49793; lane M, 0.07- to 12.2-kbp marker (Roche) (sizes are indicated on the right-hand margin); lane 6, HC63; lane 7, NU4428; lane 8, NU4714; lane 9, RMC1; lane 10, negative control (water). (B) 16S-23S rRNA ITS sequence of B. henselae Houston-1 (GenBank accession no. L35101). The tRNA, 16S, and 23S rRNA genes are underlined. The repeat region is boxed, and arrows denote base substitutions or insertions which vary from the Houston-1 sequence, as follows: 1, G-to-A substitution in HC35, HC54, HC60, HC62, HC69, NU4681, NU4714, JR2, BH2, R987, and R1073; 2, G-to-A substitution in HC54, HC69, NU4681, JR2, and BH2; 3, T-to-G substitution in HC35 and HC69; 4, C-to-T substitution in HC69; 5, C-to-T substitution in HC35, HC60, HC62, NU4428, NU4681, NU4714, RMC1, RMC8, RMC12, R987, and R1073; 6, A-to-G substitution in HC35, HC54, HC60, HC62, HC69, NU4428, NU4714, RMC1, RMC3, RMC8, RMC10, RMC12, R1073, R987, and JR2; 7, G-to-A substitution in HC60, HC62, NU4428, NU4714, RMC1, RMC8, RMC12, R987, and R1073. (C) 16S-23S ITS repeat region. There are two ATTGCTTCTAAAAAG repeats in HC54, HC69, RMC3, and JR2; three repeats in RMC10; and one repeat in BH2. The boxed region illustrates the previously described repeat sequence in B. henselae SA-2 (14).
Separate inheritance of gltA and 16S rDNA.
We observed conservative substitutions only at positions 472 (G:A) and 860 (C:T) in the gltA (citrate synthase) gene when isolate sequences for HC41, -48, -61, -63, -71, and -77, NU4423 and -4713, RMC2, -4, -5, -6, -7, -9, and -11, and Berlin-2 (all feline) and for BH4 and Marseille (human) were compared with the deposited sequence of the type strain (Houston-1; GenBank L38987). Only two gltA alleles were seen in the 59 sequences analyzed, and strong agreement was found with the 16S type (27 of 28) in human isolates. Interestingly, the Houston-variant gltA sequence was also common among feline isolates (15 of 31) and many feline strains therefore gave gltA results which appeared to conflict with the 16S type (Table 1).
AP-PCR fingerprinting correlates imperfectly with 16S type and gltA.
Single-primer (M13) AP-PCR distinguished B. henselae Houston-1 from the Marseille strain (Fig. 3), and almost all (58 of 59) isolates yielded one of these two simple patterns upon agarose gel electrophoresis (Table 1). One isolate (RMC8) exhibited altered migration of the 460-bp fragment (Fig. 3). Results for all four imported human isolates and 23 of the 24 Australian human isolates were consistent with the 16S and gltA data, with the Houston-1 (H) pattern dominant. The one exception among the 28 human isolates was again R1073 (16S type II), for which gltA and AP-PCR patterns were both H-type. Half (14 of 24) of the Sydney feline isolates were AP-PCR Marseille (M) type, with only one of these (HC69; Bergmans 16S type 1) conflicting with the (H-variant) gltA sequence. In contrast, AP-PCR correlated poorly with the gltA allele for New Zealand feline isolates, the majority of which yielded the H pattern on AP-PCR (Table 1).
FIG. 3.
AP-PCR representative gel. Lane 1, ATCC 49882 (Houston-1); lane 2, JR2; lane 3, RMC1; lane 4, BH2; lane 5, RMC8; lane 6, URLLY-8 (Marseille); lane 7, HC63; lane 8, HC77; lane 9, BH4; lane 10, NU4428; M, pGEM marker (Promega; molecular size in base pairs is given along the right border).
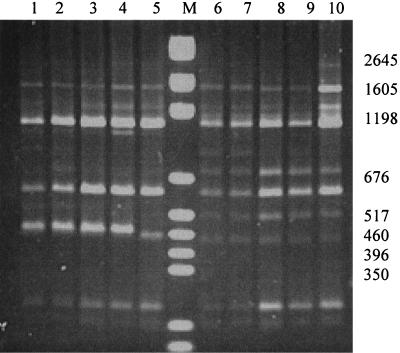
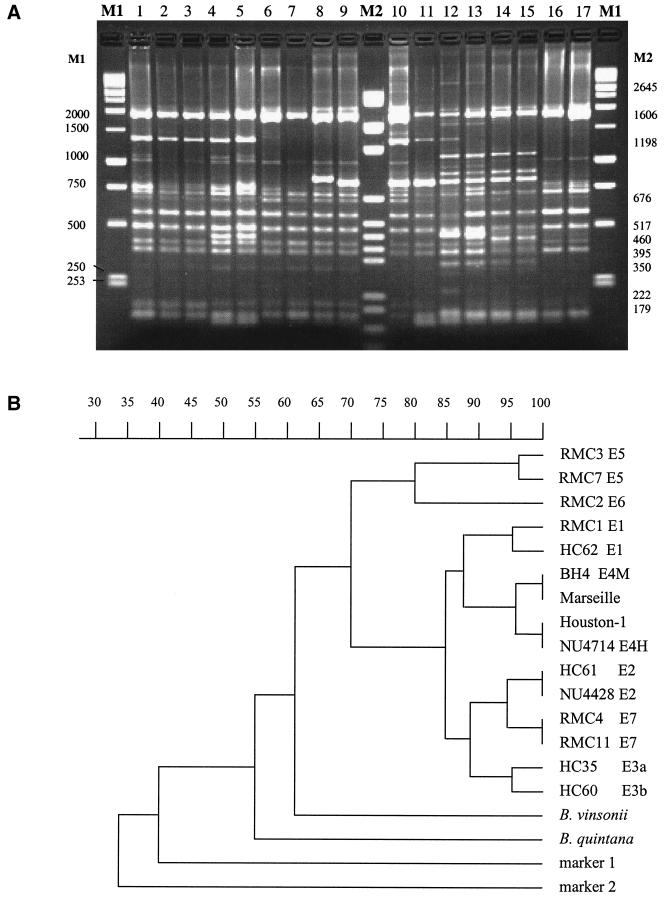
ERIC-PCR suggests a cluster containing all human isolates.
Amplification with ERIC primers generated a number of easily visible bands, which ranged from approximately 300 to 1,800 bp. Using this technique, seven reproducible patterns (E1 to E7) were characterized by the presence or absence of one to five bright bands (Fig. 4). Each isolate was subjected to this method on at least three occasions, with excellent reproducibility. We observed a greater number of easily distinguished bands than was described earlier, with Houston-1 yielding a pattern similar to that previously defined as E4 (26), with some additional bands. We also found that the pattern yielded by the Houston-1 type strain was universal among human isolates, with reproducible separation of isolates BH4 and Marseille (yielding what we termed the E4M pattern) from all other human isolates (E4H pattern) on the basis of a prominent band at approximately 460 bp (Fig. 4A) seen only for these strains (Table 1). The human strain R1073, of 16S type II but with H-variant gltA, yielded AP-PCR and ERIC patterns typical of Houston-1. E3 can be subdivided into two groups based on differing migration of the 800- to 850-bp band (Fig. 4), although this distinction may not be useful as it described only one feline isolate (HC60). Thus, all 28 human isolates clustered together by this method along with 6 of 31 feline isolates, while the remainder yielded quite diverse patterns.
FIG. 4.
ERIC-PCR. (A) Representative gel electrophoresis of ERIC-PCR. Lanes 1 to 3, pattern E4H (strains ATCC 49882 [Houston-1] and NU4714); lanes 4 and 5, pattern E4M (strains BH4 and Marseille); lanes 6 and 7, pattern E2 (strains HC61 and NU4428); lanes 8 and 9, patterns E3a and -b, respectively (strains HC35 and HC60); lanes 10 and 11, pattern E1 (strains HC62 and RMC1); lanes 12 and 13, pattern E5 (strains RMC3 and RMC7); lanes 14 and 15, pattern E6 (strain RMC2 duplicate); lanes 16 and 17, pattern E7 (strains RMC11 and RMC4); lanes M1 and M2, molecular size markers. (B) Dendrogram. B. quintana and B. vinsonii patterns are quite distinct from all others (not shown in panel A) and are included in a merged reconstructed dendrogram. The dendrogram reflects the similarity of distribution of ERIC priming sites and is derived from gels (including that shown in panel A), all of which included B. henselae and the two molecular size markers and were analyzed identically.
IRS-PCR.
HhaI/EagI IRS-PCR-generated fragments ranged in size from <100 to 1,500 bp. Certain strains possessed two additional fragments, at approximately 900 and 200 bp (Fig. 5), which have been previously proposed to distinguish Houston-1 from Marseille type strains (13). While Marseille and BH4 gave the previously predicted pattern (13), a number of strains, including R1073, Berlin-1, and our copy of the Houston-1 type strain, also lacked these fragments in repeated experiments (Table 1).
FIG. 5.
EagI-HhaI IRS-PCR. Lane 1, negative control; lane 2, NU4713; lane 3, HC61; lane 4, BH2; lane 5, R1073; lane 6, Marseille; lane 7, ATCC 49882 (Houston-1); lane 8, NU4714; lane 9, ATCC 49793; lane 10, HC35; lane 11, HC60; lane M, pGEM marker (Promega) (molecular sizes are given along the right border).
papA PCR.
All 59 isolates gave a single product of the expected size (1.1 kb) with primers specific for the papA gene, while B. vinsonii and B. quintana gave negative results (data not shown).
DISCUSSION
We have compared a large group of human disease-associated isolates (n = 28) with randomly collected feline isolates (n = 31) of B. henselae and found restricted genetic diversity among the proven human pathogens. Either of two distinct alleles of 16S rDNA or the housekeeping gene gltA can be used to divide our isolates into groups. The Houston-1 gltA allele is predominant in Australia (38 of 54 Australasian isolates), and human isolates are usually 16S type I, despite the fact that most feline isolates are 16S type II. Congruence (of 16S type I with Houston-variant gltA and of 16S type II with Marseille-variant gltA) is very high within human isolates (27 of 28), and the Houston-variant gltA-16S type I combination is predominant (23 of 24 Australian human isolates).
Our collection is dominated by CSD-related human isolates. It has been speculated that BA isolates from immunocompromised hosts might exhibit diversity similar to that of feline isolates, with more virulent types of lesser diversity seen for CSD (21). However, there has been no evidence for a distinction between BA-associated and CSD-associated isolates evinced in this or earlier studies (5, 10, 23). The slower-growing agar-pitting phenotype, lost on serial passage in vitro, is thought to be linked to invasiveness of B. henselae (3), and isolates grown from human tissues which successfully invade tissue culture monolayers may fail to grow subsequently on solid media (17). Such isolates might therefore be underrepresented, perhaps biasing our study towards a less diverse subset of easily cultured human isolates. However, the human strains grown in our own laboratory are clearly typical Houston or Marseille types with DNA fingerprints very similar to those of imported strains originally reported as agar-pitting invasive organisms, while the relatively low rate of the agar-pitting phenotype overall is roughly equivalent between feline and human isolates and best explained by the fact that most of the isolates are known to have been passaged in vitro on several occasions (3).
Regional differences may also be important (30), and it is plausible that isolates (human) from Queensland are less diverse than isolates (feline) from NSW and New Zealand, thereby distorting the data. We do not have a feline collection from Queensland to compare with the Queensland human strains, and it is likely that the NSW and Queensland cat populations are distinct. However, our imported human isolates are more similar to human isolates from NSW than are feline strains from the same region, and the human isolates differ substantially from feline isolates described in other studies (5, 7, 16, 25, 26, 30). We therefore believe that sample bias in our collection operates in the opposite direction to our findings. That is, a regional outbreak should be reflected in diminished diversity in the feline isolates, which come largely from a couple of collections which were close in space and time. Our data show the opposite to be true. If there is a characteristic human pathogenic type, it has arisen from within different 16S types and clearly has not displaced other genotypes in cats. Attributes which increase pathogenicity in humans may be relatively unimportant in the cat-flea cycle and thus be selected weakly or not at all in the main (feline) reservoir. Biological success for B. henselae need not be accompanied by serious disease in cats, and a healthy host offers distinct advantages in the host-vector cycle. The greater opportunity for genetic exchange within cats or fleas would be expected to result in greater diversity of feline isolates of B. henselae, and some or all 16S type II strains might be better adapted to cats or fleas, in which it appears to be more common (5, 7, 16, 25, 26, 30).
ERIC-PCR has also previously revealed more limited variation among U.S. human isolates (23) than was subsequently reported among European feline isolates (24, 26). All 28 of our human isolates were grouped with a minority (6 of 31) of the feline isolates by the ERIC-PCR method. In this group of 34 (E4) strains (Table 1), 16S and gltA genotypes were congruent in all but two (a feline isolate, NU4714, and a human isolate, R1073). This was significantly different for feline isolates of other ERIC types, being true in 17 of the remaining 25 (32 of 34 versus 17 of 25; χ21 = 6.981; P = 0.0082) (Table 1). Our ITS sequence data suggest a connection between HC54, HC69, RMC3, and RMC10 (NSW feline isolates), BH2 (NSW human isolates), and JR2 (Queensland human isolate) (Table 1). All of these are 16S type I, have the Houston-variant gltA sequence, and with the exception of RMC3, cluster in the E4 group by ERIC-PCR, although none are identical. The E4 group itself is unlikely to be a simple clonal expansion, as it includes all human isolates from Australia, Europe, and the United States and is strikingly underrepresented among feline isolates and since members of the group are individually separable by their gltA and 16S alleles and by 16S-23S variations.
In the first study to suggest such an association, 32 of 40 Dutch CSD-associated isolates were classified as 16S rDNA type I, as was a similar proportion (5 of 6) of U.S. (including H-1) BA-associated isolates (5) and 6 of 7 U.S. isolates (also including H-1) associated directly or indirectly with human disease (10). Despite the geographical disparity and inability of the 16S typing method to distinguish B. bacilliformis and B. quintana from B. henselae Houston-1 (5), those data suggested that 16S rDNA type I is overrepresented in clinical isolates. Consistent with this, and with our own data, almost all of 17 feline isolates from Freiburg (including four of five CSD-associated isolates) were 16S type II (26), while type II was subsequently found in a minority (9 of 32) of human lymph node samples from the same region (24).
Serotyping and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of whole-cell lysates (16), as well as gltA (6) and groEL sequencing (30), have consistently suggested only two subtypes within B. henselae. La Scola and colleagues (16) placed several well-studied human and 22 additional feline strains into one of two serotypes of B. henselae by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting, with matching genotypes evident on 16S typing and pap31 gene sequencing. Another recent study from the same laboratory (30) showed that the great majority (>96%) of 107 B. henselae-infected human lymph nodes had one of only two very closely related pap31 alleles. The sequence of a more conserved gene (groEL) revealed only two alleles. We have found that there is a predominance of 16S type II strains in feline isolates and a greater representation of type I strains in human isolates and also that the considerable diversity within 16S type II strains is much greater than that in the three human 16S type II strains. Our data are therefore quite consistent with these and a number of previous studies (5, 7, 16, 25, 26, 30).
Previous analyses of the 16S-23S intergenic spacer region revealed point mutations and a 30-bp repeat region of unknown significance in one isolate of U.S. origin when compared with Houston-1 (14). We have found similar repeats to be not infrequent in our much larger sample, along with a number of point mutations. AP-PCR patterns among human isolates all agreed with the gltA type (mostly Houston-1), and correlation with gltA type was also high among feline isolates (25 of 31). The ERIC method clearly differentiated Houston and Marseille types within the E4 cluster, which again correlated with both gltA and AP-PCR type for all human isolates and all but one feline isolate (HC69). The extra band proposed as a marker of the Houston-1 type for the EagI-HhaI IRS-PCR method (13) was detected only for a local Sydney isolate and for ATCC 49793. It was not present in the Houston-1 isolate we have, nor in the Berlin-1 isolate, previously shown to be indistinguishable from H-1 by pulsed-field gel electrophoresis analysis (2). We therefore believe that EagI-HhaI IRS-PCR and the (M13) AP-PCR are of limited value as genotyping tools.
Plasmids have not been described for B. henselae, and the extent of horizontal gene transfer is unknown. Bacteriophage particles have been associated with B. henselae and B. bacilliformis (1), and phage-mediated virulence acquisition is an appealing explanation. All isolates were positive by PCR for the papA gene, but productive bacteriophage infection was not tested for and it is not known whether other bacteriophage-related elements are present in every genome. Imperfect linking of distinct 16S rDNA and gltA alleles is consistent with the suggestion that recombination events may have occurred in the B. henselae Houston-1 genome (S. Anderson, unpublished data) and results in many feline and one human isolate falling into different types depending on the choice of sequencing target. Acquisition of specific virulence-encoding regions might well result in lowered tolerance of reduplication and/or recombination events and thus be reflected in limited diversity. In that case, we might predict a similar disequilibrium in other housekeeper genes (e.g., ribC) to that which we have shown for gltA and 16S rDNA, particularly in feline strains.
Even greater discordance is evident with DNA fingerprinting methods, although we believe that human isolates in Australia are reliably distinguished by ERIC from most feline isolates. It is also predictable that variations in DNA fingerprinting patterns of various types would be greater in feline isolates, as reported herein and previously. It seems that neither ERIC nor EagI-HhaI IRS-PCR can reliably separate serotypes or genotypes and that all such typing schemes and relationships based upon them need careful reevaluation.
We know that phase variation induced by serial passage in vitro may be accompanied by important alterations in phenotype (3), but the extent to which this affects genotype is ill-defined. It is possible that infecting clones undergo characteristic rearrangements under selection pressure in humans, but this would also sit poorly with the finding of a minority of similar strains in cats. Our data nevertheless suggest several hypotheses for testing: firstly, that detailed surveys elsewhere would show similar clustering among proven human pathogens; secondly, that those feline isolates which are similar to the human group may be intrinsically more pathogenic in humans; thirdly, that intra- and/or intergenomic genetic transfer occurs in vivo; and finally, that the discordant inheritance seen in this study will be seen for other genes. Preliminary studies in this laboratory have already confirmed the last to be true for groEL and ftsZ (unpublished data). It is likely that such discordance reflects an ability to exchange DNA between strains (and perhaps species) during coinfection, a situation which one would predict is more common in cats. The significance of the 16S type is therefore as uncertain as that of any other highly conserved genes of apparently limited allelic type, but the need for a more comprehensive study of multiple loci within a context such as multilocus sequence typing is clear.
Acknowledgments
This work was supported by grant RN084/99 from the Clive and Vera Ramaciotti Foundations and grants from the Westmead Millennium Institute. D.B. is the recipient of an Australian Postgraduate Research Award.
We thank G. Playford for assistance with statistical analysis.
REFERENCES
- 1.Anderson, B., D. Scotchlas, D. Jones, A. Johnson, T. Tzianabos, and B. Baumstark. 1997. Analysis of a 36-kilodalton protein (PapA) associated with the bacteriophage particle of Bartonella henselae. DNA Cell Biol. 16:1223-1229. [DOI] [PubMed] [Google Scholar]
- 2.Arvand, A., A. J. Klose, D. Schwartz-Porsche, H. Hahn, and C. Wendt. 2001. Genetic variability and prevalence of Bartonella henselae in cats in Berlin, Germany, and analysis of its genetic relatedness to a strain from Berlin that is pathogenic for humans. J. Clin. Microbiol. 39:743-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batterman, H. J., J. A. Peek, J. S. Loutit, S. Falkow, and L. S. Tompkins. 1995. Bartonella henselae and Bartonella quintana adherence to and entry into cultured human epithelial cells. Infect. Immun. 63:4553-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bereswill, S., S. Hinkelmann, K. Manfred, and A. Sander. 1999. Molecular analysis of riboflavin synthesis genes in Bartonella henselae and use of the ribC gene for differentiation of Bartonella species by PCR. J. Clin. Microbiol. 37:3159-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmans, A. M. C., J. F. P. Schellekens, J. D. A. van Embden, and L. M. Schouls. 1996. Predominance of two Bartonella henselae variants among cat-scratch disease patients in The Netherlands. J. Clin. Microbiol. 34:254-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birtles, R. J., and D. Raoult. 1996. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int. J. Syst. Bacteriol. 46:891-897. [DOI] [PubMed] [Google Scholar]
- 7.Box, A. T. A., A. Sander, D. Perschil, D. Goldenberger, and M. Altwegg. 1996. Cats are probably not the only reservoir for infections due to Bartonella henselae. J. Microbiol. 27:101-102. [Google Scholar]
- 8.Branley, J., C. Wolfson, P. Waters, T. Gottlieb, and R. Bradbury. 1996. Prevalence of Bartonella henselae bacteremia, the causative agent of cat-scratch disease, in an Australian cat population. Pathology 28:262-265. [DOI] [PubMed] [Google Scholar]
- 9.Drancourt, M., R. Birtles, G. Chaumentin, F. Vandenesch, J. Etienne, and D. Raoult. 1996. New serotype of Bartonella henselae in endocarditis and cat-scratch disease. Lancet 347:441-443. [DOI] [PubMed] [Google Scholar]
- 10.Ehrenborg, C., L. Wesslen, A. Jakobson, G. Friman, and M. Holmberg. 2000. Sequence variation in the ftsZ gene of Bartonella henselae isolates and clinical samples. J. Clin. Microbiol. 38:682-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flexman, J. P., S. C. Chen, D. J. Dickeson, J. W. Pearman, and G. L. Gilbert. 1997. Detection of antibodies to Bartonella henselae in clinically diagnosed cat scratch disease. Med. J. Aust. 166:532-535. [DOI] [PubMed] [Google Scholar]
- 12.Foley, J. E., B. Chomel, Y. Kikuchi, K. Yamamoto, and N. C. Pedersen. 1998. Seroprevalence of Bartonella henselae in cattery cats: association with cattery hygiene and flea infestation. Vet. Q. 20:1-5. [DOI] [PubMed] [Google Scholar]
- 12a.Fournier, P. E. J. Robson, Z. Zeaiter, R. McDougall, S. Byrne, and D. Raoult. 2002.. Improved culture from lymph nodes of patients with cat scratch disease and genotypic characterization of Bartonella henselae isolates in Australia. J. Clin. Microbiol. 40:3620-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handley, S. A., and R. L. Regnery. 2000. Differentiation of pathogenic Bartonella species by infrequent restriction site PCR. J. Clin. Microbiol. 38:3010-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houpikian, P., and D. Raoult. 2001. 16S/23S rRNA intergenic spacer regions for phylogenetic analysis, identification, and subtyping of Bartonella species. J. Clin. Microbiol. 39:2768-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph, A. K., C. W. Wood, J. M. Robson, S. L. Paul, and A. J. Morris. 1997. Bartonella henselae bacteraemia in domestic cats from Auckland. N. Z. Vet J. 45:185-187. [DOI] [PubMed] [Google Scholar]
- 16.La Scola, B., Z. Liang, Z. Zeaiter, P. Houpikian, P. A. D. Grimont, and D. Raoult. 2002. Genotypic characteristics of two serotypes of Bartonella henselae. J. Clin. Microbiol. 40:2002-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Scola, B., and D. Raoult. 1999. Culture of Bartonella quintana and Bartonella henselae from human samples: a 5-year experience (1993 to 1998). J. Clin. Microbiol. 37:1899-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marston, E. L., J. W. Sumner, and R. L. Regnery. 1999. Evaluation of intraspecies genetic variation within the 60 kDa heat-shock protein gene (groEL) of Bartonella species. Int. J. Syst. Bacteriol. 49:1015-1023. [DOI] [PubMed] [Google Scholar]
- 19.Matar, G. M., B. Swaminathan, S. B. Hunter, L. N. Slater, and D. F. Welch. 1993. Polymerase chain reaction-based restriction fragment length polymorphism analysis of a fragment of the ribosomal operon from Rochalimaea species for subtyping. J. Clin. Microbiol. 31:1730-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norman, A. F., R. Regnery, P. Jameson, C. Greene, and D. C. Krause. 1995. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J. Clin. Microbiol. 33:1797-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Relman, D. A. 1998. Are all Bartonella henselae strains created equal? Clin. Infect. Dis. 26:1300-1301. [DOI] [PubMed] [Google Scholar]
- 22.Renesto, P., J. Gouvernet, M. Drancourt, V. Roux, and D. Raoult. 2001. Use of rpoB gene analysis for detection and identification of Bartonella species. J. Clin. Microbiol. 39:430-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Barradas, M. C., R. J. Hamill, E. D. Houston, P. R. Georghiou, J. E. Clarridge, R. L. Regnery, and J. E. Koehler. 1995. Genomic fingerprinting of Bartonella species by repetitive element PCR for distinguishing species and isolates. J. Clin. Microbiol. 33:1089-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sander, A., C. Buhler, K. Pelz, E. von Cramm, and W. Bredt. 1997. Detection and identification of two Bartonella henselae variants in domestic cats in Germany. J. Clin. Microbiol. 35:584-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sander, A., M. Posselt, N. Bohm, M. Ruess, and M. Altwegg. 1999. Detection of Bartonella henselae DNA by two different PCR assays and determination of the genotypes of strains involved in histologically defined cat scratch disease. J. Clin. Microbiol. 37:993-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sander, A., M. Ruess, S. Bereswill, M. Schuppler, and B. Steinbrueckner. 1998. Comparison of different DNA fingerprinting techniques for molecular typing of Bartonella henselae isolates. J. Clin. Microbiol. 36:2973-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartzman, W. A., C. A. Nesbit, and E. J. Baron. 1993. Development and evaluation of a blood-free medium for determining growth curves and optimizing growth of Rochalimaea henselae. J. Clin. Microbiol. 31:1882-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Versalovic, J., T. Loeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeaiter, Z., P. E. Fournier, H. Ogata, and D. Raoult. 2002. Phylogenetic classification of Bartonella species by comparing groEL sequences. Int. J. Syst. Evol. Microbiol. 52:165-171. [DOI] [PubMed] [Google Scholar]
- 30.Zeaiter, Z., P. E. Fournier, and D. Raoult. 2002. Genomic variation of Bartonella henselae strains detected in lymph nodes of patients with cat scratch disease. J. Clin. Microbiol. 40:1023-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]