Abstract
While outbreaks of animal anthrax zoonoses still regularly occur in France, little is known about the epidemiology links between them. We have used the eight-locus multilocus variable-number tandem repeat analysis typing technique against a collection of 50 Bacillus anthracis isolates from France. There were eight distinct genotypes belonging to two dissimilar genetic clusters. Regional strain patterns were observed, with the B2 genotypes prevalent in southern France and the A1a genotypes found only in northern France.
Bacillus anthracis is the causal agent of anthrax, a serious infection in both livestock and humans that is often fatal. Anthrax is still endemic in many countries. Animals are infected by contact with soilborne spores. Normally, humans are infected only incidentally, via contact with diseased animals or their waste products. Unfortunately, recent events have shown that B. anthracis can also be used in bioterrorism. Virulent strains of B. anthracis produce toxins and are encapsulated. These strains have two virulence plasmids, pXO1 and pXO2, encoding the toxins and the capsule synthetic activity, respectively (3). B. anthracis is one of the most monomorphic pathogenic bacteria described. Numerous studies have shown that techniques used to discriminate between strains in other species have low discriminatory power within the B. anthracis strains. Recently, a multilocus variable-number tandem repeat (VNTR) analysis (MLVA) for the molecular typing of B. anthracis strains was developed (1). Short nucleotide sequences that are repeated multiple times vary in copy number, creating length polymorphisms. Six such fragments were defined on the chromosome, and one such fragment was defined on each of the plasmids. The MLVA has previously defined 89 genetically distinct types that can be grouped in two major clusters (1). One cluster (cluster A) is found around the world, whereas the other one (cluster B) is almost exclusively restricted to southern Africa (1). Outbreaks of animal anthrax zoonoses still occur fairly regularly in France. A collection, which includes B. anthracis isolates collected in various regions during the last 20 years, was established (7). In this paper, we report the results obtained with the MLVA on 50 independent isolates: 36 isolates are from recent episodes of animal anthrax, 13 isolates are from older, seemingly sporadic animal cases, and 1 isolate is from a human case.
Geographical and temporal distribution.
This study is based on 49 B. anthracis isolates of animal origin and one of human origin, isolated after a fatal case of meningitis. The animal isolates were collected by the CNEVA/LCRV (Central Laboratory of Veterinary Research from 1982 to 1999) or the AFSSA/LERPAZ (French Food Safety Agency/Research Laboratory for the study of Animal Pathology and Zoonoses from 1999 to 2002) during etiologic studies performed between 1982 and 2001 on herds with abnormal mortality (4, 7). The majority (36 of 49) were isolated between 1997 and 2001 during epizootic periods with many outbreaks evolving during 2 to 4 months in a given region.
The 49 animal isolates originate from 43 different farm estates located in various regions of France (Fig. 1). These isolates were mostly from bovine hosts. Only four isolates were from other species: three from sheep and one from a dog. Five isolates were from different animals on the same estate in southwestern France. Similarly, twice, in the Alps and in Burgundy, two isolates were from two animals of a given farm estate. This redundant analysis was performed to test whether the same genotype would be implicated in sick animals from the same estate.
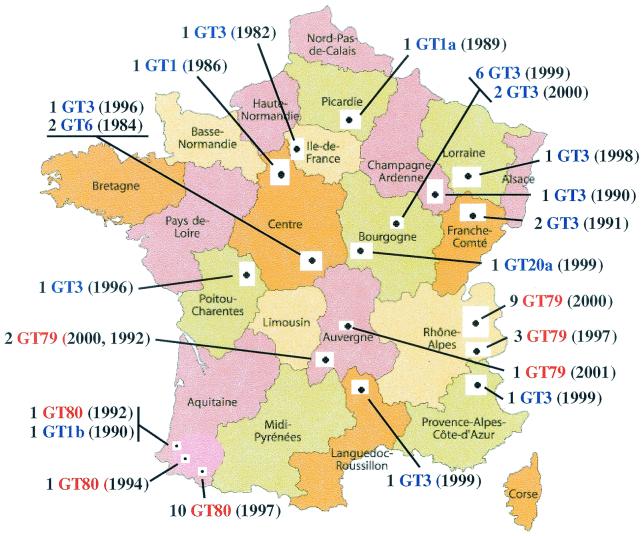
FIG. 1.
Locations of B. anthracis isolates. This map shows the regions where isolates of B. anthracis were obtained for this genetic study. The actual collection sites are indicated by the small solid black dots. The labels indicate the number of isolates from a location, the genotype (GT), and the year the isolate was collected [e.g., 1 GT3 (1985)]. The red GT numbers are from cluster B2, while the blue GT numbers are from cluster A1a in Fig. 2. Intercarto map adapted with permission.
Essentially all anthrax-affected animals were pasture-raised livestock. Only two isolates came from animals living in stalls: one from an animal with no contact with the outside and one where animals had an area to relax with a beaten-earth floor. The outbreaks occurred mainly in the spring but also occurred during the summer and fall. For many outbreaks, the origin has been correlated with work in soils that had been contaminated in the past. In other cases, the origin is unclear. However, the hydrogeological type of the soils seems of great importance in this study and has been reported previously (2, 5). Also, the appearance of anthrax is correlated with other diseases that weaken the victims or create microwounds in the mucous membranes that act as entry passages for B. anthracis spores (7).
Genetic diversity of B. anthracis isolates in France.
We examined the genetic diversity of the 50 French B. anthracis isolates described in the previous paragraph by eight-locus MLVA (1). Briefly, a single colony was “heat lysed” in Tris-EDTA (TE) buffer, and the MLVA PCR amplifications were performed after the cellular debris was removed (1, 6). Diversity group clusters were analyzed by cluster analysis as previously described (6). Genetic relationships among French B. anthracis isolates are graphically presented in Fig. 2.
FIG. 2.
Genetic relationships among French B. anthracis isolates. The dendrogram based upon eight VNTR marker loci and unweighted paired group means analysis cluster analysis illustrates the genetic relationships among isolates. The genotype (GT) designations are consistent with those of Keim et al. (1), with the exception of three novel types observed in this study (GT1a, GT1b, and GT20a), which are labeled with asterisks. The number of each genotype and the actual allele size (in nucleotides) are shown to the right of the dendrogram.
We found three dominant and eight distinct MLVA genotypes. In the A1a type, 17 strains are from the dominant genotype, genotype 3 (GT3 from reference 1). This particular genotype was found in one outbreak with many cases in the Bourgogne region in 1999 and 2000 (8 isolates) and in sporadic cases scattered over many regions between 1982 and 1999 (8 isolates). An isolate with this genotype was isolated in the human fatal meningitis case, apparently not correlated to any animal outbreak. GT3 is the most common type seen in North America, especially in the United States, and also the most common from France (Fig. 2) (1). This wide distribution also holds for the individual regions across North America and France and in South America (1). However, the French B. anthracis GT3 genotype was not found either in the Rhône-Alpes or in the Pyrénées (Aquitaine region).
The five other genotypes from the A1a cluster are extremely rare and were found only once or twice. The GT6 (two isolates) and GT1 (one isolate) genotypes were found in the Centre Province during sporadic cases in 1984 and 1986. It is noteworthy that three genotypes had not been previously described (GT1a, GT1b, and GT20a) and that each of these genotypes is represented by a single isolate from sporadic cases. They were observed at various geographical areas and times (Picardie in 1989, Aquitaine in 1990, and Bourgogne in 1999).
The two other genotypes that are dominant in France are extremely rare in the world (1) and belong to the B2 cluster. The B2 cluster is a distinct clonal lineage that has been observed only twice previously, once from Slovakia and once from Croatia. GT79 was regularly and exclusively found in anthrax episodes occurring in the Alps in 1997 and 2000 (12 isolates) and from the summer pastures in Auvergne in 1992, 2000, and 2001 (3 isolates). GT80 has been found only in outbreaks from the Pyrénées in 1992 (1 isolate), 1994 (1 isolate), and 1997 (10 isolates). Thus, the B2 genetic type is nearly exclusively French in origin. Because of the longevity of spores and the ready transport of spore-contaminated material, few genotypes are seen in a single region. The restricted distribution of the B2 cluster is unique in this regard.
The dominant French GT3 genotype is genetically quite dissimilar to the B2 types with six (A1a versus B2 GT80) and seven (A1a versus B2 GT79) different alleles among the eight MLVA markers. The only marker that does not differ between these two types is vvrB, which is in fact the same in all French isolates. In this set of isolates, similar to what was found in the worldwide study, the pXO1 marker is the most discriminatory, with a diversity index value (D) of 0.55. However, the pXO2 marker differs only in the GT79 and GT80 isolates and represents the marker that differentiates the B2 subgroup (D = 0.42). This situation is the opposite of that encountered during the other study describing type A and B B. anthracis in a limited geographical area, namely, the Kruger National Park (6). In that case, the pXO2 marker had the same allele, while the pXO1 marker differed.
While the VNTR loci examined in this study are some of the most diverse regions in the B. anthracis genome, they are relatively stable in epidemics. The large and ongoing anthrax epidemic in Canadian bison populations is an example where these eight loci are unchanged across hundreds of kilometers and decades of time (1). Laboratory studies to detect and characterize mutations at these loci have been performed, and very few mutations are observed despite extensive passages (data not presented). In one experiment, the 96 independent cultures of the Sterne strain were transferred 40 times, and only one mutation was observed (G. Zinser and P. Keim, unpublished data). Laboratory transfer experiments are limited by their logistics and will be able to characterize only mutational processes acting at very high rates (e.g., >10−5 mutation per generation). MLVA genotypes with different alleles at multiple loci must be separated by relatively great evolutionary distances.
Given the great genetic distance between the genotype A and B branches, it seems likely that B. anthracis currently active in France has been introduced in at least two events. This conclusion is based upon the two dominant types and their distinct genetic relationship. Given the great evolutionary distance between the A1a and B2 clusters, in situ differentiation seems unlikely. The prevalence of GT3 in France and its common occurrence in North America also suggest an epidemiological link between these two regions. It seems plausible that the dominant form of North American B. anthracis was introduced from Europe.
Acknowledgments
This work was supported by grants from the U.S. National Institutes of Health, General Medical Sciences, and the U.S. Department of Energy's Chemical and Biological Nonproliferation Program.
REFERENCES
- 1.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laforce, F. M. 1994. Anthrax. Clin. Infect. Dis. 19:1009-1014. [DOI] [PubMed] [Google Scholar]
- 3.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 4.Patra, G., J. Vaissaire, M. Weber-Levy, C. Le Doujet, and M. Mock. 1998. Molecular characterization of Bacillus strains involved in outbreaks of anthrax in France in 1997. J. Clin. Microbiol. 36:3412-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shafazand, S., R. Doyle, S. Ruoss, A. Weinacker, and T. A. Raffin. 1999. Inhalational anthrax epidemiology, diagnosis, and management. Chest 116:1369-1376. [DOI] [PubMed] [Google Scholar]
- 6.Smith, K. L., V. DeVos, H. Bryden, L. B. Price, M. E. Hugh-Jones, and P. Keim. 2000. Bacillus anthracis diversity in Kruger National Park. J. Clin. Microbiol. 38:3780-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaissaire, J., M. Mock, C. Le Doujet, and M. Lévy. 2001. Le charbon bactéridien. Epidémiologie de la maladie en France. Méd. Mal. Infect. 31:257-271. [DOI] [PubMed] [Google Scholar]