Abstract
Recently, the retinitis pigmentosa 3 (RP3) gene has been cloned and named retinitis pigmentosa GTPase regulator (RPGR). The amino-terminal half of RPGR is homologous to regulator of chromosome condensation (RCC1), the nucleotide exchange factor for the small GTP-binding protein Ran. In a yeast two-hybrid screen we identified the delta subunit of rod cyclic GMP phosphodiesterase (PDEδ) as interacting with the RCC1-like domain (RLD) of RPGR (RPGR392). The interaction of RPGR with PDEδ was confirmed by pull-down assays and plasmon surface resonance. The binding affinity was determined to be 90 nM. Six missense mutations at evolutionary conserved residues within the RLD, which were found in RP3 patients, were analyzed by using the two-hybrid system. All missense mutations showed reduced interaction with PDEδ. A non-RP3-associated missense substitution outside the RLD, V36F, did not abolish the interaction with PDEδ. PDEδ is widely expressed and highly conserved across evolution and is proposed to regulate the membrane insertion or solubilization of prenylated proteins, including the catalytic subunits of the PDE holoenzyme involved in phototransduction and small GTP-binding proteins of the Rab family. These results suggest that RPGR mutations give rise to retinal degeneration by dysregulation of intracellular processes that determine protein localization and protein transport.
Retinitis pigmentosa (RP) designates a heterogeneous group of hereditary retinal dystrophies characterized by impaired dark adaptation, progressive visual field defects, and severe reduction in visual acuity. X-linked RP (xlRP) is the most severe form of RP, which affects about 1 in 25,000 and leads to blindness by the third or fourth decade (1). Clinical surveys indicate that at least 16–33% of all RP patients show X-linked inheritance (2). The most common form of xlRP is RP type 3 (RP3), which affects about 70% of xlRP patients and is caused by mutations in a gene named RPGR.
The RP GTPase regulator (RPGR) gene codes for a 90-kDa protein that contains a weakly conserved nucleotide binding motif at the N terminus and a potential isoprenylation site at the carboxyl terminus (3). Most significantly, the N-terminal half of RPGR is homologous to the regulator of chromosome condensation (RCC1) (3, 4). RCC1 is a chromatin binding protein that is involved in cell cycle regulation. Biochemically, RCC1 is the guanine nucleotide exchange factor (GEF) for Ran (5, 6), a small GTP-binding protein that is essential for nucleo-cytoplasmic transport (7, 8). Recently, the three-dimensional structure of RCC1 has been determined (9). It consists of a seven-bladed propeller formed by internal repeats. This structural motif also is found in p532, a GEF for Rab and Arf proteins (10), indicating that RCC1-like proteins might be general guanine nucleotide exchange proteins. To investigate the role of the RCC1-like domain (RLD) of RPGR in RP3, we applied the yeast two-hybrid system to identify proteins interacting with the RLD of RPGR. We were able to identify the delta subunit of the rod cyclic GMP phosphodiesterase (PDEδ) (11). The purified protein products of RPGR392 and PDEδ interacted with high affinity, confirming the two-hybrid result. Mutants of RPGR found in RP3 patients showed decreased binding affinity when analyzed in the two-hybrid system. This interaction therefore provides an assay to determine the functional integrity of RPGR. PDEδ interacts with a small GTP-binding protein of the Rab family, Rab13 (12), and because RPGR is localized in the Golgi (13) it is likely that RPGR is involved in intracellular vesicle transport.
MATERIALS AND METHODS
Plasmids, Strains, and Mutagenesis.
A lexA-based two-hybrid system (provided by Stan Hollenberg, Vollum Institute, Portland, OR), containing pBTM116 as bait plasmid and pVP16 as library plasmid, together with the yeast reporter strain L40 (MATa his3–200 trp1–901 leu2–3, 112 ade2 LYS2∷(lexAop)4-HIS3 URA∷(lexAop)8-lacZ GAL4 gal80) was used. PDEδ was cloned as a BamHI–EcoRI fragment into pcDNA3 (Invitogen) for in vitro translation and into pGSTAcC5 (provided by Walter Kolch, Beatson Institute, Glasgow, U.K.) for expression in Sf9 cells (14). Full-length RPGR was cloned as a BamHI fragment into pBTM116. Single point mutants of RPGR were generated by PCR (15). The resulting DNA fragments were cloned as BamHI–PstI fragments into pBTM116.
Two-Hybrid Screening.
As bait, we used the first 392 aa of RPGR (RPGR392) generated by PCR. RPGR392 was cloned as a BamHI–PstI fragment into pBTM116 and sequenced. A size-selected mouse embryo cDNA library from 9.5- to 10.5-day embryos cloned in pVP16 was used, and yeast strain L40 containing pBTM116-RPGR392 was transformed as described (16, 17). Candidate clones were rescreened for interaction by using pBTM116, pBTM-lamin, pBTM-RCC1, and pBTM-Ran as controls.
Two-Hybrid Interaction Assay.
Saccharomyces cerevisiae reporter strain L40 was cotransformed with pVP16-PDEδ and pBTM116-RPGR containing either wild-type (WT) or mutant RPGR392. The resulting transformants were grown on medium lacking tryptophane, histidine, uracil, leucine, and lysine (THULLy dropout medium), containing 25 mM 3-aminotriazole. For qualitative interaction studies, the plates were incubated at 20°C, 26°C, 30°C, and 37°C for 6 days. Growth on THULLy plates was indicative of protein interaction. To determine protein interaction, yeast transformants were grown in medium lacking uracil, tryptophane, and leucine (UTL dropout medium) up to midlog phase (OD600 of 1.0) at 30°C. Aliquots (0.1 ml) of each culture then were used in the o-nitrophenyl β-d-galactopyranoside assay as described (18).
Detection of RPGR Expression in Yeast.
Yeast transformed with the appropriate hemagglutinin (HA)-tagged RPGR392 constructs and pVP16-PDEδ were grown for 3 days on UTL selection medium at 30°C. Cells were lysed, and 30 μg of soluble protein was used for SDS gel electrophoresis and subsequent Western blotting. HA-tagged RPGR fusion proteins were detected by using anti-HA antibody (Boehringer Mannheim) and the ECL-PLUS system (Amersham).
Protein Expression.
PGR392 was cloned as an NcoI–BamHI fragment into pET21d to generate a C-terminal His-tag fusion protein. Soluble protein was purified by using affinity purification on a Ni2+ nitrilotriacetic acid (NTA) column (Quiagen). After ammonium sulfate precipitation the protein was resuspended in 50 mM Tris, pH 7.5 and dialyzed. Glutathione S-transferase (GST)-RPGR392 and mutant derivatives were cloned BamHI–XhoI into pGEX-KG. The 100,000 × g supernatants of lysate were used for glutathione (GSH) column affinity purification. The purified GST-fusion proteins were dialyzed against PBS.
Expression and Purification of Recombinant Proteins in Sf-9 Cells.
PDEδ was cloned into pGSTAcC5 to generate a N-terminal GST-fusion protein, expressed in Sf-9 cells, and purified to near homogeneity as described (14, 19).
Protein Binding Assays.
[35S]PDEδ was synthesized by in vitro translation (TnT T7 Quick Coupled Transcription/Translation System, Promega). Ten microliters of the translation reaction, 15 μl of a 50% slurry of Ni2+NTA, and 15 μM of RPGR392-His6 in a total volume of 100 μl of PBS was incubated for 30 min at room temperature and then washed three times with PBS before analyzing on a 15% SDS gel. The stained gel was treated for fluography. Alternatively, the affinity of the GST-PDEδ–RPGR392 interaction was quantified by copurification of GST-PDEδ and RPGR392 using GSH-Sepharose (GSH pull-down experiment). GST-PDEδ (0.35 μM) was incubated with 0.12–3.0 μM RPGR392 and GSH-Sepharose for 30 min at room temperature. The GSH-Sepharose was washed three times with PBS before analyzing proteins on a 15% SDS gel together with a BSA calibration standard (Pierce). Coomassie blue-stained protein bands were scanned and quantified with densitometric software (NIH image 1.6.1). For the GST-RPGR–PDEδ pull-down assay, 3 μM of WT or mutant GST-RPGR392 (V36F, H98Q, F130C, and G215V) and 1 μM PDEδ were incubated with GSH-Sepharose in a total volume of 100 μl at 20°C or 30°C for 30 min. The GSH-Sepharose was washed three times with PBS before analyzing proteins on SDS gel. Coomassie blue-stained PDEδ protein bands were determined by densitometry. The amount of PDEδ bound to GST-RPGR392-WT was taken as 100% binding.
Surface Plasmon Resonance.
Association and dissociation kinetics were determined by surface plasmon resonance (BIACore AB, Uppsala, Sweden) in the presence of 10 mM Hepes, pH 7.4/150 mM NaCl/5 mM MgCl2/0.0005% (wt/vol) Igepal CA-630 (Sigma). Anti-GST-antibodies were covalently coupled to the sensor chip. GST-PDEδ (0.4 μM) was bound to the anti-GST antibodies, followed by successive incubations with 0.1, 0.2, 0.4, 0.6, and 1.0 μM RPGR392 as free ligand. Between the concentration changes the GST-PDEδ–RPGR392 complex was dissociated by a washing step. To regenerate the chip matrix, noncovalently bound ligands were removed by 20 mM glycine, pH 2.0 and 0.005% SDS. GST bound to the sensor chip was used as a control to verify complete dissociation of noncovalently bound ligands and to account for unspecific binding. Association and dissociation constants were calculated (biaevaluation 2.1 software) by using single exponential fit with drift (details in Results).
RESULTS
Identification of PDEδ as an Interacting Protein of RPGR.
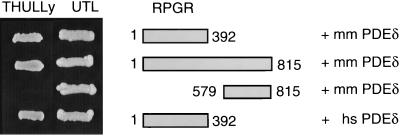
To identify proteins interacting with RPGR, a yeast two-hybrid screen was undertaken by using the RLD of RPGR (RPGR392) as bait in a lexA-based two-hybrid system together with a size-selected mouse embryo cDNA library. We obtained 40 potential candidate clones, eight of which coded for PDEδ. None of these eight clones were able to interact with control proteins lamin, Ran, or RCC1, indicating that the interaction of RPGR392 with PDEδ is specific. The mouse PDEδ clones contained the complete coding region except for the first six N-terminal amino acids. However, in a control assay, the complete coding regions of mouse and human PDEδ also were able to interact with human RPGR392 and with the full-length RPGR, but not with the carboxyl-terminal domain of RPGR (Fig. 1). Thus, the C-terminal half of RPGR is not required for PDEδ binding. Deleting as few as 23 aa from either end of PDEδ abolished the interaction with RPGR (data not shown), indicating the minimal domains necessary for this interaction.
Figure 1.
Interaction of RPGR with PDEδ in the two-hybrid system. (Right) RPGR-PDEδ combinations used for yeast transformation are shown. (Left) Yeast clones growing on medium selecting either for the presence of the two two-hybrid plasmids (UTL dropout medium) or selecting for protein–protein interaction (THULLy dropout medium). Mouse (mm) PDEδ interacts with RPGR392 and full-length RPGR, whereas the C-terminal part of RPGR (amino acids 579–815) alone does not bind nor does it contribute to the binding of PDEδ as there is no difference in binding between full-length and C terminally truncated RPGR. RPGR also interacts with the human (hs) isoform of PDEδ.
RPGR392 Binds PDEδ with High Affinity.
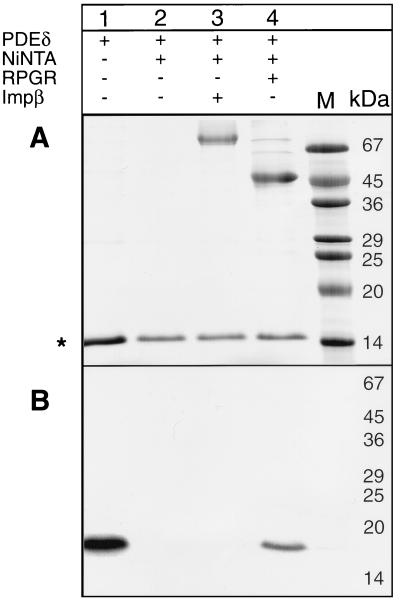
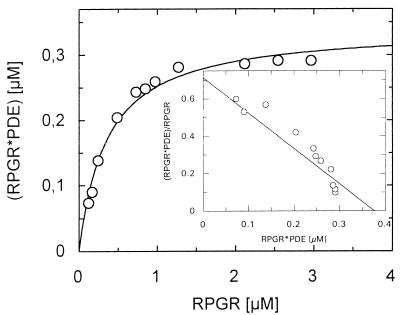
We used three independent methods to verify the interaction of RPGR392 and PDEδ. First, PDEδ was expressed in vitro by using rabbit reticulocyte translation extracts, as soluble protein could not be obtained from bacterial expression systems. Recombinant His-tagged RPGR392 (His-RPGR392) purified from bacteria was added to in vitro-translated PDEδ. The formation of a PDEδ–His-RPGR392 complex was monitored by affinity copurifying His-RPGR392 and PDEδ by using Ni2+NTA beads. The results verified the earlier two-hybrid result that PDEδ binds specifically to RPGR392 in vitro (Fig. 2). Second, we expressed a GST-PDEδ fusion protein by using the baculovirus expression system as described (14). To determine the affinity of this interaction, constant amounts of GST-PDEδ were incubated with increasing amounts of RPGR392. The resulting protein complex was copurified in batch by using GSH-Sepharose. The GST-PDEδ–RPGR392 complex was analyzed on a Coomassie-stained SDS polyacrylamide gel. The intensity of the resulting protein bands were digitally recorded and quantified by using an internal BSA standard that was present on the same gel. The affinity of the GST-PDEδ–RPGR392 interaction was calculated to be about 400 nM by using the Scatchard plot analysis (Fig. 3).
Figure 2.
PDEδ binds to His6-RPGR392 in vitro. 35S-methionine-labeled PDEδ, synthesized by in vitro translation, was incubated with purified His6-RPGR392 and repurified by using Ni2+NTA. (A) A Coomassie-stained SDS acrylamide gel containing 35S-methionine-labeled PDEδ (lane 1), proteins of the PDEδ in vitro translation reaction bound to Ni2+NTA (lane 2), 35S-labeled PDEδ and His6-tagged importin β bound to Ni2+NTA (lane 3), and 35S-labeled PDEδ and His6-RPGR392 bound to Ni2+NTA (lane 4). Proteins of the in vitro translation reaction binding unspecifically to Ni2+NTA are indicated by ∗. (B) An autoradiogram of the above SDS acrylamide gel. In lanes 1 and 4, radiolabeled PDEδ is detected.
Figure 3.
Quantitative determination of the GST-PDEδ–RPGR interaction. GST-PDEδ (0.35 μM) was incubated with RPGR392 concentrations ranging from 0.12 to 3.0 μM. The protein complex was purified by using GSH-Sepharose and analyzed on a SDS polyacrylamide gel. The protein bands were densitometrically quantified by using an internal BSA calibration standard (Pierce). The KD was determined by Scatchard plot analysis to a value of about 400 nM.
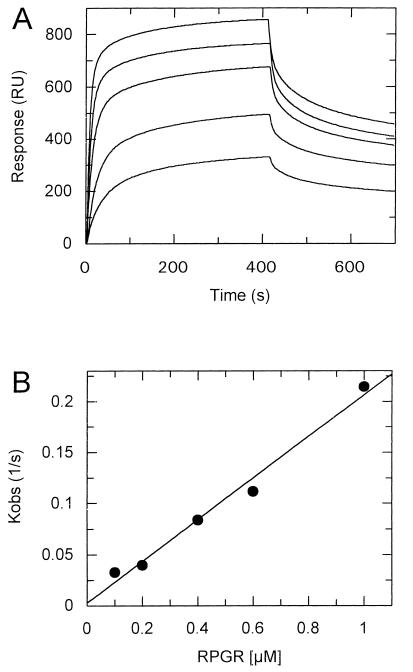
Third, the BIAcore instrument (20, 21) was used to measure the interaction of the two proteins directly. The GST-PDEδ fusion protein was attached to the dextran matrix containing an covalently bound anti-GST antibody as described (22). This method determines the on rates as well as the off rates in a single experiment. During the dissociation period, free binding sites become increasingly available, and we observed significant rebinding of the ligand, which was most pronounced at the lowest ligand concentration. The rebinding effect decreases the apparent dissociation rate and would lead to an overestimation of the affinity. The binding effect is negligible at high RPGR concentrations in the first half of the kinetic where more than 50% of the binding sites are still occupied. Therefore, the dissociation rate was calculated within a 5- to 40-sec time window of the dissociation period after which 50% of the protein complex is dissociated. We determined a koff of 0.018 sec−1 and a kon of 2.04 × 105 sec−1⋅M−1 with a calculated KD of 90 nM (Fig. 4). The BIAcore determines the affinity kinetically, whereas the pull-down assay determines the affinity at the equilibrium. The equilibrium, however, is distorted by extensive washes of the protein complex, which is why this method can be considered only semiquantitative. Given this, the 4-fold difference in affinity is relatively small and the affinity of 90 nM sufficiently high to be physiologically relevant.
Figure 4.
Direct measurement of RPGR binding to GST-PDEδ by surface plasmon resonance technique. (A) Association of 0.1, 0.2, 0.4, 0.6, and 1.0 μM RPGR392 (from bottom to top) to GST-PDEδ and subsequent dissociation of RPGR. (B) The association rate constant (kon) was calculated by fitting kobs versus the RPGR392 concentration (kon = 2.04 × 105 sec−1⋅M−1). The dissociation rate constant was calculated by fitting the data to first-order kinetics (koff = 0.018 sec−1). The resulting KD was calculated to be 8.82 × 10−8 M (for details see text).
Binding of PDEδ Decreased in RPGR Missense Mutants.
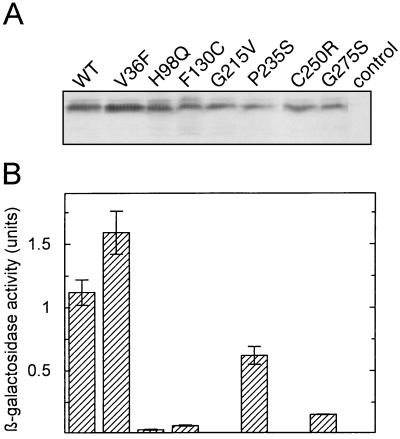
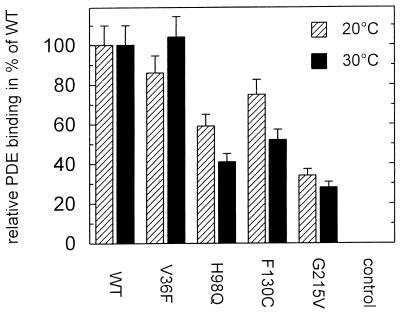
Various missense and nonsense mutations in the RPGR gene have been reported in RP3 patients (3, 23, 24). Although it is safe to assume that the nonsense mutations give rise to truncated and unstable proteins, none of the missense mutations have been shown to impair RPGR function. In the following experiments, we used the PDEδ-RPGR interaction in the two-hybrid system to investigate various RPGR missense mutations. The two-hybrid analysis demonstrates that all of the RPGR mutations found in the RLD of RPGR significantly decreased the binding to PDEδ in a thermosensitive fashion (Fig. 5). It had been shown previously that the two-hybrid system is suitable for evaluating the relative affinity of two proteins that are difficult to express or purify (25). Using the β-galactosidase activity as a quantitative measure of the interaction between RPGR and PDEδ in the two-hybrid system revealed that all RPGR mutants have severely reduced affinities. Even the weakest RPGR mutant, P235S, had a 40% reduction in activity (Fig. 6B). Moreover, a RPGR missense mutation (V36F), which shows no signs of retinal degeneration (T.M., unpublished work) did not show decreased binding to PDEδ (Fig. 6B). To exclude the possibility that the differences in β-galactosidase activity merely reflected decreased protein stability or different expression levels of the RPGR mutants, we performed Western blot analysis using an antibody against the hemagglutinin tag of the LexA-RPGR fusion protein. All LexA-RPGR derivatives were expressed to similar amounts in yeast at 30°C (Fig. 6A). As the binding appears to be thermosensitive in the two-hybrid assay and because some recombinant RPGR mutant proteins could not be purified because of stability problems, we cannot exclude the possibility that some RPGR mutations cause misfolding. Decreased protein expression, however, did not correlate with decreased binding affinity as judged by two-hybrid analysis (compare H98Q with P235S). Therefore, by using purified recombinant GST-RPGR mutant proteins in pull-down experiments, we confirmed that RPGR mutant proteins bind less efficiently to PDEδ (Fig. 7).
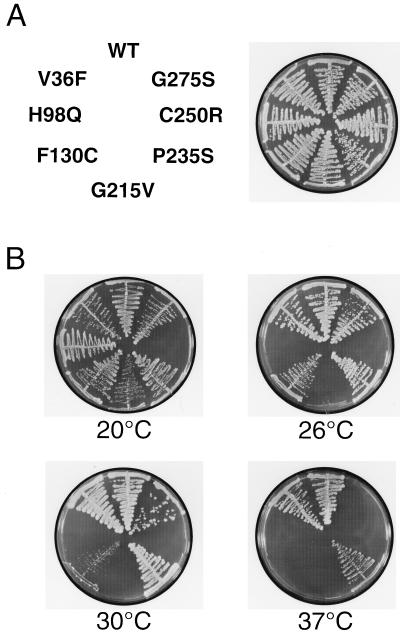
Figure 5.
Mutations within the RLD of RPGR interfere with the binding of PDEδ in a temperature-dependent manner. (A) The positions of the individual mutations are indicated (Left), and growth of the respective yeast clones under nonselective conditions (UTL) at 30°C is shown (Right). (B) Yeast clones grown on selective medium (THULLy) at 20°C, 26°C, 30°C, and 37°C were assayed for RPGR-PDEδ interaction.
Figure 6.
Quantitative determination of the RPGR392-PDEδ interaction by liquid β-galactosidase assay. (A) Yeast liquid cultures were grown at 30°C to an OD600 of 1.0. Expression of RPGR392 was followed by Western blot analysis. (B) The β-galactosidase activity, the parameter for protein–protein interaction, was detected as described (18).
Figure 7.
Relative PDEδ binding of GST-RPGR392 mutants. PDEδ (1 μM) and 3 μM of WT or mutant GST-RPGR392 were incubated with GSH-Sepharose at 20°C and 30°C for 30 min. The GSH-Sepharose bound protein complexes were applied onto a SDS/PAGE after washing with PBS. Band intensities of the PDEδ were determined by densitometric software. The intensity of the PDEδ bound to WT-RPGR392 was set to 100%.
DISCUSSION
Combining yeast two-hybrid analysis with biochemical and biophysical protein binding assays, we have shown that PDEδ binds to the RLD of RPGR. The high affinity of RPGR392 for PDEδ, which is determined to be about 90 nM by plasmon surface resonance, is sufficient to account for binding under physiological circumstances. The binding appears to be independent of the fusion moieties because native PDEδ synthesized in vitro bound to GST-RPGR392 and His-tagged RPGR392 bound to GST-tagged PDEδ. The tissue distribution of human PDEδ parallels the ubiquitous expression of RPGR (3, 26) and is greatly enhanced in retina (A.W., unpublished work). Together with the high affinity, we conclude that the RLDs of RPGR and PDEδ are likely to be genuine binding partners. The RLDs of RPGR and RCC1 are proposed to have a very similar protein fold, so the recently solved structure of RCC1 (9) was used as a model to predict the nature of the mutations in RPGR. With the exception of F130C, all mutations are within conserved parts of the structure that are necessary to maintain the protein fold and therefore are likely to destabilize the overall structure of RPGR. In this study, we have developed a two-hybrid assay that can be used to assess the functionality of RPGR. In this assay, six missense mutations found in RP3 patients do not interact with PDEδ whereas an RPGR variant not causing RP still interacts. This correlation indicates that the failure to interact with PDEδ might be the cause of or contribute to RP3 and is in keeping with the observation that the most severe forms of RP3 have mutations in the RLD of RPGR (24).
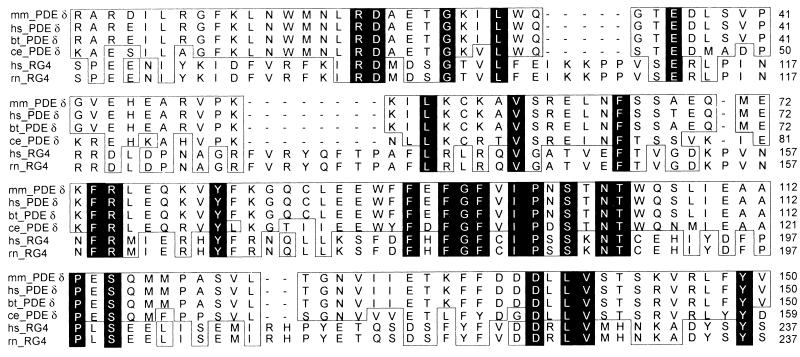
Human PDEδ codes for a 17.4-kDa protein and is strongly conserved in other species (26, 27), sharing 97–99% overall identity between mouse, bovine, and human and retaining 70% similarity to its orthologue in Caenorhabditis elegans (Fig. 8), which lacks ocular tissue. Interestingly, PDEδ shares sequence homology with the human HRG4 gene. HRG4 and its orthologues in rat and C. elegans, which encode 23- to 27-kDa proteins of unknown function (28), are localized to photoreceptor cells in mammals (29) and to the nervous system of the nematodes (30).
Figure 8.
PDEδ is highly conserved. Amino acid sequence alignment of PDEδ from bovine (bt_PDEδ; GenBank accession no. U65073), human (hs_PDEδ; GenBank accession no. AF022912), mouse (mm_PDEδ; GenBank accession no. AA726559) and C. elegans (ce_PDEδ; GenBank accession no. U14635). The sequence identity ranges from 70% to 99%. Identical residues are shaded in black and structurally conserved residues are boxed. Highly related to PDEδ is hRG4 (hs_RG4; GenBank accession no. U40998), a protein highly expressed in human retina that also has been found in rat (rn_RG4; GenBank accession no. U40999), and C. elegans (unc119; not shown; GenBank accession no. A38976).
PDEδ was isolated as a fourth subunit of bovine rod PDE (PDE6). In vertebrate phototransduction PDE6 is the effector for the trimeric G protein transducin after its activation by light-sensitive rhodopsin molecules in the retina. Mutations in the genes coding for the α and β subunit of PDE6 have been shown to cause autosomal forms of retinal degeneration (31–34) or stationary night blindness (35). However, no mutations have yet been found in the PDEδ gene in patients with autosomal recessive RP. In the case of PDEδ, this is not surprising because it is abundantly expressed in all tissues and highly conserved across evolution, so most likely PDEδ serves a more general function in tissues other than the retina.
PDEδ was shown to bind the prenylated C terminus of PDEβ (36) and the mouse orthologue of human RPGR, mRPGR, is localized in the Golgi apparatus via its C-terminal prenyl modification (13). This finding raised the possibility that PDEδ could bind to the prenylated C terminus of RPGR. However, the human RPGR392 deletion used in this study lacks the C-terminal prenylation motif and was purifed from bacteria that precludes prenyl modifications. Therefore, in addition to the prenyl-dependent binding, PDEδ also must have a second prenyl-independent binding mode that is represented by the binding to the RLD of RPGR. The complex of RPGR and PDEδ is reminiscent of the βγ complex of trimeric G proteins whose β-subunit also consists of a propeller structure, albeit unrelated to that of RCC1. The βγ complex serves as a guanine nucleotide dissociation inhibitor (GDI) for the guanine nucleotide binding α-subunit, raising the possibility that the RPGR-PDEδ complex also might bind to an α-subunit-related protein.
PDE6 is attached to the disc membranes of rod outer segments via the isoprenylated C termini of the α and β subunit (37, 38). PDEδ recognizes isoprenylated PDE6 and is able to solubilize the membrane-bound form of PDE6 under physiological conditions (36). However, as the solubilization of PDE6 does not alter its enzyme activity, it is conceivable that PDEδ serves a regulatory function by translocating PDE6 from the membrane to the cytoplasm. This function of PDEδ is reminiscent of the function of Rab escort protein REP1 and GDI. REP1 is a subunit of the geranyl transferase II essential for prenylation and a GDI of GTP-binding proteins such as Rab27 (39). A loss of function mutation in the REP1 gene is the cause for choroideremia, a form of hereditary tapeto-retinal degeneration (39, 40). The membrane association of small GTP-binding proteins also can be mediated by GDIs and accompanied by GDP/GTP exchange (41). Consistent with the idea that PDEδ might have GDI-like function are observations that PDEδ binds to modified and unmodified small GTP-binding proteins like Rab13 (12), Rho6 (J. Camonis and Y. Mahe, personal communication), and proteins of the Arl family, Arl3 and Arl184 (M.L. and J.B., unpublished work). Because RPGR is homologous to the guanine nucleotide exchange factor of the small GTP-binding protein Ran, it is conceivable that the RLD of RPGR could be a guanine nucleotide exchange factor for an as-yet-unidentified GTP-binding protein. Another protein that contains two RLDs is p532. Interestingly, like mRPGR, p532 is localized in the Golgi and in addition has guanine nucleotide exchange activity for ARF and Rab proteins (10). However, we could not detect any guanine nucleotide exchange activity of RPGR392 for Arl3, Arl184, or Ran.
Regardless of whether RPGR serves as a guanine nucleotide exchange factor or as a GDI in complex with PDEδ, both alternatives suggest that RPGR interacts with an as-yet-unidentified GTP-binding protein. At this point it is impossible to predict whether this protein will be a small Ras-related GTP-binding protein or an α-subunit of a trimeric G protein. Although we cannot exclude a role of RPGR in phototransduction, the bulk of the evidence suggests a role for RPGR in the regulation of intracellular vesicle trafficking.
Acknowledgments
We thank Caro Koerner and Beatrix Scheffer for excellent technical assistance; Walter Kolch for valuable advice with the baculovirus expression system; Jürgen Kuhlmann, Benjamin Bader, and Carolina Villa Braslavsky for discussion; Jaques Camonis and Yannick Mahe for sharing information before publication, and Alfred Wittinghofer for continuous support. This work was funded in part by a grant from the Deutsche Forschungsgemeinschaft (J.B. and T.M.), The Foundation Fighting Blindness, and British Retinitis Pigmentosa Society (A.W.).
ABBREVIATIONS
- RP
retinitis pigmentosa
- RP3
RP type 3
- RPGR
RP GTPase regulator
- RCC1
regulator of chromosome condensation
- RLD
RCC1-like domain
- PDE
phosphodiesterase
- REP1
Rab escort protein 1
- THULLy
tryptophane/histidine/uracil/leucine/lysine
- UTL
uracil/tryptophane/leucine
- GST
glutathione S-transferase
- GSH
glutathione
- NTA
nitrilotriacetic acid
- WT
wild type
- GDI
guanine nucleotide dissociation inhibitor
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Dryja T P, Li T. Hum Mol Genet. 1995;4:1739–1743. doi: 10.1093/hmg/4.suppl_1.1739. [DOI] [PubMed] [Google Scholar]
- 2.Haim M, Rosenberg T. Acta Ophthalmol. 1993;71:597–605. doi: 10.1111/j.1755-3768.1993.tb04648.x. [DOI] [PubMed] [Google Scholar]
- 3.Meindl A, Dry K, Herrmann K, Manson F, Ciccodicola A, Edgar A, Carvalho M R, Achatz H, Hellebrand H, Lennon A, et al. Nat Genet. 1996;13:35–42. doi: 10.1038/ng0596-35. [DOI] [PubMed] [Google Scholar]
- 4.Ohtsubo M, Okazaki H, Nishimoto T. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischoff F R, Ponstingl H. Nature (London) 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- 6.Klebe C, Prinz H, Wittinghofer A, Goody R S. Biochemistry. 1995;34:12543–12552. doi: 10.1021/bi00039a008. [DOI] [PubMed] [Google Scholar]
- 7.Moore M S, Blobel G. Nature (London) 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 8.Melchior F, Paschal B, Evans J, Gerace L. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. Nature (London) 1998;392:97–100. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- 10.Rosa J L, Barbacid M. Oncogene. 1997;15:1–6. doi: 10.1038/sj.onc.1201170. [DOI] [PubMed] [Google Scholar]
- 11.Gillespie P G, Prusti R K, Apel E D, Beavo J A. J Biol Chem. 1989;264:12187–12193. [PubMed] [Google Scholar]
- 12.Marzesco A M, Galli T, Louvard D, Zahraoui A. J Biol Chem. 1998;273:22340–22345. doi: 10.1074/jbc.273.35.22340. [DOI] [PubMed] [Google Scholar]
- 13.Yan D, Swain P K, Breuer D, Tucker R M, Wu W, Fujita R, Rehemtulla A, Burke D, Swaroop A. J Biol Chem. 1998;273:19656–19663. doi: 10.1074/jbc.273.31.19656. [DOI] [PubMed] [Google Scholar]
- 14.Ueffing M, Lovric J, Philipp A, Mischak H, Kolch W. Oncogene. 1997;15:2921–2927. doi: 10.1038/sj.onc.1201477. [DOI] [PubMed] [Google Scholar]
- 15.Picard V, Ersdalbadju E, Lu A Q, Bock S C. Nucleic Acids Res. 1994;22:2587–2591. doi: 10.1093/nar/22.13.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vojtek A B, Hollenberg S M, Cooper J A. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 17.Schiestl R H, Gietz R D. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 18.Bartel P L, Fields S. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 19.Hafner S, Adler H S, Mischak H, Janosch P, Heidecker G, Wolfman A, Pippig S, Lohse M, Ueffing M, Kolch W. Mol Cell Biol. 1994;14:6696–6703. doi: 10.1128/mcb.14.10.6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.VanCott T C, Loomis L D, Redfield R R, Birx D L. J Immunol Methods. 1992;146:163–176. doi: 10.1016/0022-1759(92)90225-i. [DOI] [PubMed] [Google Scholar]
- 21.Malmborg A C, Michaelsson A, Ohlin M, Jansson B, Borrebaeck C A. Scand J Immunol. 1992;35:643–650. doi: 10.1111/j.1365-3083.1992.tb02970.x. [DOI] [PubMed] [Google Scholar]
- 22.Kuhlmann J, Macara I, Wittinghofer A. Biochemistry. 1997;36:12027–12035. doi: 10.1021/bi970524k. [DOI] [PubMed] [Google Scholar]
- 23.Roepman R, van Duijnhoven G, Rosenberg T, Pinckers A J, Bleeker-Wagemakers L M, Bergen A A, Post J, Beck A, Reinhardt R, Ropers H H, et al. Hum Mol Genet. 1996;5:1035–1041. doi: 10.1093/hmg/5.7.1035. [DOI] [PubMed] [Google Scholar]
- 24.Andreasson S, Ponjavic V, Abrahamson M, Ehinger B, Wu W, Fujita R, Buraczynska M, Swaroop A. Am J Ophtalmol. 1997;124:95–102. doi: 10.1016/s0002-9394(14)71649-6. [DOI] [PubMed] [Google Scholar]
- 25.Jaitner B K, Becker J, Linnemann T, Herrmann C, Wittinghofer A, Block C. J Biol Chem. 1997;272:29927–29933. doi: 10.1074/jbc.272.47.29927. [DOI] [PubMed] [Google Scholar]
- 26.Lorenz B, Migliaccio C, Lichtner P, Meyer C, Strom T M, Durso M, Becker J, Ciccodicola A, Meitinger T. Eur J Hum Genet. 1998;6:283–290. doi: 10.1038/sj.ejhg.5200215. [DOI] [PubMed] [Google Scholar]
- 27.Li N, Florio S K, Pettenati M J, Rao P N, Beavo J A, Baehr W. Genomics. 1998;49:76–82. doi: 10.1006/geno.1998.5210. [DOI] [PubMed] [Google Scholar]
- 28.Higashide T, Murakami A, McLaren M J, Inana G. J Biol Chem. 1996;271:1797–1804. doi: 10.1074/jbc.271.3.1797. [DOI] [PubMed] [Google Scholar]
- 29.Higashide T, McLaren M J, Inana G. Invest Ophthalmol Visual Sci. 1998;39:690–698. [PubMed] [Google Scholar]
- 30.Maduro M, Pilgrim D. Gene. 1996;183:77–85. doi: 10.1016/s0378-1119(96)00491-x. [DOI] [PubMed] [Google Scholar]
- 31.Bowes C, Li T, Danciger M, Baxter L C, Applebury M L, Farber D B. Nature (London) 1990;347:677–680. doi: 10.1038/347677a0. [DOI] [PubMed] [Google Scholar]
- 32.Altherr M R, Wasmuth J J, Seldin M F, Nadeau J H, Baehr W, Pittler S J. Genomics. 1992;12:750–754. doi: 10.1016/0888-7543(92)90305-c. [DOI] [PubMed] [Google Scholar]
- 33.McLaughlin M E, Sandberg M A, Berson E L, Dryja T P. Nat Genet. 1993;4:130–134. doi: 10.1038/ng0693-130. [DOI] [PubMed] [Google Scholar]
- 34.McLaughlin M E, Ehrhart T L, Berson E L, Dryja T P. Proc Natl Acad Sci USA. 1995;92:3249–3253. doi: 10.1073/pnas.92.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gal A, Orth U, Baehr W, Schwinger E, Rosenberg T. Nat Genet. 1994;7:64–68. doi: 10.1038/ng0594-64. [DOI] [PubMed] [Google Scholar]
- 36.Florio S K, Prusti R K, Beavo J A. J Biol Chem. 1996;271:24036–24047. doi: 10.1074/jbc.271.39.24036. [DOI] [PubMed] [Google Scholar]
- 37.Qin N, Pittler S J, Baehr W. J Biol Chem. 1992;267:8458–8463. [PubMed] [Google Scholar]
- 38.Anant J S, Ong O C, Xie H Y, Clarke S, O’Brien P J, Fung B K. J Biol Chem. 1992;267:687–690. [PubMed] [Google Scholar]
- 39.Seabra M C, Ho Y K, Anant J S. J Biol Chem. 1995;270:24420–24427. doi: 10.1074/jbc.270.41.24420. [DOI] [PubMed] [Google Scholar]
- 40.Seabra M C, Brown M S, Goldstein J L. Science. 1993;259:377–381. doi: 10.1126/science.8380507. [DOI] [PubMed] [Google Scholar]
- 41.Ullrich O, Horiuchi H, Bucci C, Zerial M. Nature (London) 1994;368:157–160. doi: 10.1038/368157a0. [DOI] [PubMed] [Google Scholar]