Abstract
The use of real-time quantitative PCR (5′ nuclease PCR assay) as a tool to study the gastrointestinal microflora that adheres to the colonic mucosa was evaluated. We developed primers and probes based on the 16S ribosomal DNA gene sequences for the detection of Escherichia coli and Bacteroides vulgatus. DNA was isolated from pure cultures and from gut biopsy specimens and quantified by the 5′ nuclease PCR assay. The assay showed a very high sensitivity: as little as 1 CFU of E. coli and 9 CFU of B. vulgatus could be detected. The specificities of the primer-probe combinations were evaluated with samples that were spiked with the species most closely related to E. coli and B. vulgatus and with eight other gut microflora species. Mucosal samples spiked with known amounts of E. coli or B. vulgatus DNA showed no PCR inhibition. We conclude that the 5′ nuclease PCR assay may be a useful alternative to conventional culture techniques to study the actual in vivo composition of a complex microbial community like the gut microflora.
The human intestinal tract harbors a large and complex community of microbes which is involved in maintaining human health by preventing colonization by pathogens and by producing nutrients. At least 400 bacterial species are thought to be present in the human gastrointestinal tract (4, 9, 14, 19). Microorganisms are not randomly distributed throughout the intestine. The flora of the stomach and proximal small intestine differs significantly from that of the terminal ileum and colon. The numerically predominant species of bacteria are obligate anaerobes and are represented by both gram-positive and gram-negative genera. The most frequently identified anaerobic microorganisms are Bacteroides spp., Bifidobacterium spp., Eubacterium spp., Peptostreptococcus spp., and Fusobacterium spp. (17). The total number of bacterial cells, as determined by fecal culture methods, increases gradually from the small intestine to the large bowel and reaches 1011 to 1012 cells/ml of contents. Conventional bacteriological methods like microscopy, culture, and identification are used for the analysis and/or quantification of the intestinal microflora (7, 16, 20). Limitations of conventional methods are their low sensitivities (5), their inability to detect noncultivatable bacteria and unknown species, their time-consuming aspects, and their low levels of reproducibility due to the multitude of species to be identified and quantified. In addition, the large differences in growth rates and growth requirements of the different species present in the human gut indicate that quantification by culture is bound to be inaccurate. To overcome the problems of culture, techniques based on 16S ribosomal DNA (rDNA) genes were developed (2, 22). These include fluorescent in situ hybridization (8, 11, 12, 13, 21), denaturing gradient gel electrophoresis (15, 18), and temperature gradient gel electrophoresis (6, 23). These techniques have low sensitivities and are laborious and technically demanding. We evaluated the suitability of a real-time quantitative PCR (5′ nuclease PCR assay) to identify and quantify microbial species present in complex microbial communities, like the colonic flora. We are particularly interested in the part of this flora which adheres to the colonic mucosa. For this evaluation, we selected two bacteria that are prominent in the normal gut and that play an important role in maintenance of a healthy gut microflora: the facultative aerobic species Escherichia coli and the anaerobic species Bacteroides vulgatus. We tested colonic mucosal biopsy specimens for adherent flora of these two species. We demonstrate that real-time quantitative PCR is a promising method for characterization of complex microbial communities like the gastrointestinal microflora.
MATERIALS AND METHODS
Bacterial strains and culture.
The bacterial strains used in this study included Actinomyces viscosus (a clinical isolate), Bacteroides fragilis (ATCC 23745), B. vulgatus (a clinical isolate), Bifidobacterium adolescentis (a clinical isolate), Clostridium difficile (ATCC 9685), Enterococcus faecalis (a clinical isolate), E. coli (ATCC 35218), Fusobacterium nucleatum (ATCC 10953), Lactobacillus acidophilus (ATCC 4356), Propionibacterium acnes (ATCC 6919), and Salmonella enterica subsp. enterica serovar Typhimurium (ATCC 14128). Anaerobic and aerobic bacteria were cultured at 37°C in brain heart infusion medium (Difco Laboratories, Detroit, Mich.) under anaerobic (10% CO2) or aerobic (no CO2) conditions. The numbers of CFU were determined by culture of appropriate dilutions.
Clinical specimens.
Biopsy samples of the mucosa of the sigmoid colon were obtained from 25 individuals undergoing routine sigmoidoscopy for various reasons, mainly irritable bowel syndrome. Samples from patients with inflammatory bowel disease, mucosal inflammation, diverticulitis, or cancer were excluded. Biopsy specimens were collected with a flexible videoendoscope (Olympus, GmbH, Hamburg, Germany) and a flexible biopsy forceps (Wilson-Cook, European Endoscopy Group, Fujinon Medical Holland, Veenendaal, The Netherlands). Biopsy samples were immediately snap frozen in liquid nitrogen and stored at −20°C.
DNA extraction.
Chromosomal DNA was isolated from bacterial cultures and from 25 gastrointestinal mucosal biopsy specimens. Biopsy specimens and E. coli bacteria used for comparison of conventional culture and real-time PCR were pretreated with collagenase (see “Real-Time PCR versus Conventional Culture” below). After addition of 100 μl of Tris-EDTA to the biopsy specimens, DNA from both cultures and biopsy specimens was extracted with the DNeasy tissue kit (Qiagen, Hilden, Germany), according to the instructions of the manufacturer.
Design of primers and probes.
The primers and probes for E. coli and B. vulgatus were based on 16S rDNA sequences, available in the National Center for Biotechnology Information databases. Alignment of 16S rDNA sequences with those from closely related bacteria revealed sequences specific for each species. The sequences of the primer and probe combinations optimal for each species were selected from these specific sequences with Primer Express software, provided with the ABI Prism 7700 sequence detector (Applied Biosystems, Foster City, Calif.). To check for specificity, the selected primer target sites were compared to all available 16S rDNA sequences by using the BLAST database search program (www.ncbi.nlm.nih.gov/BLAST) (1). For E. coli the forward primer sequence was 5′-CATGCCGCGTGTATGAAGAA-3′ (base pairs 395 to 414), the reverse primer sequence was 5′-CGGGTAACGTCAATGAGCAAA-3′ (base pairs 470 to 490), and the Taqman probe sequence was 5′-TATTAACTTTACTCCCTTCCTCCCCGCTGAA-3′ (base pairs 437 to 467). For B. vulgatus the forward primer sequence was 5′-AAGGGAGCGTAGATGGATGTTTA-3′ (base pairs 577 to 599), the reverse primer sequence was 5′-CGAGCCTCAATGTCAGTTGC-3′ (base pairs 750 to 769), and the probe sequence was 5′-CCTGCCTCAACTGCACTCAAGATATCCAGTA-3′ (base pairs 645 to 675). Both Taqman fluorescent probes were 5′ labeled with 6-carboxyfluorescein as the reporter dye and 6-carboxytetramethylrhodamine as the 3′ quencher dye. The real-time quantitative fluorescent Taqman assay has been described by Heid et al. (10).
5′ nuclease PCR assay conditions.
The amplification reactions were carried out in a total volume of 50 μl, and reaction mixtures contained 1× Taqman universal PCR master mixture (PCR buffer, deoxynucleoside triphosphates, AmpliTaq Gold, internal reference signal ][(6-carboxy-′x′-rhodamine], uracil N-glycosylase [UNG], MgCl2; Applied Biosystems, Foster City, Calif.), 300 nM each E. coli-specific oligonucleotide primer, 100 nM fluorescence-labeled E. coli-specific probe, and 5 μl of E. coli DNA in serial 10-fold dilutions per reaction mixture. For B. vulgatus, 900 nM primers, 200 nM probe, and 5 μl of B. vulgatus DNA in serial dilutions were used in each reaction mixture. The standard curve was based on five replicate reactions.
Amplification reactions were performed on an ABI Prism 7700 sequence detector (Applied Biosystems). Amplification was carried out after 2 min at 50°C (activation of the UNG) and 10 min at 95°C (activation of the AmpliTaq Gold DNA polymerase), followed by 15 s at 95°C and 1 min at 60°C for 40 cycles. Data analysis was carried out with sequence detection system software (Applied Biosystems).
Specificity and inhibition.
The specificities of the primer-probe combinations were determined by performing the real-time PCR assay with samples that contained DNA isolated from Salmonella serovar Typhimurium or B. fragilis. The specificities were also tested with samples that contained a mixture of DNA isolated from eight other representative gastrointestinal bacterial species (8 pg of DNA of each of the species A. viscosus, B. adolescentis, C. difficile, E. faecalis, F. nucleatum, L. acidophilus, P. acnes, and either E. coli or B. vulgatus. We used sterile water as a negative control.
To determine whether the presence of mucosal tissue inhibited the PCR assay, after DNA extraction we compared the results of assays performed with unspiked biopsy specimens, biopsy specimens spiked with bacteria in numbers close to the cutoff value, biopsy specimens spiked with 5 CFU of E. coli, and a pure E. coli culture.
Real-time PCR versus conventional culture.
Twelve mucosal biopsy specimens were incubated with 5 mg of collagenase (Roche Diagnostics, Mannheim, Germany) per ml for 5 h at 37°C. The resulting cell suspension was equally divided; one part was used for DNA isolation, and the other part was used for determination of the viable counts of E. coli on MacConkey III plates (Oxoid, Basingstoke, England). An E. coli culture was also incubated for 5 h at 37°C in the presence of 5 mg of collagenase per ml to determine the effect of collagenase on the recovery of E. coli CFU.
DNase I treatment of PCR master mixture.
A universal PCR master mixture (Applied Biosystems) was treated with DNase I (Amersham Pharmacia Biotech, Roosendaal, The Netherlands) at 1.16 × 10−5 U of DNase I per μl of universal PCR master mixture to remove possible traces of E. coli DNA contamination that occurred during manufacture. Primers, probe, and DNA were added after incubation of the universal PCR master mixture with DNase I at room temperature for 5 min and heat denaturation at 99°C for 10 min.
RESULTS
Specificity of 5′ nuclease PCR assay.
The specificities of the 5′ nuclease PCR assays with the primer and probe combinations that we developed for detection of E. coli and B. vulgatus were examined by comparing the quantification of pure E. coli DNA (8 pg) or B. vulgatus DNA (8 pg) with the quantification of DNA from E. coli or B. vulgatus mixed with DNA isolated from representative bacterial species (see above) which belong to the major groups of bacteria present in the colon. Both primer-probe sets were specific, and no difference in amplification or quantification was obtained in the presence of the mixture of gut bacteria. Both of the PCRs for E. coli and the mixture of gut bacteria, including E. coli, had a cycle threshold (Ct) value of 27, and the PCR with the mixture without E. coli and the negative control had a Ct value of 35 (Fig. 1). For B. vulgatus, the Ct values were 28, while no signals were obtained with the mixture of gut bacteria without B. vulgatus and the negative control. The homology between the 16S rDNA sequences of E. coli and Escherichia vulneris and Shigella species is so close (100% for the sequences to which the primers and probes are specific) that no distinction can be made between these bacteria. As a consequence, we used the primer-probe combinations with DNA isolated from Salmonella serovar Typhimurium, which is the species next most closely related to E. coli, and with DNA isolated from B. fragilis, which is the species most closely related to B. vulgatus. No signals were obtained with these bacteria.
FIG. 1.
Amplification sensitivity of 5′ nuclease PCR assay for E. coli. DNA isolated from log-phase bacteria was used in serial 10-fold dilutions from 135,000 bacteria to 1 bacterium per reaction mixture. NTC, no-template control; ΔRn, fluorescence intensity after subtraction of background signal.
With the negative control samples (the mixture of gut bacteria without E. coli and sterile water), we noted a fluorescent signal with a Ct value of 35. Because we hypothesized that this might be due to E. coli DNA contamination of the Taq polymerase, we repeated the assay after treatment of the master mixture with DNase I. After such treatment, no signal was obtained with the negative control samples.
Sensitivity of 5′ nuclease PCR assay.
To determine the sensitivities of both primer-probe combinations, serial dilutions of cultures of E. coli and B. vulgatus were used for determination of the numbers of CFU and genomic DNA isolation. Six dilutions for quantitative amplification were created, with a dynamic range from 0.5 to 135,000 CFU for E. coli (Fig. 1) and 1 to 985,000 CFU for B. vulgatus. The reproducibility of the quantitative experiments was approximately 99%, based on the Ct values for five replicate PCR runs. The average Ct value for the dilution containing 135,000 CFU of E. coli was 24.2, with a range of Ct values from 23.9 to 24.5. We obtained the same reproducibility with the other bacterial concentrations tested.
The detection limit for E. coli was 1 CFU, and the detection limit for B. vulgatus was 9 CFU. No signals above that for the negative control were obtained when 0.5 CFU of E. coli or 1, 2.5, 3, or 5 CFU of B. vulgatus was used. These results were confirmed by at least three independent PCR amplifications.
Mean equivalents of 4.2 × 104 of E. coli CFU (range, 0 to 4.2 × 105) and 4.2 × 106 of B. vulgatus CFU (range, 0 to 3.0 × 107) per specimen were detected in the biopsy samples (Fig. 2).
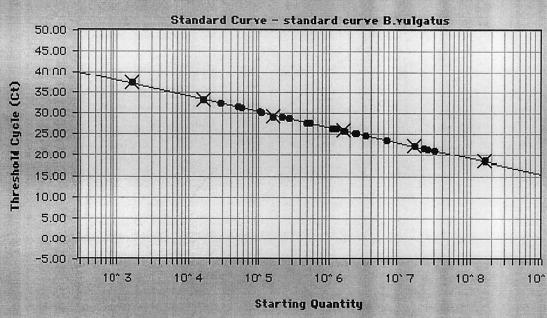
FIG. 2.
Quantification of B. vulgatus adherent to mucosal biopsy specimens from healthy persons. DNA isolated from biopsy specimens was used in a 5′ nuclease PCR assay with the primer-probe set for B. vulgatus. A range of known concentrations of bacteria was amplified and plotted (black crosses) as a standard curve as a reference for the quantification of B. vulgatus in mucosal biopsy specimens. For quantification, a standard curve was obtained by plotting the points of the fluorescent signals that cross the threshold line (Ct values). Each dot represents the result of an amplification of DNA from a mucosal biopsy specimen. A mean equivalent of 4.2 × 106 B. vulgatus CFU was found in mucosal biopsy specimens. R was equal to 0.999, and the slope was −3.412.
Experiments with biopsy specimens spiked with known amounts of E. coli DNA (equivalent to the amount of DNA from 5 CFU) showed that mucosal tissue did not inhibit the PCR: the fluorescent signal in the presence or absence of a biopsy sample was the same, with an average Ct value of 34.0 (range, 33.4 to 34.6).
Real-time PCR versus culture.
Collagenase treatment did not affect the viability or the growth rate of E. coli. The results of the comparison of real-time PCR and culture are provided in Table 1. Real-time PCR generated Ct values that were used to calculate the equivalent number of CFU for determination of viable counts. In every sample tested (n = 12), the number of CFU calculated from the fluorescent signal obtained by real-time PCR was higher than the number of bacteria found on quantification by culture. On average, this value was 100-fold higher. The percentage of cultivatable bacteria was calculated by dividing the number of CFU extrapolated from the Ct values (by use of a standard curve). The median percentage of cultivatable bacteria was 1%.
TABLE 1.
Results of real-time PCR versus those of culture
| Sample no. | Cta | PCR resultb | Culture resultc | % Cultivatable |
|---|---|---|---|---|
| 1 | 25.1 | 5.5 × 107 | 4.5 × 106 | 8.2 |
| 2 | 31.7 | 6.5 × 105 | 1.9 × 104 | 2.9 |
| 3 | 32.2 | 4.8 × 105 | 1.0 × 103 | 0.2 |
| 4 | 34.4 | 1.1 × 105 | 2.0 × 103 | 1.8 |
| 5 | 23.2 | 1.9 × 108 | 5.4 × 105 | 0.3 |
| 6 | 30.5 | 1.5 × 106 | 9.0 × 103 | 0.6 |
| 7 | 28.6 | 5.3 × 106 | 2.8 × 104 | 0.5 |
| 8 | 35.5 | 5.3 × 104 | 0 | 0 |
| 9 | 31.4 | 7.8 × 105 | 3.1 × 105 | 40 |
| 10 | 35.0 | 7.5 × 104 | 0 | 0 |
| 11 | 26.1 | 2.8 × 107 | 3.6 × 105 | 1.3 |
| 12 | 22.7 | 2.8 × 108 | 6.8 × 106 | 2.4 |
Ct values were used for calculation of CFU equivalent.
CFU equivalent based on real-time PCR.
CFU of E. coli.
DISCUSSION
In this study an approach that combines the specificity of fluorescent oligonucleotide probes with the sensitivity of PCR was used. E. coli and B. vulgatus were used for evaluation of the 5′ nuclease PCR assay as a tool to identify and quantify intestinal bacteria. Both bacteria are prominent normal gut bacteria that play an important role in the maintenance of a healthy gut microflora.
Conventional PCR has several disadvantages. These include the sensitivity of the assay to inhibition by substances present in the sample to be analyzed, the limitation of a small sample input, and the possibility of nonspecific binding of the primers or the probe. Finally, the PCR assay is very susceptible to contamination. The 5′ nuclease PCR assay (real-time PCR) solves several of these problems. In real-time PCR two primers and one probe are used, and a fluorescent signal can be generated only when all three are bound to the DNA at the correct primer and probe locations, which greatly reduces the risk of nonspecific binding. The assay is performed in a closed-tube system, and no post-PCR handling is necessary. This reduces the risk of contamination.
Specificity, a key factor for accurate quantification of the bacteria of interest, was determined by performing the reaction with a sample consisting of a mixture of genomic DNA isolated from eight different bacterial species commonly found in the intestinal microflora. In addition, the specificity was also tested by using DNA purified from closely related species: Salmonella serovar Typhimurium for E. coli and B. fragilis for B. vulgatus. Nonspecific signals were not detected in these reactions. The specificity of our approach is hampered only by the availability of species-specific DNA sequences. In this respect, 16S rDNA sequences are not always satisfactory. This is illustrated by the homology between the 16S rDNA sequences of E. coli, E. vulneris, and Shigella. This homology is so close that we were not able to design a primer-probe combination that would distinguish these bacteria. More and more sequence information is continuously becoming available, however, and sequences which are truly specific for each bacterial species will be available in the near future. Such specific sequences can be used, like we used the 16S rDNA sequences, to design primers and probes in cases in which 16S rDNA-based discrimination is not possible.
In the first set of experiments, a positive signal was also obtained with samples that in principle did not contain E. coli DNA. We were able to show that this was due to contamination, probably caused by traces of E. coli DNA present in the Taq polymerase and/or the UNG enzyme. DNase I treatment of the universal PCR master mixture eliminated this signal, which confirmed this hypothesis. Corless et al. (3) previously mentioned the E. coli contamination of Taq DNA polymerase.
The assay proved very sensitive: as little as 1 CFU of E. coli and 9 CFU of B. vulgatus bacteria could be detected. The difference in sensitivity cannot be explained by differences in the numbers of copies of the 16S rRNA genes, since both species have seven copies. It is possible that the DNA extraction efficiency or anaerobic culture conditions may have influenced the sensitivities of the assays for E. coli and B. vulgatus and resulted in differences in sensitivities.
Compared to quantification by determination of viable counts, the quantification of bacteria by real-time PCR yielded approximately 100-fold higher numbers of E. coli. This appears plausible, since all DNA is amplified by PCR, including DNA from dead bacteria. The median percent cultivatable bacteria was 1%, indicating that a high percentage of the bacteria that adhere to the intestinal mucosa are dead or viable but not cultivatable.
We used the real-time PCR to investigate bacteria that adhere to the gastrointestinal mucosa, not the bacteria present in fecal samples (6, 8, 11, 12, 13, 15, 18, 21, 23). We did this because in future studies we aim to evaluate the role of this adherent flora in the inflammation of the mucosa that is characteristic of inflammatory bowel diseases.
A problem with PCR-based assays is that the reaction is very susceptible to inhibitory factors that can be present in the samples under study. To determine whether inhibition was a problem, we spiked biopsy samples with as little as 5 CFU of E. coli. These low numbers were accurately detected. This indicated that no inhibition factors seem to be influencing the quantitative PCR results for mucosal biopsy specimens or that inhibitory factors were removed with the DNeasy tissue kit (Qiagen) used for DNA isolation.
We found that the reproducibility of the quantitative experiments was over 99%, based on multiple replicate PCR runs. The 5′ nuclease PCR assay is therefore an accurate method that can be used to gain a better insight into the actual in vivo composition of the microflora that adheres to the intestinal mucosa.
Investigation of this microflora is crucial for obtaining an understanding of the role of the microflora in gut health but also an understanding of the role of the microflora in inflammatory bowel diseases like Crohn's disease and ulcerative colitis.
Many factors, such as differences in nutrition or use of antibiotics, can have an effect on the gastrointestinal microflora. The 5′ nuclease PCR assay in combination with a high-throughput screening system makes the study of these effects on the microflora of each individual possible.
Acknowledgments
This study was supported by NWO grant 901-14-204. X.W.H. was supported by MLDS grant WS98-22, and R.K.L. was supported by Byk.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corless, C. E., M. Guiver, R. Borrow, V. Edwards-Jones, E. B. Kaczmarski, and A. J. Fox. 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J. Clin. Microbiol. 38:1747-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn, D. 1990. Autochthonous microflora of the gastrointestinal tract. Perspect. Colon Rectal Surg. 2:105. [Google Scholar]
- 5.Dutta, S., A. Chatterjee, P. Dutta, K. Rajendran, S. Roy, K. C. Pramanik, and S. K. Bhattacharya. 2001. Sensitivity and performance characteristics of a direct PCR with stool samples in comparison to conventional techniques for diagnosis of Shigella and enteroinvasive Escherichia coli infection in children with acute diarrhoea in Calcutta, India. J. Med. Microbiol. 50:667-674. [DOI] [PubMed] [Google Scholar]
- 6.Felske, A., A. D. Akkermans, and W. M. de Vos. 1998. Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl. Environ. Microbiol. 64:4581-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finegold, S. M., V. L. Sutter, and G. E. Mathisen. 1983. Normal indigenous intestinal flora, p. 3-31. In D. J. Hentgens (ed.), Human intestinal microflora in health and disease. Academic Press, London, United Kingdom.
- 8.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorbach, S. L., L. Nakas, P. I. Lerner, and L. Weinstein. 1967. Effects of diet, age and periodic sampling on numbers of faecal microorganisms in man. Gastroenterology 53:845-855. [PubMed] [Google Scholar]
- 10.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 11.Jansen, G. J., A. C. Wildeboer-Veloo, R. H. Tonk, A. H. Franks, and G. W. Welling. 1999. Development and validation of an automated, microscopy-based method for enumeration of groups of intestinal bacteria. J. Microbiol. Methods 37:215-221. [DOI] [PubMed] [Google Scholar]
- 12.Jansen, G. J., M. Mooibroek, J. Idema, H. J. Harmsen, G. W. Welling, and J. E. Degener. 2000. Rapid identification of bacteria in blood cultures by using fluorescently labeled oligonucleotide probes. J. Clin. Microbiol. 38:814-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore, W. E., and L. V. Holdeman. 1974. Special problems associated with the isolation and identification of intestinal bacteria in fecal flora studies. Am. J. Clin. Nutr. 27:1450-1455. [DOI] [PubMed] [Google Scholar]
- 15.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 16.O'Sullivan, D. J. 1999. Methods of analysis of the intestinal microflora, p. 23-44. In G. W. Tannock (ed.), Probiotics: a critical review. Horizon Scientific Press, Wymondham, United Kingdom.
- 17.Simon, G. L., and S. L. Gorbach. 1986. The human intestinal microflora. Dig. Dis. Sci. 31:147S-162S. [DOI] [PubMed] [Google Scholar]
- 18.Simpson, J. M., V. J. McCracken, B. A. White, H. R. Gaskins, and R. I. Mackie. 1999. Application of denaturant gradient gel electrophoresis for the analysis of the porcine gastrointestinal microbiota. J. Microbiol. Methods 36:167-179. [DOI] [PubMed] [Google Scholar]
- 19.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tannock, G. W. 1999. Analysis of the intestinal microflora: a renaissance. Antonie Leeuwenhoek 76:265-278. [PubMed] [Google Scholar]
- 21.Welling, G. W., P. Elfferich, G. C. Raangs, A. C. Wildeboer-Veloo, G. J. Jansen, and J. E. Degener. 1997. 16S ribosomal RNA-targeted oligonucleotide probes for monitoring of intestinal tract bacteria. Scand. J. Gastroenterol. Suppl. 222:17-19. [DOI] [PubMed] [Google Scholar]
- 22.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoetendal, E. G., A. D. Akkermans, and W. M. de Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]