Abstract
Using reverse transcriptase–PCR and degenerate oligonucleotides derived from the active-site residues of subtilisin/kexin-like serine proteinases, we have identified a highly conserved and phylogenetically ancestral human, rat, and mouse type I membrane-bound proteinase called subtilisin/kexin-isozyme-1 (SKI-1). Computer databank searches reveal that human SKI-1 was cloned previously but with no identified function. In situ hybridization demonstrates that SKI-1 mRNA is present in most tissues and cells. Cleavage specificity studies show that SKI-1 generates a 28-kDa product from the 32-kDa brain-derived neurotrophic factor precursor, cleaving at an RGLT↓SL bond. In the endoplasmic reticulum of either LoVo or HK293 cells, proSKI-1 is processed into two membrane-bound forms of SKI-1 (120 and 106 kDa) differing by the nature of their N-glycosylation. Late along the secretory pathway some of the membrane-bound enzyme is shed into the medium as a 98-kDa form. Immunocytochemical analysis of stably transfected HK293 cells shows that SKI-1 is present in the Golgi apparatus and within small punctate structures reminiscent of endosomes. In vitro studies suggest that SKI-1 is a Ca2+-dependent serine proteinase exhibiting a wide pH optimum for cleavage of pro-brain-derived neurotrophic factor.
Limited proteolysis of inactive precursors to produce active peptides and proteins generates biologically diverse products from a finite set of genes. Most often, such processing occurs at either monobasic or dibasic residues as a result of cleavage by mammalian serine proteinases related to bacterial subtilisin and yeast kexin (1, 2). These enzymes, known as proprotein convertases (PCs), cleave a variety of precursors at the consensus (R/K)-(Xaa)n-R↓ sequence, where Xaa is any amino acid except Cys and n = 0, 2, 4, or 6 (1–3).
Less commonly than cleavage at basic residues, bioactive products also can be produced by limited proteolysis at amino acids such as L, V, M, A, T, and S (3). This type of cellular processing has been implicated in the generation of bioactive peptides such as α- and γ-endorphin (4), the C-terminal glycopeptide fragment 1–19 of provasopressin (5), platelet factor 4 (6), the metalloprotease ADAM-10 (7), site 1 cleavage of the sterol regulatory element-binding proteins (SREBPs) (8), as well as in the production of the Alzheimer’s amyloidogenic peptides Aβ40, -42, and -43 (9). Processing of this type occurs either in the endoplasmic reticulum (ER) (8), late along the secretory pathway, within secretory granules (4, 5), at the cell surface, or in endosomes (6, 7, 9). The proteinases responsible for these cleavages are not yet identified.
We hypothesized that an enzyme (or enzymes) distinct from, but related to, PCs may generate polypeptides by cleavage at nonbasic residues. To test that idea, we employed a reverse transcriptase–PCR (RT-PCR) strategy similar to the one used to identify the PCs (10), except that we used degenerate oligonucleotides closer to bacterial subtilisin than to yeast kexin. This resulted in the isolation of a cDNA fragment encoding a putative subtilisin-like enzyme from human cell lines. This partial sequence was identical to a segment of a human myeloid cell-derived cDNA reported by Nagase et al. (11). Preliminary results demonstrated that this putative proteinase cleaves pro-brain-derived neurotrophic factor (proBDNF) (ref. 12; S.J.M. N.G.S., and R.A.M., unpublished results).
In this paper, we show that the sequences of rat, mouse, and human orthologues of this putative type I membrane-bound subtilisin-kexin-isoenzyme, which we called SKI-1, exhibit a high degree of sequence conservation. Tissue distribution analysis by both Northern blots and in situ hybridization revealed that SKI-1 mRNA is widely expressed. A vaccinia virus recombinant and a stable transfectant of human SKI-1 in HK293 cells allowed the analysis of its biosynthesis and intracellular localization. Finally, we present data demonstrating that SKI-1 cleaves at a specific T↓ residue within the N-terminal segment of proBDNF. SKI-1 is thus identified as a mammalian secretory subtilisin/kexin-like enzyme capable of cleaving a proprotein at nonbasic residues.
MATERIALS AND METHODS
PCR and Sequencing.
Most RT-PCRs were performed using a Titan One Tube RT-PCR system (Boehringer Mannheim) on 1 μg of total RNA isolated from a human neuronal cell line (IMR-32), mouse corticotrophic cells (AtT20), or rat adrenal glands. The active-site degenerate primers were as follows: His (sense) 5′-GGICA(C,T)GGIACI(C,T)(A,T)(C,T)(G,T)(T,G)IGCIGG-3′ and Ser (antisense) 5′-CCIG(C,T)IACI(T,A)(G,C)IGGI(G,C)(T,A)IGCIACI(G,C)(A,T)IGTICC-3′, based on the sequences GHGT(H,F)(V,C)AG and GTS(V,M)A(T,S)P(H,V)V(A,T)G, respectively. The amplified 525-bp products were sequenced on an automated laser fluorescence DNA sequencer (Pharmacia). To obtain the full-length sequence of rat and mouse SKI-1, we used PCR primers based on the human (11) and mouse sequences, in addition to 5′ (13) and 3′ (14) RACE amplifications. At least three clones of the amplified cDNAs were sequenced. The GenBank accession numbers of the 3,788-bp mouse mSKI-1 cDNA and 3,895-bp rat rSKI-1 are AF094820 and AF094821, respectively.
Infection, Transfection, and Metabolic Labeling.
Human SKI-1 (nucleotides 1–4338) (11) in Bluescript (a generous gift from N. Nomura, Kazusa DNA Research Institute, Chiba, Japan; accession no. D42053) was digested with SacII (nucleotides 122-4338) and inserted into the vector PMJ602, and a vaccinia virus recombinant was isolated. The PMJ602 construct was also digested with 5′ KpnI/3′ NheI and cloned into the KpnI/XbaI sites of pcDNA3 (Invitrogen), and the cDNA was transfected into HK293 cells by using Lipofectin. A number of stable transfectants resistant to G418 and positive on Western blots using an SKI-1 antiserum (see below) were isolated, and one of them (clone 9), was investigated further. Either vaccinia virus-infected or -transfected cells were pulsed for 20 min with [35S]cysteine and then chased for various times in the presence or absence of either tunicamycin (5 μg/ml) or brefeldin A (2.5 μg/ml). Media and cell lysates were immunoprecipitated with SKI-1 antisera directed against either amino acids 634–651 or 217–233, or a pro-SKI-1 antiserum directed against the prosegment comprising amino acids 18–188 (Fig. 1). Immune complexes were resolved by SDS/PAGE on an 8% polyacrylamide/N-[tris(hydroxymethyl)methyl]glycine gel (15).
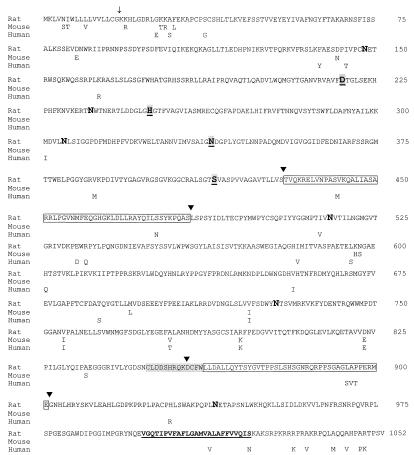
Figure 1.
Comparative protein sequences of SKI-1 deduced from rat, mouse, and human cDNAs. The position of the predicted end of the 17-aa signal peptide is shown by an arrow. The active sites Asp218, His249, and Ser414 as well as the oxyanion hole Asn338 are shown as bold, shaded, and underlined characters. The positions of the six potential N-glycosylation sites are emphasized in bold. The conserved shaded sequence fits the consensus signature for growth factors and cytokine receptor family. Each of the two boxed sequences was absent (▾) in a number of rat clones. The predicted transmembrane segment is in bold and underlined.
Northern Blots, in Situ Hybridizations, and Immunocytochemistry.
Northern blot analyses (16) were done on total RNA from adult male rat tissues by using either a TRIzol reagent kit (Life Technologies, Gaithersburg, MD) or a Quick Prep RNA kit (Pharmacia) and on poly(A)+ RNA of (male + female) rat adult tissues (BIO/CAN, Montreal). The blots were hybridized overnight at 68°C in the presence of [32P]UTP SKI-1 cRNA probes, which consisted of the antisense of nucleotides 655-1249 of rat SKI-1. For in situ hybridization on newborn rats, the same rat sense and antisense cRNA probes were labeled with uridine and cytosine 5′-[γ-[35S]thio]triphosphate (1, 16). For immunofluorescence staining we used a rabbit anti-SKI-1 antiserum at a 1:100 dilution and rhodamine-labeled goat anti-rabbit IgGs diluted 1:20 (16). Red SKI-1 immunostaining was compared with green-staining patterns of both fluorescein-labeled concanavalin A (Con A; Molecular Probes), an ER marker, or fluorescein-conjugated wheat germ agglutinin (WGA; Molecular Probes), a Golgi marker (17).
Ex Vivo and in Vitro proBDNF Processing.
A vaccinia virus recombinant of human SKI-1 (vv:SKI-1) was isolated as described for human proBDNF (vv:BDNF) (15). The vaccinia virus recombinants of α1-antitrypsin Pittsburgh (α1-PIT; vv:PIT) and α1-antitrypsin Portland (α1-PDX; vv:PDX) (18) were generous gifts from G. Thomas (Vollum Institute, Portland, OR). COS-7 cells (4 × 106) were coinfected with 1 plaque-forming unit (pfu) per cell of vv:BDNF and either the wild-type virus (vv:WT) alone at 2 pfu per cell or with 1 pfu per cell of each virus in the combinations [vv:SKI-1+vv:WT], [vv:SKI-1+vv:PIT], and [vv:SKI-1+vv:PDX]. At 10 h postinfection, cells were pulsed for 4 h with 0.2 mCi of [35S]cysteine/[35S]methionine (DuPont). Media and cell extracts were immunoprecipitated with a BDNF antiserum (ref. 19; provided by Amgen) at 0.5 μg/ml, and the proteins were resolved on SDS/PAGE 13–22% gradient gels (15). [35S]Met-labeled 32-kDa proBDNF and [3H]Leu-labeled 28-kDa BDNF were sequenced as described (20). For in vitro analysis, 32-kDa proBDNF obtained from the media of LoVo cells infected with vv:BDNF was incubated overnight with the shed form of SKI-1 obtained from cells coinfected with vv:SKI-1 and vv:PDX, either at different pH values or at pH 6.5 in the presence of selected inhibitors: pepstatin (1 μM), antipain (50 μM), cystatin (5 μM), E64 (5 μM), soybean trypsin inhibitor (SBTI, 5 μM), 0.5 M phenylmethylsulfonyl fluoride (PMSF) + 50 μM para-amino-PMSF (pAPMSF), o-phenanthroline (5 mM), and EDTA (10 mM). The products were resolved by SDS/PAGE on a 15% polyacrylamide gel, blotted, and then probed with a BDNF antiserum (Santa Cruz Biotechnology) at a dilution of 1:1,000.
RESULTS
Protein Sequence Analysis of SKI-1.
We first aligned the protein sequences within the catalytic domain of PC7 (21), yeast subtilases, and bacterial subtilisins, together with that of a novel Plasmodium falciparum subtilisin-like enzyme called pf-SUB2 (J.-C.B., unpublished results). This led to the choice of conserved amino acids GHGT(H/F)(V/C)AG and GTS(M/V)A(T/S)P(H/V)V(A/T)G around the active sites His and Ser, respectively. Thus, using degenerate oligonucleotides coding for the sense His and antisense Ser consensus sequences, we initiated a series of RT-PCRs on total RNA and isolated a 525-bp cDNA fragment from the human neuronal cell line IMR-32. This sequence was found to be 100% identical to that reported for a human cDNA called KIAA0091 obtained from a myeloid KG-1 cell line (11) and 88% identical to that of a 324-bp expressed sequence tag (accession no. H31838) from rat PC12 cells. The full-length rat and mouse cDNA sequences were obtained after RT-PCR amplifications of total RNA isolated from rat adrenal glands and PC12 cells and from mouse AtT20 cells. As shown in Fig. 1, alignment of the protein sequence deduced from the cDNAs revealed that rat and mouse SKI-1 share 98% sequence identity and a 96% identity to human SKI-1. Interestingly, within the catalytic domain (Asp218 → Ser414) the sequence similarity between the three species is 100%. Analysis of the predicted amino acid sequence suggests a 17-aa signal peptide, followed by a putative prosegment beginning at Lys18 and extending for some 160–180 aa. The proposed catalytic domain encompasses the typical active sites Asp218, His249, and Ser414 and the oxyanion hole Asn338. This domain is followed by an extended C-terminal sequence characterized by the presence of a conserved growth factor/cytokine receptor family motif C849LDDSHRQKDCFW861. This sequence is followed by a potential 24-aa hydrophobic transmembrane segment and a less-conserved 31-aa cytosolic tail that, remarkably, consists of 35% basic residues. Some of the clones isolated from rat adrenal glands suggested the existence of alternatively spliced rSKI-1 mRNAs in which the segments coding for amino acids 430–483 or 858–901 are absent. Finally, the phylogenetic tree derived from the alignment of the catalytic domain of SKI-1 with subtilases (22) suggests that it is an ancestral protein that is closer to plant and bacterial subtilases than to either yeast or mammalian homologues (not shown).
Tissue Distribution of SKI-1 mRNA.
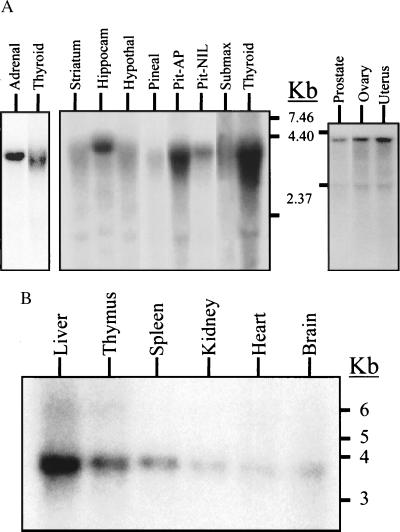
Northern blot analyses of SKI-1 mRNA in adult male rat reveal that rSKI-1 mRNA is widely expressed and is particularly rich in anterior pituitary, thyroid, and adrenal glands (Fig. 2A). A Northern blot of poly(A)+ RNA obtained from mixed adult male and female rat tissues also showed a wide distribution and a particular enrichment in liver (Fig. 2B). Similarly, analysis of 24 different cell lines (23) revealed a ubiquitous expression of SKI-1 mRNA (not shown).
Figure 2.
Northern blot analysis of the expression of SKI-1 in adult rat tissues. (A) Five micrograms of male rat total RNA was loaded in each lane: pituitary anterior (AP) and neurointermediate (NIL) lobes and submaxillary gland (submax). (B) Two micrograms of poly(A)+ RNA of (male + female) Sprague–Dawley rat adult tissues. The estimated size of rat SKI-1 mRNA is about 3.9 kb.
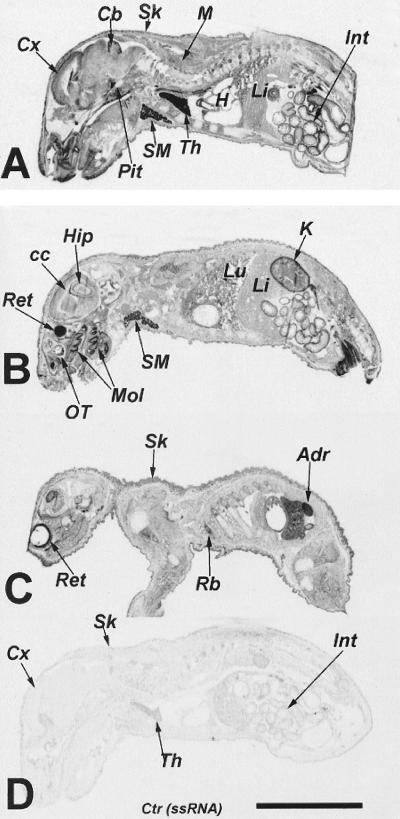
In situ hybridization data obtained in a day 2 postnatal rat also provided evidence of a widespread, if not ubiquitous, distribution of rSKI-1 mRNA. Fig. 3 shows at the anatomical level the presence of SKI-1 mRNA in developing skin, striated muscles, cardiac muscles, bones, and teeth as well as brain and many internal organs. Strong hybridization signals were detectable in the retina, cerebellum, pituitary, submaxillary, thyroid, and adrenal glands, molars, thymus, kidney, and intestine. Evidence for the cellular expression of rSKI-1 mRNA was obtained from analysis of the relative labeling densities per cell in selected tissues, based on a semiquantitative analysis of emulsion autoradiographs (not shown). In the central nervous system rSKI-1 mRNA labeling was mostly confined to neurons, whereas ependymal cells and supportive glial cells, such as presumed astrocytes, oligodendrocytes, and microglia, exhibited 5- to 30-fold-less labeling per cell. In addition, within the peripheral nervous system, trigeminal ganglia revealed a 5- to 10-fold greater expression in neurons as compared with presumptive Schwann cells. Labeling was observed in most of the glandular cells in the anterior and intermediate lobes of the pituitary as well as in the pituicytes of the pars nervosa. A semiquantitative comparison in the adult and newborn rat pituitary gland, submaxillary gland, thymus, and kidney demonstrated an overall 2-fold-decreased labeling of rSKI-1 mRNA with age (not shown).
Figure 3.
In situ hybridization of rSKI-1 mRNA in a 2-day-old rat. In situ hybridization is shown at anatomical resolution on x-ray film using an 35S-labeled antisense riboprobe (A–C) and sense control riboprobe (D). Adr, adrenal gland; Cb, cerebellum; cc, corpus callosum; Cx, cerebral cortex; H, heart; Int, intestine; K, kidney; Li, liver; Lu, lungs; M, muscles; Mol, molars; OT, olfactory turbinates; Pit, pituitary gland; Rb, ribs; Ret, retina; Sk, skin, SM, submaxillary gland; Th, thymus. [×4; bar (D) = 1 cm.]
Biosynthesis of hSKI-1.
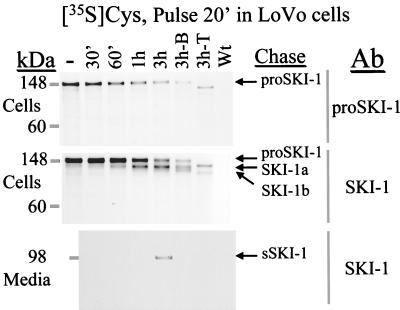
To define the molecular forms of human SKI-1 we generated both a vaccinia virus recombinant (vv:SKI-1) and a stable transfectant in HK293 cells. Three antisera were produced against amino acids 18–188 (prosegment), 217–233, and 634–651 of SKI-1. Expression of vv:SKI-1 in four different cell lines revealed that the enzyme is synthesized as a 148-kDa proSKI-1 zymogen, which is processed progressively into 120-, 106-, and 98-kDa proteins (Fig. 4). Only the 148-kDa form is recognized by the prodomain antiserum, whereas all four forms react with the other two antisera. Processing of the 148-kDa proSKI-1 into the 120- and 106-kDa forms occurs in the ER based on the presence of these proteins in cells preincubated with the fungal metabolite brefeldin A (Fig. 4; ref. 24). Furthermore, preincubation with tunicamycin revealed only two bands (Fig. 4), suggesting that the presumably membrane-bound 106- and 120-kDa forms differ by their N-glycosylation. At the 3-h chase time, results reveal the secretion of a 98-kDa shed form (sSKI-1) recognized by both SKI-1 antisera (Fig. 4) but not by the proSKI-1 antiserum (not shown). Similar SKI-1-related forms were seen in stably transfected HK293 cells after a 4-h pulse labeling with [35S]methionine (not shown).
Figure 4.
Biosynthetic analysis of SKI-1 in LoVo-C5 cells overexpressing vv:SKI-1. Cells were pulsed for 20 min with [35S]cysteine and chased for 30 min, 1 h, and 3 h in the absence or presence of either brefeldin A (3h-B) or tunicamycin (3h-T). The control represents the 3-h chase period for cells infected with the wild-type virus (Wt). Media and cell lysates were immunoprecipitated with either a SKI-1 antiserum (Ab: SKI-1; against amino acids 634–651) or a proSKI-1 antiserum. The arrows point to the 148-, 120-, 106-, and 98-kDa forms immunoprecipitated.
Intracellular Localization of SKI-1.
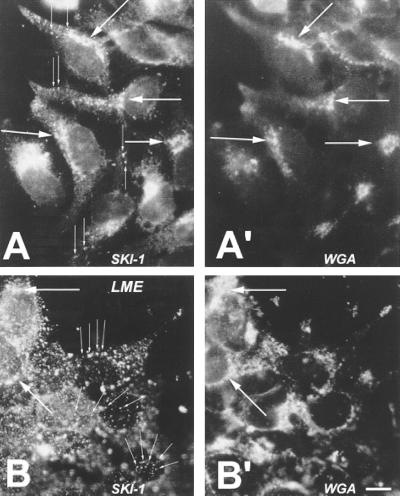
Double-staining immunofluorescence was used to compare the intracellular localization of the stably transfected human SKI-1 in HK293 cells with that of either the ER or Golgi markers Con A and WGA (17), respectively. The data show that SKI-1 exhibits (i) perinuclear staining, colocalizing with Con A fluorescence, presumably corresponding to the ER (not shown); (ii) paranuclear staining colocalizing with WGA fluorescence, suggesting the presence of SKI-1 in the Golgi (Fig. 5 A and B), and (iii) punctate staining observed in the cytoplasm and within extensions of a few cells (Fig. 5A). Some, but not all, of the punctate immunostaining matched that observed with WGA. This suggests that SKI-1 localizes in the Golgi but may sort to other organelles, including lysosomal and/or endosomal compartments. An indication of lysosomal/endosomal localization was provided by the analysis of SKI-1 immunofluorescence within cells preincubated for 4 h with 10 mM leucine methyl ester, a specific lysosomal/endosomal protease inhibitor (25). The results showed a net increase in the proportion of cells exhibiting punctate staining as compared with control cells (Fig. 5 A and B). The relative proportions of SKI-1 in cellular organelles and their dependence on culture conditions are now amenable to evaluation by subcellular fractionation and electron microscopy.
Figure 5.
hSKI-1 immunoreactivity in stably transfected HK293 cells. Black and white representation of the comparative double (red and green) fluorescence staining using an SKI-1 antiserum (directed against amino acids 634–651) (A and B) and fluorescein isothiocyanate-labeled WGA (A′ and B′) in control (A and A′) and leucine methyl ester (LME)-treated (B and B′) cells. Thin arrows emphasize the observed punctate staining, which is enhanced in the presence of LME. Large arrows point to the coincident staining of SKI-1 and WGA. [×900; bar (B′) = 10 μm.]
Enzymatic Activity and Cleavage Specificity of SKI-1.
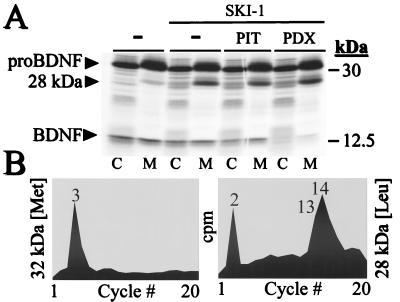
To prove that SKI-1 is a proteolytic enzyme we examined its ability to cleave five different potential precursor substrates, including pro-opiomelanocortin, pro-atrial natriuretic factor, HIV gp160, pro-nerve growth factor, and proBDNF. Cellular coexpression of vv:SKI-1 with the vaccinia virus recombinants of each of the above precursors revealed that only proBDNF was cleaved intracellularly by SKI-1. Thus, upon expression of vv:BDNF alone in COS-7 cells we observed a partial processing of proBDNF (32 kDa) into the known, major 14-kDa BDNF product (15) and the minor production of a previously observed (ref. 16; S.J.M., N.G.S., and R.A.M., unpublished results) but still undefined 28-kDa product (Fig. 6A). Upon coexpression of proBDNF and SKI-1, a net increase in the level of the secreted 28-kDa BDNF is evident, without significant alteration in the amount of 14-kDa BDNF (Fig. 6A). To examine whether the 28-kDa product results from cleavage at a basic residue or at an alternative site, we first coexpressed proBDNF, SKI-1, and either α1-PIT or α1-PDX, which are inhibitors of thrombin and PC cleavages, respectively (18, 26). The results show that different from α1-PIT, the serpin α1-PDX selectively blocks the production of the 14-kDa BDNF and that neither α1-PIT nor α1-PDX affects the level of the 28-kDa product. This finding shows that α1-PDX effectively inhibits the endogenous furin-like enzyme(s) responsible for the production of the 14-kDa BDNF (15), but does not inhibit the ability of SKI-1 to generate the 28-kDa product. Thus, it is likely that the generation of the 28-kDa BDNF takes place via an alternate cleavage. Incubation of the cells with brefeldin A or the Ca2+ ionophore A23187 revealed that the 28-kDa proBDNF is formed in the ER and that this cleavage is Ca2+-dependent (not shown).
Figure 6.
Processing of proBDNF by SKI-1. (A) COS-7 cells were infected with vv:BDNF and either vv:WT (−) or vv:SKI-1 in the presence of either vv:PIT or vv:PDX. The cells were labeled metabolically with [35S]cysteine/[35S]methionine for 4 h, and the media (M) and cell lysates (C) were immunoprecipitated with a BDNF antiserum before SDS/PAGE analysis. The autoradiogram shows the migration positions of proBDNF (32 kDa), the 28-kDa BDNF produced by SKI-1, and the 14-kDa BDNF. (B) Microsequence analysis of the [35S]Met-labeled 32-kDa proBDNF (maximal scale, 1,000 cpm) and [3H]Leu-labeled 28-kDa BDNF (maximal scale, 250 cpm).
In Fig. 6B, we present the N-terminal microsequence analysis of [35S]Met-labeled 32-kDa proBDNF and [3H]Leu-labeled 28-kDa BDNF. The sequence of the 32-kDa form revealed the presence of an [35S]Met at position 3 (Fig. 6B), which is in agreement with the proposed sequence of human proBDNF (27) resulting from the removal of an 18-aa signal peptide cleaved at GCMLA18↓APMK site. The N-terminal sequence of the 28-kDa product revealed a [3H]Leu at positions 2, 13, and 14 (Fig. 6B). This result demonstrates that the 28-kDa BDNF is generated by a unique cleavage at Thr57 in the sequence RGLT57↓SLADTFEHVIEELL (27).
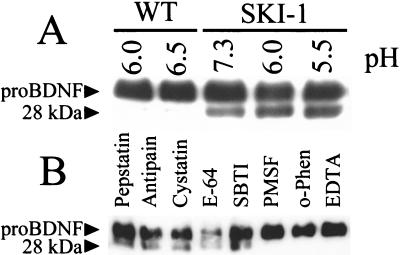
To prove that SKI-1 is directly responsible for the production of the 28-kDa BDNF at the novel Thr-directed cleavage, proBDNF was incubated at various pH values with concentrated medium of vv:SKI-1-infected Schwann cells. A similar preparation obtained from wild-type vaccinia virus-infected cells served as control. The data show that SKI-1 exhibits a wide pH-dependence profile revealing activity at both acidic and neutral pH values from pH 5.5 to 7.3 (Fig. 7A). Analysis of the inhibitory profile of this reaction revealed that metal chelators such as EDTA and o-phenanthroline or a mixture of the serine proteinase inhibitors PMSF and pAPMSF effectively inhibits the processing of proBDNF by SKI-1. The inhibition by EDTA is expected because, like all PCs, SKI-1 is a Ca2+-dependent enzyme. The unexpected inhibition by 5 mM o-phenanthroline may be a result of excess reagent because at 1 mM only 25% inhibition is observed (not shown). All other class-specific proteinase inhibitors (aspartyl-, cysteinyl-, and serine proteases- of the trypsin type) proved to be inactive.
Figure 7.
In vitro processing profile of proBDNF by SKI-1. (A) pH dependence of the processing of proBDNF by SKI-1. (B) Inhibitor profile of the processing of proBDNF to the 28-kDa BDNF by the same SKI-1 preparation as in A. The reaction was performed overnight at 37°C, pH 6.0.
DISCUSSION
This work provides evidence for the existence of a mammalian secretory Ca2+-dependent serine proteinase of the subtilisin/kexin type that selectively cleaves at nonbasic residues. Thus, SKI-1 processes the 32-kDa human proBDNF at an KAGSRGLT↓SL sequence, generating a 28-kDa form, which may have its own biological activity (S.J.M., N.G.S., and R.A.M., unpublished results). Such a cleavage site is close to the consensus site deduced from a large body of work done with the PCs, whereby an (R/K)-(X)n-R↓X-(L/I/V), (where n = 0, 2, 4, or 6) motif is favored by most PCs (1–3, 28). Note that in the SKI-1 site, P1 Arg is replaced by Thr and an aliphatic Leu is present at P2′, an amino acid also favored by PCs (1–3, 28). Several proteins are known to be cleaved after Thr. These include human antiangiogenic platelet factor 4 (ref. 6; QCLCVKTT↓SQ), the neuroendocrine α-endorphin (ref. 4; KSQTPLVT↓LF), and ADAM-10 metalloprotease (ref. 7; LLRKKRTT↓SA).
Interestingly, comparison of the phylogenetically highly conserved sequence of proBDNF revealed an insertion of hydroxylated amino acids (threonine and serine) just after the identified SKI-1 cleavage site of human proBDNF. Thus, in rat and mouse proBDNF, two threonines are inserted (RGLTTT—SL), and in porcine proBDNF, five serines are added (RGLTSSSSS—SL) (27). These observations raise a number of questions: (i) Do these insertions affect the kinetics of proBDNF cleavage by SKI-1? In that context, it was published recently that rat proBDNF is also cleaved into a 28-kDa protein (29). (ii) Does SKI-1 recognize both single and pairs of Thr and Ser and combinations thereof? (iii) Is the presence of a basic residue at P4, P6, or P8 critical for cleavage?
Another question that arises is whether SKI-1 can cleave at residues other than Thr. In that context, after submission of this manuscript, Sakai et al. demonstrated that the sequence of hamster S1p responsible for the site 1 cleavage of SREBPs is almost identical to the presently reported human, mouse, and rat SKI-1 (30). In this model, within the lumen of the ER, S1p cleaves SREBP-2 at an RSVL↓SF sequence, where Arg at P4 is very critical, whereas the P1 Leu could be replaced by a number of other amino acids (8). Our in vitro data show that sSKI-1 does not cleave small fluorogenic substrates of sequence RGLT-MCA, RGLTTT-MCA, or RSVL-MCA (MCA is 4-methylcoumaryl-1-amide), suggesting that it has an extended substrate-specificity pocket. In agreement, preliminary data show that SKI-1 specifically cleaves at neutral pH a 27-mer synthetic peptide of sequence GGAHDSDQHPHSGSGRSVL↓SFESGSGG, representing the luminal amino acids 504–530 of human SREBP-2 (8) (B.B.T. and N.G.S., unpublished data). We have shown that this synthetic peptide is efficiently processed by SKI-1 in vitro, paving the way for refined kinetic analyses.
Biosynthetic analysis of the zymogen processing of proSKI-1 demonstrated an ER-associated removal of the prosegment (Fig. 4). Furthermore, analysis of the 35SO4-labeled SKI-1 demonstrated the presence of only sulfated 106- and 98-kDa forms but not that of either the 148 proSKI-1 or the 120-kDa SKI-1a forms (not shown). Because sulfation occurs in the trans-Golgi network, this confirms that the removal of the prosegment occurs in the ER. As with furin and PC5-B (1–3, 24) the membrane-bound 106-kDa SKI-1 is transformed into a soluble 98-kDa form. The secreted 98-kDa sSKI-1 is enzymatically active because it processes proBDNF in vitro (Fig. 7). Attempts to sequence the SDS/PAGE-purified [3H]Leu- and Val-labeled 148- and 98-kDa forms resulted in ambiguous results, suggesting that SKI-1 is refractory to N-terminal Edman degradation. Presently, we are unable to define the zymogen cleavage site leading to the formation of the 120-kDa SKI-1a and 106-kDa SKI-1b deduced by pulse–chase studies (Fig. 4). Examination of the prosegment sequence (Fig. 1), the species-specific proBDNF motif potentially recognized by SKI-1, the sequence of the luminal portion of SREBP-2 (see above), and the alignment of SKI-1 with other subtilases (22) suggests three possible conserved sites: RASL167↓SLGS, RHSS182↓RRLL, and RRLL186↓RAIP. These predict cleavages at motifs containing a P4 Arg and a P1 either Leu or Ser.
Phylogenetic structural analysis of the predicted amino acid sequence of SKI-1 reveals that this serine proteinase is closer to plant and bacterial subtilases than it is to yeast and mammalian PCs. The 100% conservation of the catalytic domain sequence, although striking and suggestive of an important function, is not far from the 98% similarity between human and rat PC7 (3, 21). The sequence C-terminal to the catalytic domain of SKI-1 is very different from that of any of the known PCs. In fact, although PCs have a typical P-domain critical for the folding of these enzymes (for reviews see refs. 1–3), we did not find the hallmark sequences (3, 31) of the P-domain within the SKI-1 structure. Instead, different from the PCs, we find a conserved growth factor/cytokine receptor motif of which functional importance will need to be addressed, especially because this motif is partly missing in alternatively spliced forms (Fig. 1). Finally, the highly basic nature of the cytosolic tail of SKI-1 (Fig. 1) may be critical for its probable cellular localization within endosomal/lysosomal compartments (Fig. 5), similar to the importance of basic residues for the accumulation of the α-amidation enzyme PAM in endosomal compartments (S. L. Milgram, personal communication).
The wide tissue distribution of SKI-1 mRNA transcripts suggests that this enzyme processes numerous precursors in various tissues. Furthermore, the observed developmental down-regulation of the level of its transcripts also suggests a functional importance during embryonic development. That SKI-1 can cleave C-terminal to Thr, Leu, and, possibly, Ser residues suggests that, like the combination of PCs and carboxypeptidases E and D (32), a specific carboxypeptidase also may be required to trim out the newly exposed C-terminal hydroxylated or Leu residues.
SKI-1 is closest to the pyrolysin branch of the six-membered family of subtilisin-like proteinases (22) and we believe is the first known mammalian subtilase cleaving at sites other than basic amino acids. That other eukaryotic subtilases exist is supported by a recent report on the structure of a soluble subtilisin-like enzyme called PfSUB-1 found in Plasmodium falciparum (33) and exhibiting a 29% sequence identity to SKI-1. This enzyme, which is closest to the subtilisin branch of subtilases (22), localizes to granular-like compartments and presumably cleaves at a Leu↓Asn bond (33). Therefore, because only mammalian members of the kexin (PCs) and pyrolysin (SKI-1) subfamilies have been identified, could it be that the other four subtilase subfamilies (22) have their mammalian counterparts?
Acknowledgments
We thank Dr. N. Nomura (Kazusa DNA Research Institute, Chiba, Japan) for the generous gift of human SKI-1 cDNA. We acknowledge the technical help of A. Lemieux, O. Théberge, J. Rochemont, A. Chen, M. V. Seidah, A. V. Seidah, C. Lefebvre, and S. Emond. We thank A. Prat, J. Cromlish, and M. Mbikay for reading the manuscript and G. Langsley (Pasteur Institute) for discussions. This work was supported in part by group Grant GR11474 (to N.G.S., M.C., and M.M.) and Grants MT-14766 (to C.L.) and MT-14216 (to R.A.M.) from the Medical Research Council of Canada and from the Protein Engineering Network Centres of Excellence (PENCE). S.J.M. was supported by a studentship from the Iranian Ministry of Culture and Higher Education. The Clinical Research Institute of Montreal is affiliated with the University of Montreal and PENCE.
ABBREVIATIONS
- SKI-1
subtilisin/kexin-isozyme-1
- PC
proprotein convertase
- ER
endoplasmic reticulum
- BDNF
brain-derived neurotrophic factor
- SREBP
sterol regulatory element-binding protein
- RT-PCR
reverse transcriptase–PCR
- vv
vaccinia virus
- PDX
α1-antitrypsin Portland
- WGA
wheat germ agglutinin
- PMSF
phenylmethylsulfonyl fluoride
Footnotes
References
- 1.Seidah N G, Day R, Marcinkiewicz M, Chrétien M. Ann N Y Acad Sci. 1998;839:9–24. doi: 10.1111/j.1749-6632.1998.tb10727.x. [DOI] [PubMed] [Google Scholar]
- 2.Steiner D F. Curr Opin Chem Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 3.Seidah N G, Mbikay M, Marcinkiewicz M, Chrétien M. In: Proteolytic and Cellular Mechanisms in Prohormone and Neuropeptide Precursor Processing. Hook V Y H, editor. Georgetown, TX: Landes; 1998. pp. 49–76. [Google Scholar]
- 4.Ling N, Burgus R, Guillemin R. Proc Natl Acad Sci USA. 1976;73:3042–3046. doi: 10.1073/pnas.73.11.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burbach J P H, Seidah N G, Chrétien M. Eur J Biochem. 1986;156:137–142. doi: 10.1111/j.1432-1033.1986.tb09558.x. [DOI] [PubMed] [Google Scholar]
- 6.Gupta S K, Hassel T, Singh J P. Proc Natl Acad Sci USA. 1995;92:7799–7803. doi: 10.1073/pnas.92.17.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosendahl M S, Ko S C, Long D L, Brewer M T, Rosenzweig B, Hedl E, Anderson L, Pyle S M, Moreland J, Meyers M A, et al. J Biol Chem. 1997;272:24588–24593. doi: 10.1074/jbc.272.39.24588. [DOI] [PubMed] [Google Scholar]
- 8.Duncan E A, Brown M S, Goldstein J L, Sakai J. J Biol Chem. 1997;272:12778–12785. doi: 10.1074/jbc.272.19.12778. [DOI] [PubMed] [Google Scholar]
- 9.Checler F. J Neurochem. 1995;65:1431–1444. doi: 10.1046/j.1471-4159.1995.65041431.x. [DOI] [PubMed] [Google Scholar]
- 10.Seidah N G. Methods Neurosci. 1995;23:3–15. [Google Scholar]
- 11.Nagase T, Miyajima N, Tanaka A, Sazuka T, Seki N, Sato S, Tabata S, Ishikawa K-I, Kawarabayasi Y, Kotani H, Nomura N. DNA Res. 1995;2:37–43. doi: 10.1093/dnares/2.1.37. [DOI] [PubMed] [Google Scholar]
- 12.Seidah, N. G., Benjannet, S., Hamelin, J., Mamarbachi, A. M., Basak, A., Marcinkiewicz, J., Mbikay, M., Chrétien, M. & Marcinkiewicz, M. (1999) Ann. N. Y. Acad. Sci., in press. [DOI] [PubMed]
- 13.Edwards J B D M, Delort J, Mallet J. Nucleic Acids Res. 1991;19:5227–5232. doi: 10.1093/nar/19.19.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lusson J, Vieau D, Hamelin J, Day R, Chrétien M, Seidah N G. Proc Natl Acad Sci USA. 1993;90:6691–6695. doi: 10.1073/pnas.90.14.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seidah N G, Benjannet S, Pareek S, Chrétien M, Murphy R A. FEBS Lett. 1996;379:247–250. doi: 10.1016/0014-5793(95)01520-5. [DOI] [PubMed] [Google Scholar]
- 16.Marcinkiewicz M, Savaria D, Marcinkiewicz J. Mol Brain Res. 1998;59:229–246. doi: 10.1016/s0169-328x(98)00141-7. [DOI] [PubMed] [Google Scholar]
- 17.Lippincott-Schwartz J, Youan L C, Bonifacino J S, Klausner R D. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson E D, Thomas L, Hayflick J S, Thomas G. J Biol Chem. 1993;268:24887–24891. [PubMed] [Google Scholar]
- 19.Yan Q, Rosenfeld R D, Matheson C R, Hawkins N, Lopez O T, Bennett L, Welcher A A. Neuroscience. 1997;78:431–448. doi: 10.1016/s0306-4522(96)00613-6. [DOI] [PubMed] [Google Scholar]
- 20.Paquet L, Bergeron F, Seidah N G, Chrétien M, Mbikay M, Lazure C. J Biol Chem. 1994;269:19279–19285. [PubMed] [Google Scholar]
- 21.Seidah N G, Hamelin J, Mamarbachi M, Dong W, Tadros H, Mbikay M, Chrétien M, Day R. Proc Natl Acad Sci USA. 1996;93:3388–3393. doi: 10.1073/pnas.93.8.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siezen R J, Leunissen J A M. Protein Sci. 1997;6:501–523. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seidah N G, Day R, Chrétien M. Biochimie. 1994;76:197–209. doi: 10.1016/0300-9084(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 24.de Bie I, Marcinkiewicz M, Malide D, Lazure C, Nakayama K, Bendayan M, Seidah N G. J Cell Biol. 1996;135:1261–1275. doi: 10.1083/jcb.135.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reeves J P, Decker R S, Crie J S, Wildenthal K. Proc Natl Acad Sci USA. 1981;78:4426–4429. doi: 10.1073/pnas.78.7.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjannet S, Savaria D, Laslop A, Chrétien M, Marcinkiewicz M, Seidah N G. J Biol Chem. 1997;272:26210–26218. doi: 10.1074/jbc.272.42.26210. [DOI] [PubMed] [Google Scholar]
- 27.Maisonpierre P C, Le Beau M M, Espinosa R, Ip N Y, Belluscio L, de la Monte S M, Squinto S, Furth M E, Yancoupolos G D. Genomics. 1991;10:558–568. doi: 10.1016/0888-7543(91)90436-i. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama K. Biochem J. 1997;327:625–635. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haubensak W, Narz F, Heumann R, Lessmann V. J Cell Sci. 1998;111:1483–1493. doi: 10.1242/jcs.111.11.1483. [DOI] [PubMed] [Google Scholar]
- 30.Sakai J, Rawson R B, Espenshade P J, Cheng D, Seegmiller A C, Goldstein J L, Brown M S. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 31.Lipkind G M, Zhou A, Steiner D F. Proc Natl Acad Sci USA. 1998;95:7310–7315. doi: 10.1073/pnas.95.13.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varlamov O, Fricker L D. J Cell Sci. 1998;111:78–85. doi: 10.1242/jcs.111.7.877. [DOI] [PubMed] [Google Scholar]
- 33.Blackman M J, Fujioka H, Stafford W H L, Sajid M, Clough B, Fleck S L, Aikawa M, Grainger M, Hackett F. J Biol Chem. 1998;273:23398–23409. doi: 10.1074/jbc.273.36.23398. [DOI] [PubMed] [Google Scholar]