Abstract
Altogether, 173 Shiga toxin-producing Escherichia coli (STEC) serotype O157 (n = 111) and non-O157 (n = 62) isolates from 170 subjects were screened by PCR-restriction fragment length polymorphism for eight different stx genes. The results were compiled according to serotypes, phage types of O157, production of Stx toxin and enterohemolysin, and the presence of eae. The stx genes occurred in 11 combinations; the most common were stx2 with stx2c (42%), stx2 alone (21%), and stx1 alone (16%). Of the O157 strains, 64% carried stx2 with stx2c versus 2% of the non-O157 strains (P < 0.001). In the non-O157 strains, the prevailing gene was stx1 (99% versus 1% in O157 strains; P < 0.001). In addition, one strain (O Rough:H4:stx2c) which has not previously been described as associated with hemolytic-uremic syndrome (HUS) was found. Ten stx-positive virulence profiles were responsible for 71% of all STEC infections. Of these profiles, five accounted for 71% of the 21 strains isolated from 20 patients with HUS or thrombotic thrombocytopenic purpura (TTP). The strains having the virulence profile that caused mainly HUS or TTP or bloody diarrhea produced Stx with titers of ≥1:128 (90%) more commonly than did other strains (51%; P < 0.001). These strains were also more commonly enterohemolytic (98% versus 68% for other strains; P < 0.001) and possessed the eae gene (100%) more commonly than did other strains (74%; P < 0.001). A particular virulence profile, O157:H7:PT2:stx2:stx2c:eae:Ehly, was significantly more frequently associated with HUS and bloody diarrhea than were other profiles (P = 0.02) and also caused the deaths of two children. In this study, the risk factors for severe symptoms were an age of <5 years and infection by the strain of O157:H7:PT2 mentioned above.
Shiga toxin-producing Escherichia coli (STEC) cells have emerged as new food-borne pathogens of clinical and public health concern (35). The most epidemic STEC serogroup has been O157, but about 200 other STEC serogroups have beenidentified (http://www.microbionet.com.au/frames/feature/vtec/brief01.html). Shiga toxins (Stx1 and/or Stx2) have a prominent role in the pathogenesis of STEC bacteria (15, 20, 26). The clinical picture of a STEC infection may vary from an asymptomatic state to bloody diarrhea and severe life-threatening complications such as hemolytic-uremic syndrome (HUS) or thrombotic thrombocytopenic purpura (TTP) (35). Children and elderly people have been more susceptible to STEC infections than healthy adults (32). It is estimated that the incidence of HUS varies from 2.0 to 3.0 cases per 100,000 children under 5 years of age (15) and from 0.9 to 1.2 cases per 100,000 in children under 18 years of age (5, 43).
The pathogenesis of STEC infection in humans is not fully understood. It is considered to be multifactorial and dependent on several bacterial virulence factors such as enterohemolysin (Ehly) and the eae gene, in addition to host factors (6, 35). A recent discovery of a quorum-sensing system, mediated by self-produced extracellular factors, may play an important role in the control of the colonization of STEC O157 on human cells (17, 18, 44). The main bacterial properties in the pathogenesis of HUS, however, are considered to be the Stx toxins encoded by the stx genes. It is known that STEC strains frequently carry more than one stx gene, and several variants of these genes (e.g., stx1c, stx1vO111, stx2vha, stx2vhb, stx2vOX392, stx2vOX393, stx2c, stx2d-Ount, stx2d-OX3a, stx2e, stx2ev, stx2f) have been found (1, 16, 27, 30, 37, 41, 42, 46, 48). In the literature, different designations have been used for the same genes, causing confusion. However, recently, Scheutz et al. (40) compiled data on the designations of the stx genes. In the Stx2 toxin family, the amino acid homologies in the A subunit to the mature Stx2 are 93, 99, and 100% for Stx2e, Stx2d (also designated as Stx2vh [40]), and Stx2c, respectively (31). The corresponding percentages in the B subunit are 84, 97, and 97% (31).
Previously obtained data (13, 16, 37) have suggested that not only the main toxin type but also the toxin variant type could be important in determining the probability of developing HUS. For example, humans infected by strains producing Stx2 have developed HUS more frequently than those infected by strains producing Stx1 only (7, 34). Of the stx2 variants, stx2c has been associated with HUS but the risk of developing HUS after infection with STEC of the stx2c genotype has been significantly lower than that after infection with STEC of the stx2 genotype only (13, 16). The strains carrying the stx2d genes encoding VT2d-Ount or VT2d-OX3a have been concluded to be less pathogenic for humans than other Stx2-class-toxin-producing strains (37). stx2d has been associated with various forms of diarrhea but not with HUS (13). Of the other stx variants, stx2e has been associated with edema disease in pigs (30, 46) though it has been detected rarely in human infections (13). stx2f has been isolated in STEC isolates from pigeons, and only a single STEC human isolate harboring a similar stx gene has been found (41).
By combining PCR with restriction fragment length polymorphism (PCR-RFLP) (37), we investigated all the 173 human STEC strains isolated during the period from 1990 to 2000 from Finns with STEC infection to evaluate the prevalence of the stx2d genes for VT2d-Ount (hereafter referred to as stx2d-Ount) or VT2d-OX3a (hereafter stx2d-OX3a) among these strains. We also subtyped all STEC isolates for the possession of the stx1/stx1vO111 (hereafter stx1), stx2, stx2c/stx2vha/stx2vOX393 (hereafter stx2c), stx2vhb, stx2vO111/stx2vOX392 (hereafter stx2vO111 and stx2vOX392), stx2e, and stx2ev genes (1, 27). In addition, the association of these stx genes with the clinical pictures of the subjects and with several other bacterial characteristics (the serotype, the possession of the eae gene, the titers of the Stx toxins, the production of Ehly, and the phage types [PTs] of the serogroup O157 strains) was studied. We also searched for specific virulence profiles potentially associated with the clinical picture of STEC infection.
MATERIALS AND METHODS
Subjects.
The age data and the clinical pictures (HUS or TTP, bloody diarrhea, nonbloody diarrhea, or asymptomatic carriage) of the 170 subjects were indicated on a special form that always accompanied the isolate that was received from the clinical microbiology laboratory, or the diagnosis was inquired from the hospital over the telephone. Also, the relationship between asymptomatic carriers and STEC-infected patients was indicated. The subjects were divided into seven age groups: <1, 1 to 5, 6 to 15, 16 to 25, 26 to 55, 56 to 65, and >65 years old (38).
STEC strains.
One hundred seventy-three STEC strains (O157 isolates, n = 111; non-O157 isolates, n = 62) were isolated from Finns (n = 170) during an 11-year period (1990 to 2000). The pure cultures of the isolates or fecal cultures of the STEC-infected patients were referred to the Finnish routine microbiological laboratories for microbiological verification and detection of the stx1, stx2, and eae genes by PCR. All STEC isolates were also serotyped and their enterohemolytic activity was investigated, and the STEC O157 isolates were phage typed as previously described (12, 25, 39). Certain single characteristics of 161 strains have been reported previously (12, 22, 23, 24, 25, 39). Of the 173 strains, 15 were derived from one major outbreak (36) caused by strains of serotype O157:H7:PT2:stx2:stx2c (39). In addition, 65 strains were associated with small family clusters in 24 families.
Shiga toxin production.
The production of Stx1 and Stx2 by the isolates was determined by using a reversed-passive latex agglutination kit (VTEC-RPLA; Denka Seiken Co., Ltd., Tokyo, Japan) after having been grown and shaken in 5 ml of Casamino Acids-yeast extract broth overnight at 37°C. Of this suspension, 1 ml was shaken with 1 ml of polymyxin B (5,000 U) solution (2) for 1 h at 37°C and was then centrifuged for 30 min at 3,000 rpm (BiofugeA instrument Heraeus, Sepatech, West Germany). The titer of the supernatant was determined in the VTEC-RPLA test according to the manufacturer's instructions up to 1:128. All strains were tested for the production of Stx1 and Stx2. Titers lower than 1:4 were interpreted as negative.
Detection of stx genes by PCR-RFLP.
PCRs for the stx1, stx2, and stx2 variants (1, 27, 37) (Table 1) were executed with minor modifications: boiled bacterial supernatant (1.0 μl) was used as a template, and a final elongation step (10 min at 72°C) was added to each PCR run (39). Amplified DNA fragments of specific sizes were located in electrophoresis gels by UV fluorescence after staining with ethidium bromide. Strain ATCC 43895 (RH 4270) was used as a positive control in the run according to Lin et al. (27) and Bastian et al. (1). Strain LMG 18459 (RH 4872) (the Belgian Coordinated Collections of Microorganisms-Laboratorium voor Microbiologie Universiteit Gent bacterium collection, Universiteit Gent, Ghent, Belgium) was used as a positive control in the run according to Piérard et al. (37). In all runs, strain ATCC 25922 (RH 1484) was used as a negative control. The stx-positive PCR products were restricted with 20 U of four restriction enzymes (New England Biolabs Inc., Beverly, Mass.): a ca. 900-bp fragment with HincII and AccI (1, 27) and a 348-bp fragment with HaeIII and PvuII (37).
TABLE 1.
stx genes expected to be detected by PCR or PCR-RFLP methods used in this study
| Primer pairs for indicated stx genesa | Size (bp) of PCR product | stx gene | Sizes (bp)b of restricted PCR-RFLP products | Reference(s) |
|---|---|---|---|---|
| All (HincII and AccI) | ||||
| LIN (5′ GAACGAAATAATTTATATGT) and LIN (3′ TTTGATTGTTACAGTCAT) | ca. 900 | stx1 | 705, 158, 32; 768, 127 | 1, 27 |
| stx1vO111 | 705, 158, 32; 768, 127 | |||
| stx2 | 555, 262, 62; 544, 351 | |||
| stx2c | 555, 324, 16; 544, 351 | |||
| stx2vha | 555, 324, 16; 544, 351 | |||
| stx2vOX393 | 555, 324, 16; 544, 351 | |||
| stx2vbb | 555, 340; 544, 351 | |||
| stx2vO111 | 880, 15; 544, 351 | |||
| stx2vOX392 | 880, 15; 544, 351 | |||
| stx2e | 555, 340; 900 | |||
| stx2ev | 521, 374; 900 | |||
| stx2d | ||||
| VT2-cm (AAGAAGATATTTGTAGCGG) and VT2-f (TAAACTGCACTTCAGCAAAT) | 256 | stx2d-Ount | —c | 37 |
| stx2d-OX3a | —c | |||
| stx2d (HaeIII and PvuII) | ||||
| VT2-e (AATACATTATGGGAAAGTAATA) and VT2-f (TAAACTGCACTTCAGCAAAT) | 348 | stx2d-Ount | 216, 132; 200, 120 (28)d | 37 |
| stx2d-OX3a | 167, 132 (49)d; 200, 120 (28)d |
Restriction enzymes are given in parentheses after gene designations.
Sizes are grouped according to the order in which the restriction enzymes are given in column 1.
—, detection of stx2d-Ount (256 bp) and stx2d-OX3a (256 bp) by PCR with no restriction enzymes.
This fragment did not resolve or was too small to be clearly visible under the electrophoretic conditions used.
Statistical methods.
Epi-Info 2000 version 1.1.2 was used for Fisher's exact two-tailed test. A P of <0.05 indicated statistical significance.
RESULTS
stx genotypes.
Of the 173 STEC strains studied, 168 (97%) gave stx-positive results in PCR for the HincII and AccI restriction enzymes. Three strains were repeatedly negative in PCR, and two showed very weak positive reactions. The 168 STEC strains that were clearly positive in PCR were divided into 11 groups after HincII and AccI endonuclease restriction (Table 2). Seventy-two (42%) of all 173 strains possessed stx2 in combination with stx2c. The stx2 gene alone was found in 36 strains (21%); the stx1 gene was found alone in 28 strains (16%) and in combination with either stx2 or some stx2 variants (stx2c and stx2vhb) in 10 strains (6%). Of the latter strains, one possessed three stx genes: stx1, stx2, and stx2vhb. Nine strains (5%) were positive for stx2c only, five strains were positive for stx2vhb, and three strains were positive for both stx2 and stx2vhb. Five strains were positive in PCR but were undigestible by the HincII or AccI restriction enzymes (Table 2).
TABLE 2.
stx genes in 173 STEC isolates and toxin production of the strains
| Method and related stx genes | No. (%) of strains [n = 173] | No. of samples with indicated titers of Stx toxin |
|||
|---|---|---|---|---|---|
| ≤1:2 | 1:4-1:16 | 1:32-1:64 | ≥1:128 | ||
| PCR for stx according to Bastian et al. (1) and Lin et al. (27) | |||||
| Positive strains | 168 (97) | 3 | 11 | 47 | 117 |
| Weak positive strainsa | 3 (2) | 0 | 0 | 1 | 2 |
| Negative strainsa | 2 (1) | 1 | 0 | 1 | 0 |
| stx genes detected after restriction (HincII and AccI)b | |||||
| stx1 | 28 (16) | 0 | 1 | 18 | 9 |
| stx1 and stx2 or stx2 variant | 10b (6) | ||||
| Stx1 titers | 0 | 1 | 9 | 0 | |
| Stx2 titersc | 0 | 2 | 2 | 6 | |
| stx2 and stx2c | 72 (42) | 0 | 0 | 0 | 72 |
| stx2 | 36 (21) | 0 | 1 | 13 | 22 |
| stx2c | 9 (5) | 0 | 4 | 3 | 2 |
| stx2vhb | 5 (3) | 1 | 0 | 0 | 4 |
| stx2 and stx2vhb | 3 (2) | 0 | 0 | 1 | 2 |
| Undigestible | 5d (3) | 2 | 2 | 1 | 0 |
| PCR for stx2d according to Piérard et al. (37) | |||||
| Positive strains | 2d (1) | 1 | 1 | 0 | 0 |
| Negative strains | 171 (99) | 3 | 11 | 49 | 119 |
| stx genes detected after restriction (HaeIII and PvuII)e | |||||
| stx2d-Ount | 2d (1) | 1 | 1 | 0 | 0 |
This result was obtained repeatedly.
No stx2e, stx2ev, stx2vO111, or stx2vOX392 genes were detected.
The corresponding stx2 genes were stx2c (titers of 1:4 to 1:16 [two strains], 1:32 to 1:64 [two strains], and ≥1:128 [two strains]), stx2 (titer of ≥1:128 [one strain]), stx2vhb (titers of ≥1:128 [two strains]), and stx2 and stx2vhb (titer of ≥1:128 [one strain]).
Two undigestible strains were positive for stx2d-Ount.
No stx2d-OX3a genes were detected.
In PCR for screening stx2d, only two strains of the 173 were positive (1%). Both of them were confirmed as the stx2d-Ount variant type after HaeIII and PvuII restriction (Table 2). These strains were undigestible by the HincII or AccI restriction enzymes in the PCR according to Lin et al. (27) and Bastian et al. (1).
Stx production.
Of the 173 strains, 169 (98%) produced Stx1 or Stx2 or both (Table 2). The majority of the strains (119 of 173 [69%]) produced Stx with high titers (≥1:128) not depending on their stx type. However, of the 168 PCR-positive strains, 98 (87%) of the 113 strains possessing the stx2 gene with or without any other genes produced Stx with titers of ≥1:128 whereas the strains without the stx2 gene did not (24 of 55 strains [44%]; P < 0.001). Lower titers (<1:128) were observed especially in the strains possessing stx2c (7 of 9 [78%]) and in the strains possessing stx1 with or without other genes (29 of 38 strains [76%]). Of the strains producing Stx1 only, 32% (9 of 28) produced the Stx toxin with high titers (≥1:128) compared with 78% (102 of 130) of the strains producing Stx2 only (P < 0.001). Four strains (2%), three positive and one negative for stx in PCR, did not produce Stx. Of the two strains carrying stx2d, one did not produce detectable levels of Stx2 and the Stx titer of the other was low (1:4). However, both strains produced Stx1 with titers of 1:16 although stx1 was not detected by the method used.
Serotypes, presence of eae, and Ehly production.
Of all the strains, 111 (64%) belonged to the O157 serogroup. Within this O group, the most common stx gene combination was stx2 with stx2c (71 strains [64%]) whereas among the 62 non-O157 strains, this gene combination was found in only one strain (2%; P < 0.001) (Table 3). In contrast, among non-O157 strains, the most common stx gene was stx1 (27 of 62 strains [44%]), which, among the 111 O157 strains, was found in one strain only (1%; P < 0.001). The stx2 gene alone was equally common among the O157 (21 of 111 [19%]) and non-O157 (15 of 62 [24%]) strains. Almost all the strains carrying these stx genes also had the eae gene, and they produced Ehly (Table 3). On the other hand, of the nine strains carrying the stx2c gene only, six (67%) strains of the non-O157 group were negative for both eae and Ehly. The two strains carrying stx2d-Ount were both of serogroup non-O157 (O Rough:H− and O76:H19); neither of them carried eae, but both produced Ehly.
TABLE 3.
stx genes in 173 STEC isolates and other characteristics of the strains
| stx gene(s) | Total no. of strains (n = 173) | No. of strains possessing the indicated characteristic (%): |
|||||
|---|---|---|---|---|---|---|---|
| O157 (n = 111) | Non-O157 (n = 62) | eae positive (n = 151) | eae negative (n = 22) | Ehly positive (n = 144) | Ehly negative (n = 29) | ||
| stx1 | 28 (16) | 1a (1) | 27 (44) | 26 (17) | 2 (9) | 27 (19) | 1 (3) |
| stx1 and stx2c | 6 (3) | 5 (5) | 1 (2) | 6 (4) | 0 (0) | 5 (3) | 1 (3) |
| stx1 and stx2 | 1 (1) | 1 (1) | 0 (0) | 1 (1) | 0 (0) | 1 (1) | 0 (0) |
| stx1 and stx2vhb | 2 (1) | 2 (2) | 0 (0) | 2 (1) | 0 (0) | 1 (1) | 1 (3) |
| stx1, stx2, and stx2vhb | 1 (1) | 1 (1) | 0 (0) | 1 (1) | 0 (0) | 0 (0) | 1 (3) |
| stx2 and stx2c | 72 (42) | 71a (64) | 1 (2) | 71 (47) | 1 (5) | 72 (50) | 0 (0) |
| stx2 | 36 (21) | 21 (19) | 15 (24) | 35 (23) | 1 (5) | 28 (19) | 8 (28) |
| stx2c | 9 (5) | 3 (3) | 6 (9) | 3 (2) | 6 (27) | 3 (2) | 6 (21) |
| stx2vhb | 5 (3) | 3 (3) | 2 (3) | 3 (2) | 2 (9) | 3 (2) | 2 (7) |
| stx2 and stx2vhb | 3 (2) | 1 (1) | 2 (3) | 1 (1) | 2 (9) | 2 (1) | 1 (3) |
| stx2d-Ount | 2 (1) | 0 (0) | 2 (3) | 0 (0) | 2 (9) | 2 (1) | 0 (0) |
| Undigestibleb | 3 (2) | 0 (0) | 3 (5) | 0 (0) | 3 (14) | 0 (0) | 3 (10) |
| Weak positive strainsc in PCR for stxb | 3 (2) | 1 (1) | 2 (3) | 1 (1) | 2 (9) | 0 (0) | 3 (10) |
| Negative strainsd in PCR for stxb | 2 (1) | 1 (1) | 1 (2) | 1 (1) | 1 (5) | 0 (0) | 2 (7) |
PTs.
Distribution of the stx genes by PTs among the strains of the O157 serogroup showed that 55 of 59 (93%) of the PT2 strains and all 10 PT49 strains had the stx2 and stx2c genes (Table 4). The PT4 strains had stx2 only (7 of 13 [54%]) almost as often as stx2 with stx2c (5 of 13 [38%]), whereas the PT8 strains had most commonly stx1 (8 of 9 [89%]) with the stx2 genes (stx2c [five strains], stx2vhb [two strains], and stx2 with stx2vhb [one strain]). The nine strains belonging to the reacts- but-does-not-conform phenotype group had a variable set of the stx genes.
TABLE 4.
stx genes among sorbitol-negative and sorbitol-positive STEC O157 strains belonging to different PTs
| PT (n) | stx gene(s) | Sorbitol fermentation of strains (n = 111) |
Total no. of strains (%) | |
|---|---|---|---|---|
| Negative (n = 103) | Positive (n = 8) | |||
| PT2 (59) | stx2 and stx2c | 55 | 0 | 55 (93) |
| stx2 | 4 | 0 | 4 (7) | |
| PT4 (13) | stx2 and stx2c | 5 | 0 | 5 (38) |
| stx2 | 7 | 0 | 7 (54) | |
| stx2 and stx2vhb | 1 | 0 | 1 (8) | |
| PT8 (9) | stx1 and stx2 varianta | 8 | 0 | 8 (89) |
| stx2c | 1 | 0 | 1 (11) | |
| PT14 (1) | stx2 | 1 | 0 | 1 (100) |
| PT21/28 (1) | stx1 and stx2 | 1 | 0 | 1 (100) |
| PT34 (1) | stx1 | 1 | 0 | 1 (100) |
| PT49 (10) | stx2 and stx2c | 10 | 0 | 10 (100) |
| PT50 (2) | stx2 | 2 | 0 | 2 (100) |
| PT88 (6) | stx2 | 0 | 4 | 4 (67) |
| Weak positiveb | 0 | 1 | 1 (17) | |
| Negativeb | 0 | 1 | 1 (17) | |
| RDNCc (9) | stx2 | 1 | 2 | 3 (33) |
| stx2 and stx2c | 1 | 0 | 1 (11) | |
| stx2vhb | 3 | 0 | 3 (33) | |
| stx2c | 2 | 0 | 2 (22) | |
Association of stx with clinical symptoms.
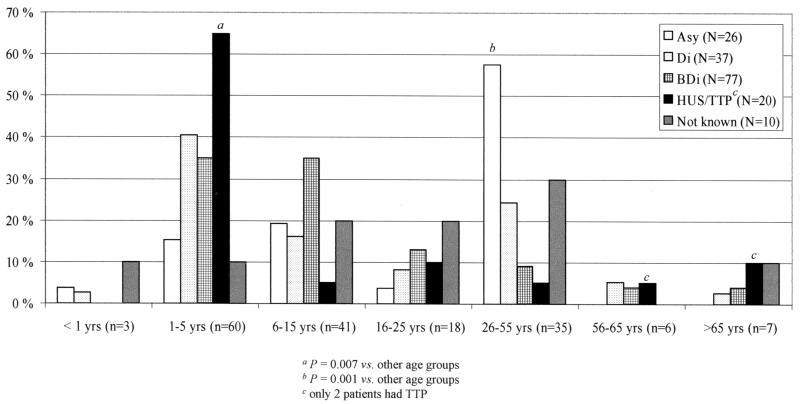
The clinical diagnosis was available for 160 (94%) of the 170 patients (Fig. 1). Of them, three patients were simultaneously infected by two different STEC strains (O157 and non-O157 strains). HUS or TTP was diagnosed in 20 (12%) patients, bloody diarrhea occurred in 77 (45%) patients, nonbloody diarrhea occurred in 37 (22%) patients, and asymptomatic carriage was determined in 26 (16%) subjects. Twenty-four subjects were family members of the children with HUS (11 subjects), bloody diarrhea (6 subjects), nonbloody diarrhea (6 subjects), or an unknown clinical picture (1 subject). However, of the 11 patients with HUS, a STEC strain could be isolated from four of the subjects. Among the asymptomatic subjects, two findings were connected to enhanced screening of travelers (22). Of the 20 patients with HUS, most were 1- to 5-year-old children (13 of 20 subjects [65%] versus 47 of 150 [31%] for other clinical groups; P = 0.007), and of the asymptomatic carriers, most were 26- to 55-year-old subjects (15 of 26 subjects [58%] versus 20 of 144 [14%] for other clinical groups; P < 0.001).
FIG. 1.
Percentage of STEC-infected subjects in different age groups according to the clinical picture of the infection (asymptomatic carriage [Asy], nonbloody diarrhea [Di], bloody diarrhea [BDi], HUS or TTP, or clinical picture not known).
In the strains found in subjects with HUS or TTP, bloody diarrhea, nonbloody diarrhea, or asymptomatic carriage, the most common stx genes were stx2 with stx2c (detected in 50, 53, 30, and 35% of patients, respectively) and stx2 only (detected in 25, 14, 22, and 31% of patients, respectively) (Table 5). Of the nine asymptomatic persons infected by STEC harboring stx2 and stx2c, all were associated with a patient with HUS, bloody diarrhea, or nonbloody diarrhea (two, three, and four subjects, respectively). Five were 16- to 55-year-old family members of the STEC-infected patients and four were <1- to 15 year-old siblings. Of the eight asymptomatic carriers from whom a STEC strain possessing stx2 was isolated, six were connected to patients with HUS or to a patient with nonbloody diarrhea (one subject). One infection was sporadic. Of these eight subjects, five were 16- to 55-year-old adults and three were siblings under 10 years old.
TABLE 5.
Distribution of different stx genes in STEC strains according to the clinical pictures of the 160 patients for whom clinical data were available
| stx gene(s) (n) | No. of patients with symptom or diagnosis (%): |
||||
|---|---|---|---|---|---|
| HUS or TTPf (n = 20)a | Bloody diarrhea (n = 77)b | Nonbloody diarrhea (n = 37) | Asymptomatic (n = 26)c | Not known (n = 10) | |
| stx2 only (34) | 5a (25) | 11b (14) | 8 (22) | 8 (31) | 2 (20) |
| stx2 and stx2c (72) | 10 (50) | 41 (53) | 11 (30) | 9 (35) | 1 (10) |
| stx2c (9) | 3 (15) | 2 (3) | 1 (3) | 2 (8) | 1 (10) |
| Undigestible (2) | 1f (5) | 0 (0) | 0 (0) | 1c (4) | 0 (0) |
| PCR negativee (2) | 1f (5) | 1b (1) | 0 (0) | 0 (0) | 0 (0) |
| stx2 and stx2vhb (3) | 0 (0) | 1 (1) | 2 (5) | 0 (0) | 0 (0) |
| stx2 or stx2 variant and stx1 (10) | 0 (0) | 5 (6) | 3 (8) | 1 (4) | 1 (10) |
| stx2vhb (5) | 0 (0) | 3 (4) | 2 (5) | 0 (0) | 0 (0) |
| stx2d-Ount (2) | 0 (0) | 1 (1) | 1 (3) | 0 (0) | 0 (0) |
| stx1 (28) | 0 (0) | 12 (16) | 7 (19) | 4 (15) | 5 (50) |
| Weak positived (3) | 0 (0) | 0 (0) | 2 (5) | 1 (4) | 0 (0) |
One patient had a simultaneous infection with O157:H−:PT88:stx2 and O145:H28:stx2 strains (the number of isolates is 21 when including the O145:H28:stx2 strain).
One patient had a simultaneous infection with O157:H−:PT88 lacking stx and O145:H28:stx2 strains (the number of isolates is 78 when including the O145:H28/H−:stx2 strain).
One patient had a consecutive infection with O102:H7:undigestible and O Rough:H18:undigestible (the number of isolates is 27 when including the O Rough:H18:undigestible strain).
Two patients had TTP.
In addition, among the strains associated with HUS or TTP, there were three (15%) strains possessing stx2c alone, an undigestible strain, and a PCR-negative strain. None of the 38 stx1-positive strains (28 stx1 only, 10 with stx2 or the stx2 variant) were associated with HUS or TTP, although they were the third most common genes of all the STEC strains and were equally common in all other clinical groups. The only strain possessing three distinct stx genes (stx1, stx2, and stx2vhb) was isolated from a patient with bloody diarrhea.
The strains of 10 different stx-positive virulence profiles (P1 through P10) were responsible for the majority (120 of 170 [71%]) of all STEC infections (Table 6). The strains belonging to five virulence profiles (P1 through P5) accounted for 71% (15 of 21) of all the strains found in patients with HUS or TTP but only 4% (6 of 152) of all remaining strains associated with HUS or TTP (P = 0.03). Of the strains associated with HUS or TTP, nine (43%) were of the non-O157 serogroup: three strains of O145:H28/H−:stx2 and one strain each of OX174:H21:stx2c, O Rough:H4:stx2c, O101:H−:stx2, O Rough:H49:stx2c, O107:H27 lacking stx, and O2:H29:undigestible. P1 was the most prevalent virulence profile, with 55 strains, and these strains more often caused HUS and bloody diarrhea (39 of 55 [71%]) than other clinical symptoms (16 of 55 [29%]). This association with HUS and bloody diarrhea (71%) was statistically significant compared with that for all other strains (60 of 118 strains [51%]; P = 0.02). The P1 virulence profile also caused the deaths of two children under 5 years of age. The strains belonging to profiles P1 through P10 (95 of 120 strains [79%]) produced Stx toxin with titers of ≥1:128 more commonly than did other strains (24 of 53 strains [45%]; P < 0.001). These strains were also enterohemolytic (112 of 120 [93%] versus 32 of 53 [60%]; P < 0.001) and possessed the eae gene more often (119 of 120 [99%] versus 32 of 53 [60%]; P < 0.001) than the other strains. Three patients, one with HUS, one with bloody diarrhea, and one who was an asymptomatic carrier, had infections caused simultaneously by two strains belonging to the different virulence profiles (of P2 and P3, of P2 and other O157 groups, or of other non-O157 groups, correspondingly).
TABLE 6.
Most common virulence profiles of STEC strains and their distribution by the clinical pictures of 170 subjects
| Virulence profile | Phenotype and genotype | Total no. of subjects (n = 170a) | No. of subjects with: |
Other bacterial characteristics |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HUS or TTP (n = 20b) | Bloody diarrhea (n = 77c) | Nonbloody diarrhea (n = 37) | Asymptomatic (n = 26d) | Not known (n = 10) | eae positive | Ehly positive | Stx titer of ≥128 | |||
| P1 | O157:H7:PT2:stx2:stx2c | 55 | 9e | 30 | 8 | 7 | 1 | 55 | 55 | 55 |
| P2 | O145:H28/H−:stx2 | 7 | 3b | 3c | 0 | 1 | 0 | 7 | 6 | 3 |
| P3 | O157:H7/H−:PT88:stx2 | 4 | 1b | 0 | 0 | 1 | 2 | 4 | 0 | 0 |
| P4 | O157:H7:PT49:stx2:stx2c | 10 | 1 | 5 | 2 | 2 | 0 | 10 | 10 | 10 |
| P5 | O157:H7:PT2:stx2 | 4 | 1 | 3 | 0 | 0 | 0 | 4 | 4 | 4 |
| P6 | O103:H2/H−:stx1 | 13 | 0 | 5 | 6 | 1 | 1 | 12 | 13 | 5 |
| P7 | O157:H7/H−:PT8:stx1 and stx2 or stx2 variantf | 8 | 0 | 4 | 2 | 1 | 1 | 8 | 6 | 5 |
| P8 | O157:H7:PT4:stx2 | 7 | 0 | 3 | 3 | 1 | 0 | 7 | 7 | 7 |
| P9 | O26:H11/H−:stx1 | 7 | 0 | 4 | 0 | 2 | 1 | 7 | 6 | 1 |
| P10 | O157:H7:PT4:stx2:stx2c | 5 | 0 | 4 | 1 | 0 | 0 | 5 | 5 | 5 |
| Other O157 | 18 | 0 | 5c | 10 | 1 | 2 | 18 | 14 | 13 | |
| Other non-O157 | 35 | 6g | 12h | 5i | 10d,j | 2k | 14 | 18 | 11 | |
Three patients had infections with STEC O157 and non-O157 strains (the number of isolates is then 173).
One patient had a simultaneous infection with O157:H−:PT88:stx2 and O145:H28:stx2 (the number of isolates is then 21).
One patient had a simultaneous infection with O157:H−:PT88 lacking stx and O145:H28:stx2 (the number of isolates is then 78).
One patient had consecutive infection with O102:H7:undigestible and O Rough:H18:undigestible (the number of isolates is then 27).
Two patients in the age group of 1 to 5 years died.
stx2 variants were stx2c (five strains), stx2vhb (two strains), and stx2 and stx2vhb (one strain).
The corresponding virulence profiles were (one strain each) OX174:H21:stx2c, O Rough:H4:stx2c, O101:H−:stx2, O Rough:H49:stx2c, O107:H27 lackin g stx, and O2:H29:undigestible.
The corresponding virulence profiles were (one strain each) O156:H25:stx1, O15:H−:stx1, OX181:H49:stx2:stx2c, OX174:H21:stx2vhb, OX174:H2:stx2vhb, O165:H25:stx2, O103:H2:stx2, O91:H40:stx1:stx2c, O Rough:H−:stx1, O Rough:H−:stx2d-Ount, O8:H9:stx2vhb, and O20:H7:stx2c.
The corresponding virulence profiles were (one strain each) O76:H19:stx2d-Ount, O15:H−:stx2, O91:H21:stx2:stx2vhb, O111:H8:stx1 , ONT:H33:weak positive in PCR.
The corresponding virulence profiles were O Rough:H2:stx2 (one strain), O15:H−:stx1 (one strain), O103:H2:stx2 (two strains), O Rough:H21:weak positive in PCR (one strain), O Rough:H4:stx2 (two strains), O116:H21:stx2 (one strain), O102:H7:undigestible (one strain), and O Rough:H18:undigestible (one strain).
The corresponding virulence profiles were O43:H2:stx1 and O Rough:H−:stx1.
DISCUSSION
To date, only a few studies have described the stx genes and their variants in STEC strains in relation to the symptoms of STEC-infected patients and the relative contributions of the various forms of Stx toxin to pathogenesis are not fully known (13, 14, 37). In a study concerning Stx production as a single microbial factor, the most pathogenic strains for humans have been found to produce Stx2 only (9, 34). The Stx2 toxin has been described as being 1,000 times more cytotoxic than Stx1 towards human renal microvascular endothelial cells, the putative target of Shiga toxins in the development of HUS (28). The STEC strains isolated from HUS patients have produced mostly Stx2 and/or Stx2c (19), but the risk of developing HUS after infection with STEC harboring stx2 alone has been higher than that after infection with any of the strains harboring stx2c alone (13). However, there are numerous variants of Stx2, differing by only 1 or 2 amino acids in either the A or B subunit (35) and the similarity of the sequences of different Stx2 toxins may vary from 80 to 99% (16). This may affect their catalytic activity or receptor binding, and pathogenicity. The ability of STEC bacteria to produce the Stx toxin only, however, is suspected to be insufficient for an organism to cause disease without appropriate complementary virulence factors (26, 45). The present situation has created a need for studies concerning the simultaneous data of several microbial characteristics combined with the data of the clinical pictures of the patients.
In our study, 111 STEC O157 and 62 non-O157 isolates from 170 Finns with STEC infection were investigated for the possession of various genes by PCR-RFLP. Also, data on other bacterial characteristics, namely, serotype, PT of O157, eae, and Ehly and Stx production, were included. In order to search for the virulence profiles among the strains, the bacterial data were combined and their potential correlation with the clinical diagnosis (HUS, bloody diarrhea, nonbloody diarrhea, or asymptomatic infection) of the subjects in different age groups was investigated. In the strains studied, stx genes occurred in 11 different combinations. In addition, 10 virulence profiles accounted for 71% of all 173 STEC strains isolated in Finland from humans since 1990. Moreover, a STEC serotype (O Rough:H4:stx2c) which had never before been described as being associated with HUS was found.
In PCR-RFLP according to Lin et al. and Bastian et al. (1, 27), the HincII and AccI enzymes were expected to produce fragments of variant-specific size for the stx genes. However, some of our strains were negative, showed weak positive results, or were undigestible. The PCR products which the HincII and AccI enzymes were unable to cut possibly lacked the required specific site for the restriction enzymes to cut the DNA. Either these undigestible stx genes may have been rare stx variants which remained undetectable by the HincII and AccI enzymes or the strains may have changed during storage and cultivation.
A considerable share (21 or 42%) of the strains carried stx2 only or stx2 with stx2c. Also, in Belgium and Germany, stx2 alone and stx2 with either stx2vha or stx2c have been prevalent (7, 13, 37). In our strains, the third most common gene was stx1, which was present alone in 28 strains and in combination with stx2, stx2c, or stx2vhb in 10 strains. In Germany, of the 212 human isolates harboring stx1, almost half also carried stx2, stx2c, or stx2d (48). Of our strains, only two (1%) were of the stx2d-Ount variant type. In Belgium and Germany, the stx2d gene has been more prevalent and has been found in 7% of 359 human or animal isolates (37) and in 4% of 626 human isolates (13). These data indicate similar distributions of the stx genes in different European countries.
By the VTEC-RPLA assay, the strains carrying the stx2 gene with or without any other genes produced Stx significantly more than did the strains without the stx2 gene. Lower titers were observed especially within the strains possessing stx1, stx2c, or stx2d. The VTEC -RPLA assay has previously been found to be a reliable method for the detection of Stx1, Stx2, and Stx2c but not for the Stx2e porcine or Stx2d-Ount variant toxins (2, 3, 21). Our results might indicate a weak capability of the gene to express the Stx1 toxin or a putative carriage of the stx1c gene and thus a possible weak ability to detect the Stx1c toxin by the VTEC-RPLA, as suggested by Zhang et al. (48). Of our two strains carrying stx2d-Ount, both produced Stx1. However, these strains were negative for stx1 although they were positive in PCR (12) with primers described by Olsvik and Strockbine (33). Similarly, four strains (O107:H27 lacking stx, O157:H− lacking stx, O157:H7:stx1, and O43:H2:stx1) negative for any stx2 gene on the basis of the method of Lin et al. (27) and Bastian et al. (1) had previously been positive for stx2 either as a sole gene (O107:H27 lacking stx and O157:H− lacking stx) or in combination with stx1 (O157:H7:stx1 and O43:H2:stx1) (12, 25, 39). In addition, one strain (O91:H40) possessed stx1 with stx2c in this study, although previously it has been shown to possess only stx2 and to produce Stx2 toxin only (12, 25). These results clearly demonstrate the differences among different protocols in the detection of the stx genes.
The comparison of the distribution of the stx genes in strains of the O157 and non-O157 serogroups showed that stx2 with stx2c was statistically significantly associated with strains of the O157 serogroup. Also in Belgium, stx2 with stx2vha was prevalent among the human STEC O157 isolates, unlike stx1, which was more prevalent in other O serogroups (37). Also among our non-O157 strains, the significant association with stx1 was seen. This gene was most common in strains of serotypes O26:H11/H− and O103:H2/H−. In France, the serotype O103:H2 has commonly possessed stx1 (29).
stx2 with the stx2c gene was found in strains associated with all symptoms but mostly in strains associated with bloody diarrhea or HUS. In contrast, none of the strains carrying stx1 alone or in combination with another gene were associated with HUS or TTP. However, in France, strains of the serotype O103:H2 carrying stx1 have been found in HUS patients (29). Also, in Germany, a child infected by STEC O118:H− positive for stx1 suffered from HUS (47). Neither of our stx2d-Ount-infected patients had HUS or TTP; this supports other studies describing the presence of the stx2d gene in isolates from patients with symptoms milder than HUS or TTP (13, 35, 47, 48).
One of our strains isolated from a patient with bloody diarrhea carried three distinct genes simultaneously. This finding is an example that the infection caused by STEC bacteria possessing several stx genes does not always cause a severe clinical picture, such as HUS, as observed previously also by others (14).
Most of the patients with HUS were 1- to 5-year-old children, and most of the asymptomatic carriers were 26- to 55-year-old adults. Thus, our data are in accordance with other studies concerning the age as a risk factor for STEC infection (8, 11, 13, 32).
Three of the 170 subjects, one with HUS, one with bloody diarrhea, and one an asymptomatic carrier, had infections caused by two STEC strains of different serotypes: correspondingly, O157:H−:PT88:stx2 and O145:H28:stx2, O157:H−:PT88 lacking stx and O145:H28:stx2, or O102:H7:undigestible and O Rough:H18:undigestible. Similarly, in the Czech Republic, some HUS patients were infected with two different STEC serotypes: O157:H7 and O5:H−, O157:H7 and O55H?, O157:H7 and O111:H−, O26:H11 and O5:H−, or O111:H− and O1:H− (4). These findings support the possibility of people being infected by several STEC strains simultaneously.
The strains belonging to five virulence profiles (from P1 to P5) accounted for 71% of all strains found in patients with HUS. In addition, of all the strains studied, those carrying stx2 with stx2c mainly belonged to the virulence profile P1 (O157:H7:PT2:stx2:stx2c) and, among them, the majority (71%) were associated with HUS or bloody diarrhea. All these strains possessed eae, were enterohemolytic, and produced Stx2 with high titers. Strains of the virulence profile P1 also caused the deaths of two children under 5 years of age. Our results are also supported by other studies concerning the carriage of stx2 with stx2c (7, 13). Friedrich et al. (13) found that 68 (25%) of 268 STEC isolates from HUS patients harbored stx2 with stx2c, and among all the 626 STEC isolates investigated, these 68 HUS-associated strains accounted for 11% of the cases of HUS infection. Also, in a study by Cornu et al. (7), nine STEC isolates of serogroup O157 and four STEC non-O157 isolates carrying stx2 only or stx2 with stx2vha were identified in 13 cases of HUS.
In our study, seven virulence profiles of STEC non-O157 were associated with HUS or TTP (O145:H28/H−:stx2, OX174:H21:stx2c, O Rough:H4:stx2c, O101:H−:stx2, O Rough:H49:stx2c, O107:H27 lacking stx, and O2:H29:undigestable). In the liter-ature (10; http://www.microbionet.com.au/frames/feature/vtec/brief01.html; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi), serotype O Rough:H4 has never been published as being associated with patients suffering from HUS.
In this study, the risk factor for HUS in STEC-infected subjects was young age (1 to 5 years) and an infection especially by STEC O157:H7:PT2:stx2:stx2c:eae:Ehly but also by O145:H28/H−:stx2:eae:Ehly. Of the 21 strains causing HUS or TTP, almost half (43%) belonged to the non-O157 serogroup. This high prevalence of the non-O157 STEC strains indicates the importance of the detection of all STEC serogroups in clinical diagnostics. PCR and Stx toxin detection assays have proven to be essential methods worldwide (10), and they should be used routinely in all hospital laboratories. However, the development of molecular methods of high capacity for detecting all stx genes should be considered in future studies. In addition, the further characterization of the stx genes can be exploited for predicting the clinical outcome of STEC infection, especially in young children.
Acknowledgments
We thank Sirkku Waarala and Tarja Heiskanen for their excellent technical assistance and Susanna Lukinmaa for her helpful advice on PCR-RFLP methodology.
This study was financially supported by the Finnish Association of Academic Agronomists, the ABS Graduate School (the Finnish Graduate School on Applied Bioscience Bioengineering, Food & Nutrition, Environment), and the Finnish Cultural Foundation.
REFERENCES
- 1.Bastian, S. N., I. Carle, and F. Grimont. 1998. Comparison of 14 PCR systems for the detection and subtyping of stx genes in Shiga-toxin-producing Escherichia coli. Res. Microbiol. 149:457-472. [DOI] [PubMed] [Google Scholar]
- 2.Beutin, L., S. Zimmerman, and K. Gleier. 1996. Rapid detection and isolation of Shiga-like toxin (verocytotoxin)-producing Escherichia coli by direct testing of individual enterohemolytic colonies from washed sheep blood agar plates in the VTEC-RPLA assay. J. Clin. Microbiol. 34:2812-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutin, L., K. Gleier, and S. Zimmermann. 1999. The new VT2 variant, VT2d-Ount, is not detected by the VTEC-RPLA assay. Notizziario dell'instituto Superiore di Sanita. IVC News 12:3. [Google Scholar]
- 4.Bielaszewska, M., J. Janda, K. Blahova, J. Feber, V. Potuznik, and A. Souckova. 1996. Verocytotoxin-producing Escherichia coli in children with hemolytic uremic syndrome in the Czech Republic. Clin. Nephrol. 46:42-44. [PubMed] [Google Scholar]
- 5.Bitzan, M., K. Ludwig, M. Klemt, H. König, J. Büren, and D. E. Müller-Wiefel. 1993. The role of Escherichia coli O157 infections in the classical (enteropathic) haemolytic uraemic syndrome: results of a central European, multicentre study. Epidemiol. Infect. 110:183-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boerling, P., S. A. McEwen, F. Boerling-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornu, G., W. Proesmans, A. Dediste, F. Jacobs, J. Van De Walle, A. Mertens, J. Ramet, and S. Lauwers. 1999. Hemolytic uremic syndrome in Belgium: incidence and association with verocytotoxin-producing Escherichia coli infection. Clin. Microbiol. Infect. 5:16-22. [DOI] [PubMed] [Google Scholar]
- 8.Decludt, B., P. Bouvet, P. Mariani-Kurkdjian, F. Grimont, P. A. D. Grimont, B. Hubert, and C. Loirat. 2000. Haemolytic uraemic syndrome and Shiga toxin-producing Escherichia coli infection in children in France. Epidemiol. Infect. 124:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donohue-Rolfe, A., I. Kondova, S. Oswald, D. Hutto, and S. Tzipori. 2000. Escherichia coli 0157:H7 strains that express Shiga toxin (Stx) 2 alone are more neurotropic for gnotobiotic piglets than are isotypes producing only Stx1 or both Stx1 and Stx2. J. Infect. Dis. 181:1825-1829. [DOI] [PubMed] [Google Scholar]
- 10.Duffy, G., P. Garvey, and D. McDovell (ed.). 2002. Verocytotoxigenic E. coli. Food & Nutrition Press, Inc., Trumbull, Conn.
- 11.Dundas, S., W. T. A. Todd, A. I. Stewart, P. S. Murdoch, A. K. R. Chaudhuri, and S. J. Hutchinson. 2001. The central Scotland Escherichia coli O157:H7 outbreak: risk factors for the hemolytic uremic syndrome and death among hospitalized patients. Clin. Infect. Dis. 33:923-931. [DOI] [PubMed] [Google Scholar]
- 12.Eklund, M., F. Scheutz, and A. Siitonen. 2001. Clinical isolates of non-O157 Shiga toxin-producing Escherichia coli: serotypes, virulence characteristics and molecular profiles of strains of the same serotype. J. Clin. Microbiol. 39:2829-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich, A. W., M. Bielaszewska, W.-L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 14.Fürst, S., J. Scheef, M. Bielaszewska, H. Russmann, H. Schmidt, and H. Karch. 2000. Identification and characterisation of Escherichia coli strains of O157 and non-O157 serogroups containing three distinct Shiga toxin genes. J. Med. Microbiol. 49:383-386. [DOI] [PubMed] [Google Scholar]
- 15.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 16.Ito, H., A. Terai, H. Kurazono, Y. Takeda, and M. Nishibuchi. 1990. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb. Pathog. 8:47-60. [DOI] [PubMed] [Google Scholar]
- 17.Kanamaru, K., K. Kanamaru, I. Tatsuno, T. Tobe, and C. Sasakawa. 2000. Regulation of virulence factors of enterohemorrhagic Escherichia coli O157:H7 by self-produced extracellular factors. Biosci. Biotechnol. Biochem. 64:2508-2511. [DOI] [PubMed] [Google Scholar]
- 18.Kanamaru, K., K. Kanamaru, I. Tatsuno, T. Tobe, and C. Sasakawa. 2000. SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38:805-816. [DOI] [PubMed] [Google Scholar]
- 19.Karch, H. 2001. The role of virulence factors in enterohemorrhagic Escherichia coli (EHEC)-associated hemolytic-uremic syndrome. Semin. Thromb. Hemost. 27:207-213. [DOI] [PubMed] [Google Scholar]
- 20.Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karmali, M. A., M. Petric, and M. Bielaszewska. 1999. Evaluation of a microplate latex agglutination method (Verotox-F assay) for detecting and characterizing verotoxins (Shiga toxins) in Escherichia coli. J. Clin. Microbiol. 37:396-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keskimäki, M., M. Eklund, H. Pesonen, T. Heiskanen, A. Siitonen, et al. 2001. EPEC, EAEC, and STEC in stool specimens: prevalence and molecular epidemiology of isolates. Diagn. Microbiol. Infect. Dis. 40:151-156. [DOI] [PubMed] [Google Scholar]
- 23.Keskimäki, M., R. Ikäheimo, P. Kärkkäinen, F. Scheutz, Y. Ratiner, R. Puohiniemi, and A. Siitonen. 1997. Shiga toxin-producing Escherichia coli serotype OX3:H21 as a cause of hemolytic-uremic syndrome. Clin. Infect. Dis. 24:1278-1279. [DOI] [PubMed] [Google Scholar]
- 24.Keskimäki, M., Y. Ratiner, S. Oinonen, E. Leijala, M. Nurminen, M. Saari, and A. Siitonen. 1999. Haemolytic-uraemic syndrome caused by vero toxin-producing Escherichia coli serotype Rough:K−:H49. Scand. J. Infect. Dis. 31:141-144. [DOI] [PubMed] [Google Scholar]
- 25.Keskimäki, M., M. Saari, T. Heiskanen, and A. Siitonen. 1998. Shiga toxin-producing Escherichia coli in Finland from 1990 through 1997: prevalence and characteristics of isolates. J. Clin. Microbiol. 36:3641-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Law, D. 2000. Virulence factors of Escherichia coli O157 and other Shiga toxin-producing E. coli. J. Appl. Microbiol. 88:729-745. [DOI] [PubMed] [Google Scholar]
- 27.Lin, Z., H. Kurazono, S. Yamasaki, and Y. Takeda. 1993. Detection of various variant verotoxin genes in Escherichia coli by polymerase chain reaction. Microbiol. Immunol. 37:543-548. [DOI] [PubMed] [Google Scholar]
- 28.Louise, C. B., and T. G. O'Brig. 1995. Specific interaction of Escherichia coli O157:H7-derived Shiga-like toxin II with human renal endothelial cells. J. Infect. Dis. 172:1397-1401. [DOI] [PubMed] [Google Scholar]
- 29.Mariani-Kurkdjian, P., E. Denamur, A. Milon, B. Picard, H. Cave, N. Lambert-Zechovsky, C. Loirat, P. Goullet, P. J. Sansonetti, and J. Elion. 1993. Identification of a clone of Escherichia coli O103:H2 as a potential agent of hemolytic-uremic syndrome in France. J. Clin. Microbiol. 31:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marques, L. R. M., J. S. M. Peiris, S. J. Cryz, and A. D. O'Brien. 1987. Escherichia coli strains isolated from pigs with edema disease produce a variant of Shiga-like toxin II. FEMS Microbiol. Lett. 44:33-38. [Google Scholar]
- 31.Melton-Celsa, A. R., and A. D. O'Brien. 1998. Structure, biology, and relative toxicity of Shiga toxin family members for cells and animals, p. 121-128. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, D.C.
- 32.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsvik, Ø., and N. A. Strockbine. 1993. PCR detection of heat-stable, heat-labile and shiga-like toxin genes in Escherichia coli, p. 271-276. In D. H. Persing, T. H. Smith, F. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. ASM Press, Washington, D.C.
- 34.Ostroff, S. M., P. I. Tarr, M. A. Neill, J. H. Lewis, N. Hargrett Bean, and J. M. Kobayashi. 1989. Toxin genotypes and plasmid profiles as determinants of systemic sequale in Escherichia coli O157:H7 infections. J. Infect. Dis. 160:994-998. [DOI] [PubMed] [Google Scholar]
- 35.Paton, J., and A. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paunio, M., R. Pebody, M. Keskimäki, M. Kokki, P. Ruutu, S. Oinonen, V. Vuotari, A. Siitonen, E. Lahti, and P. Leinikki. 1999. Swimming associated outbreak of enterohemorrhagic Escherichia coli (EHEC) O157:H7. Epidemiol. Infect. 122:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piérard, D., G. Muyldermans, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piérard, D., N. Crowcroft, S. De Bock, D. Potters, G. Crabbe, F. van Loock, and S. Lauwers. 1999. A case-control study of sporadic infection with O157 and non-O157 verocytotoxin-producing Escherichia coli. Epidemiol. Infect. 122:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saari, M., T. Cheasty, K. Leino, and A. Siitonen. 2001. Phage types and genotypes of Shiga toxin-producing Escherichia coli O157 in Finland. J. Clin. Microbiol. 39:1140-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheutz, F., L. Beutin, D. Piérard, and H. R. Smith. 2001. Appendix: Nomencalture of verocytotoxins, p. 447-452. In G. Duffy, P. Garvey, and D. A. McDowell (ed.), Verocytotoxigenic E. coli. Food & Nutrition Press, Inc., Trumbull, Conn.
- 41.Schmidt, H., J. Scheef, S. Morabito, A. Caprioli, L. H. Wieler, and H. Karch. 2000. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt, C. K., M. L. McKee, and A. D. O' Brien. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strains E32511. Infect. Immun. 59:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siegler, R. L., A. T. Pavia, R. D. Christofferson, and M. K. Milligan. 1994. A 20-year population-based study of postdiarrheal hemolytic uremic syndrome in Utah. Pediatrics 94:35-40. [PubMed] [Google Scholar]
- 44.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarr, P. I., and M. A. Neill. 1996. Perspective: the problem of non-O157:H7 Shiga toxin (verocytotoxin)-producing Escherichia coli. J. Infect. Dis. 174:1136-1139. [DOI] [PubMed] [Google Scholar]
- 46.Weinstein, D. L., M. P. Jackson, J. E. Samuel, R. K. Holmes, and A. D. O'Brien. 1988. Cloning and sequencing of a shiga-like toxin type II variant from an Escherichia coli strain responsible for edema disease of swine. J. Bacteriol. 170:4223-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wieler, L. H., B. Busse, H. Steinruck, L. Beutin, A. Weber, H. Karch, and G. Baljer. 2000. Enterohemorrhagic Escherichia coli (EHEC) strains of serogroup O118 display three distinctive clonal groups of EHEC pathogens. J. Clin. Microbiol. 38:2162-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, W., M. Bielaszewska, T. Kuczius, and H. Karch. 2002. Identification, characterization, and distribution of a Shiga toxin 1 gene variant (stx1c) in Escherichia coli strains isolated from humans. J. Clin. Microbiol. 40:1441-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]