Abstract
Early diagnosis of tuberculosis and screening of other mycobacteria is required for the appropriate management of patients. We have therefore developed a 5′-exonuclease fluorogenic PCR assay in a single-tube balanced heminested format that simultaneously detects Mycobacterium tuberculosis complex (MTC) and members of the Mycobacterium genus (MYC) using the 16S ribosomal DNA target directly on clinical samples. One hundred twenty-seven clinical samples (65 smear negative and 62 smear positive) with a positive culture result from 127 patients were tested, including 40 negative control specimens. The finding of both a positive MTC and probe value and a positive MYC probe value confirmed the presence of MTC or mycobacteria with a 100% positive predictive value. However, a negative value for MTC or MYC did not discount the presence of mycobacteria in the specimen. Interestingly, the addition of the MYC probe allowed the diagnosis of an additional 7% of patients with tuberculosis and rapid screening of nontuberculous mycobacteria (NTM). Thus, over 75% of the patients were diagnosed with mycobacterial disease by PCR. The sensitivity was much higher on smear-positive samples (90.3%) than smear-negative samples (49.2%) and was slightly higher for MTC than NTM samples. With regard to the origin of the sample, MTC pulmonary samples gave better results than others. In conclusion, we believe this test may be useful for the rapid detection of mycobacteria in clinical samples and may be a valuable tool when used together with conventional methods and the clinical data available.
During recent years the prevalence of tuberculosis (TB) and mycobacterial infections has risen. Worldwide, the Mycobacterium tuberculosis complex (MTC) is responsible for more than 2 million deaths and 8 million new cases of disease every year (41). On the other hand, nontuberculous mycobacteria (NTM) are a major cause of opportunistic infections in immunocompromised individuals, such as AIDS patients (11, 25, 32). Consequently, early detection and identification of the type of mycobacteria are required for achieving optimal therapy and correct patient management. Nonetheless, culture-based methods require several weeks for a positive result, and acid-fast bacillus smears are not sensitive. Thus, new alternatives are clearly needed.
Despite their limitations, nucleic acid amplification-based methods allow rapid and sensitive identification of mycobacteria from clinical specimens before culture results. Even though results are sometimes not conclusive, they are useful in orienting medical decisions. Therefore, simultaneous detection and rapid differentiation of MTC from other NTM species provide an added value. Accordingly, several studies have been described (17, 18, 33). The use of multiplex PCR may be indicated for this objective, although it may not always be effective due to the inconveniences of design, competition among targets, and overall sensitivity and specificity. The choice of multiple probes within a single target is preferable. Among the sequences used (the 32-kDa gene, recA, hsp65, rpoB, dnaJ, sodA, and the 16S-23S rRNA internal transcriber spacer) the 16S ribosomal DNA (rDNA) is the most commonly studied (3, 8, 16-18, 31, 38). It has been found in all bacteria but has been found as a single copy in the MTC genome (7). It contains highly conserved Mycobacterium genus (MYC) regions different from those of other bacteria groups and specific hypervariable zones that allow differentiation and identification of most Mycobacterium species.
Recently, the introduction of new fluorescent detection systems, such as Molecular Beacons (39), Sunrise primers (26), Scorpion primers (36), TaqMan probes (9, 34, 40), or LightCycler methodology (35, 37), have simplified routine performance. These systems rely on quenching fluorescence energy transfer among molecules (4), and most have proven to be sensitive, specific, and highly discriminative, enabling automated, multiple simultaneous and real-time detection in a closed-tube format that avoids the disadvantages of ethidium bromide-stained gels or other time-consuming methods. At present, fluorogenic detection methods allow the possibility of less difficult real-time PCR in a closed-tube format for multiple targets. Accordingly, several laboratories have already tested them in the diagnosis (35), quantification (10), or drug resistance testing (28, 37) of mycobacteria.
In the present study we developed a single-tube balanced heminested PCR (12) coupled to a 5′-exonuclease assay by two different fluorescent probes within the 16S rDNA target, allowing simultaneous detection of MYC and MTC in clinical samples.
MATERIALS AND METHODS
Study patients.
One hundred twenty-seven prospectively recruited patients who were human immunodeficiency virus (HIV) seropositive (n = 27) or seronegative (n = 100) and who had TB or mycobacteriosis, diagnosed by positive culture, and were admitted to hospital during the study period were included in the study.
Samples.
Four hundred twenty-nine clinical samples from 127 TB patients were processed for culture and routine procedures. Only the first sample (one per patient) from those patients with a positive culture was selected for PCR procedures. One hundred fourteen samples (89.8%) were of pulmonary origin, mostly sputum, and 13 had other origins.
To check the specificity and sensitivity of primers and fluorogenic probes, aqueous dilutions of several mycobacterial and bacterial strains of clinical origin were included: one M. tuberculosis strain, one M. kansasii strain, one M. avium strain, one M. intracellulare strain, one M. xenopi strain, one M. fortuitum strain, five coagulase-negative staphylococcus strains, two Staphylococcus aureus strains, three viridans group streptococcus strains, two Pseudomonas aeruginosa strains, two Corynebacterium sp. strains, two Escherichia coli strains, one Moraxella catarrhalis strain, one Citrobacter diversus strain, and one Morganella morganii strain. The concentrations tested contained 101, 102, 103, and 104 CFU/ml for mycobacterial strains and 103 and 105 CFU/ml for other bacterial strains.
Forty negative controls were also included: 20 culture- and stain-negative saliva specimens from healthy students with no history of TB and who were negative for TB by skin test and sputum samples from 20 patients with chronic obstructive pulmonary disease admitted because of an acute episode of exacerbation without clinical signs, radiological lesions, or a history of TB.
All series of PCR also included two negative controls from each batch of extraction, one positive control of amplification consisting of MTC DNA and three negative controls of amplification.
Routine sample procedures.
All the clinical samples were processed in the Microbiology Laboratory of the Hospital Clínic (Barcelona, Spain). Nonsterile samples were digested and decontaminated by the standard N-acetyl-l-cysteine-NaOH method before culture (21). The resulting pellet was resuspended in 2 ml of phosphate-buffered saline (140 mM NaCl, 2.6 mM KCl, 10.1 mM Na2HPO4, 1.7 mM KH2PO4 [pH 7.4]). Auramine staining was done, and a semiquantitative grade (scale, 1 to 4) was made (30). Two hundred microliters of the pellet was inoculated in Löwenstein-Jensen slants and incubated at 37°C up to 8 weeks for colony confirmation. In addition, 500 μl of the sediment was cultured in radiometric Bactec 12B medium (Becton Dickinson) at 37°C for 6 weeks, except for blood samples, which were cultured directly by adding 5 ml to a Bactec 13A medium. For skin samples, an additional set of culture media was incubated at 30°C for 6 weeks. A growth index of 200 was considered positive and further confirmed by Ziehl-Neelsen stain. Strains were identified by the morphological appearance on Bactec smears and the AccuProbe (Gen Probe) method (14). When identification was inconclusive, gas chromatography and routine biochemical methods were performed. The remaining pellet was stored at 4°C for immediate DNA extraction or at −20°C until DNA extraction was carried out.
In blood samples white cells were extracted by the Ficoll-Hypaque (1119) gradient, washed, and resuspended in Tris-EDTA buffer (100 mM Tris-HCl, 10 mM EDTA [pH 8.0]).
Cell lysis and DNA extraction.
The selection of a DNA extraction approach compatible with fluorescent reading was especially important in order to avoid undesired fluorescent molecules naturally found in samples or bacteria, which could lead to misidentification (15). Altogether, the choice of the modified Kulski et al. (22) alkali wash and heat lysis method seemed to fulfill the necessary criteria. Briefly, 500 μl of sample was added to 700 μl of alkaline wash solution (0.5 M NaOH, 0.05 M sodium citrate) and incubated for 10 min at room temperature with agitation and thereafter was centrifuged at 13,000 × g for 5 min. The pellet was resuspended in 500 μl of 0.5 M Tris-HCl (pH 8.0) and centrifuged again. This step was done twice. The pellet was resuspended in 100 μl of distilled water, heated to 95°C for 30 min, centrifuged briefly, and frozen at −20°C until PCR processing was carried out. The whole protocol was performed in a single tube, thus reducing the risk of cross-contamination.
Fluorogenic PCR.
A single-tube balanced heminested PCR coupled to a 5′-exonuclease assay was performed.
(i) Previous setup.
The conventional PCR conditions were modified and optimized in previous experiments (A. García G. Tudó, E. Dern, M. Navarro, J. González, and M. T. Jiménez de Anta, Abstr. VIIIth Reunión Grupo Espan. Micobacteriol., abstr. C2-01, 1998) to be compatible with fluorogenic detection. Improvements were directed in three ways: first, to favor the quenching of the intact probes; second, to increase the efficiency of probe hybridization; and finally, to enhance the breaking of probes by the 5′-exonuclease activity. In accordance with these criteria, two different pairs of fluorescent molecules were chosen for labeling the probes, thus reducing the loss of sensitivity that is observed when an additional probe is incorporated into the test (data not shown). The MgCl2 concentration was increased up to 3 mM to achieve a good fluorescent signal. The distance between the fluorogenic probes and upstream primers was reduced to a maximum of 10 residues, achieving a significant increase in the fluorescent signal intensity. The melting temperature of the probes was several degrees higher than that of the upstream primers, as cited in the literature (13, 40) and recommended by manufacturer.
Briefly, primers and probes were chosen from those described in the literature or newly designed with Oligo 4.01 software (National Biosciences, Inc., Plymouth, Minn.). The theoretical specificity of primers and probes was checked by searching for homology with the BLAST service of the GenBank (http://www.ncbi.nlm.nih.gov). In addition, secondary structures of target DNA were taken into account at different PCR stages (http://www.ibc.wustl.edu). Initially, only outer primers were added. After choosing the best pair candidates, different inner primers were tested. The best combination of three primers was selected. The heminested reaction was then balanced by attaching the sequence of the inner primer at the 5′ end of the opposite outer primer, avoiding asymmetric amplification and increasing the yield of the reaction (12). Probe candidates for MTC and MYC were tested separately and then mixed together for use in all the experiments. Optimized conditions and reagents are described below.
(ii) Target.
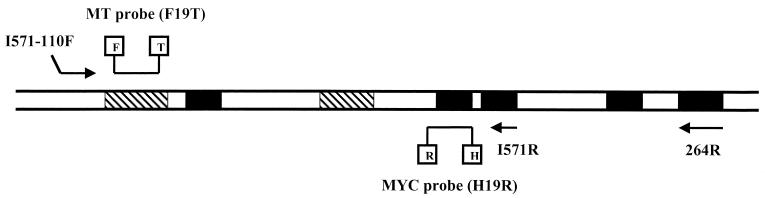
The primers and probes belonged to the 16S rDNA gene called rrn (GenBank accession number X52917). This gene is present in all bacteria and has highly conserved and highly hypervariable sequences (3). Whenever possible, primers were placed in MYC-specific sequences. MYC probe was also placed in a genus-conserved sequence. The MTC probe belonged to an MTC-specific region. A map of the target is shown in Fig. 1.
FIG. 1.
Map of the amplified region from the 16S rDNA gene. Mycobacterium genus-conserved regions are shown in black. Hypervariable regions are striped. Primers and probes are also shown. Abbreviations: F, FAM; T, TAMRA; H, HEX; R, ROX.
(iii) Composition.
Amplification was performed in 0.2-ml thin-walled PCR tubes with a total reaction volume of 53 μl by using a Gene Amp PCR system 2400 thermal cycler (Perkin-Elmer).
All the reagents were added at the beginning of the reaction, therefore not requiring that the tubes be opened to add the nested primer or fluorogenic probes.
Each reaction tube contained 20 μl of sample; 2.5 U of Taq DNA polymerase (Boehringer Mannheim); 0.5 U of uracil-N-glycosylase (Boehringer Mannheim); 200 μl (each) of dATP, dCTP, and dGTP; 600 μl of dUTP (Boehringer Mannheim); 1× final buffer (20 mM Tris-HCl, 50 mM KCl [pH 8.4]); 3 mM MgCl2; 10 nM 264R reverse outer primer (3, 17, 23), 100 nM I571-110F balanced forward primer, 1 μM I571R reverse inner primer (Boehringer Mannheim); 100 nM F19T MTC probe labeled with 6-carboxyfluorescein (FAM) as the reporter (R) and 6-carboxy-tetramethyl-rhodamine (TAMRA) as the quencher (Q); and 100 nM H19R MYC probe labeled with hexachloro-fluorescein (HEX) as the R and 6-carboxy-X-rhodamine (ROX) as the Q (Boehringer Mannheim). Both probes were blocked in their 3′-end with a phosphate in order to avoid primer elongation. Fluorescent probes were handled in the dark. Since working dilutions of fluorogenic probes can be easily degraded by freeze-thawing processes and light exposure, fresh dilutions, consisting of a known MTC DNA dilution were prepared each time we observed a weaker fluorogenic signal other than those expected in positive controls. This way we could minimize false-negative results due to this cause. Sequences of the primers and probes are shown in Table 1.
TABLE 1.
Primers and probes used in the 16S rDNA fluorogenic PCR
| Primer or probe name | Sequencea (5′→3′) | Tmc (°C) | Strand | Positionsb |
|---|---|---|---|---|
| Primers | ||||
| 264R | TGCACACAGGCCACAAGGGA | 61.4 | − | 978-997 |
| I571R | CGCACGCTCACAGTTA | 51.7 | − | 571-586 |
| I571-110F | cgcacgctcacagttaCCTGGGAAACTGGGTCTAAT | 57.3 | + | 110-129 |
| Probes | ||||
| H19R | (HEX)TTTCACGAACAACGCGACA(ROX)AACt(P) | 58.4 | − | 560-539 |
| F19T | (FAM)GACCACGGGATGCATGTCT(TAMRA)TGT(P) | 62.1 | + | 139-160 |
Underlining, labeling site (molecules are indicated in parentheses); boldface type, MYC-conserved sequence; italics, MTC-specific sequence; lowercase, nucleotide not present or different from that in the original target.
Numbering according to GenBank accession number X52917.
Tm, melting temperature.
(iv) Running conditions.
After 15 min at 25°C to allow the uracil-N-glycosylase to work (24), the temperature was raised to 94°C for 5 min to deactivate the enzyme. The first stage of amplification involved 30 cycles of denaturation at 94°C for 45 s, with primer annealing at 65°C for 30 s, and the extension was carried out at 72°C for 1 min. The second stage included 30 cycles of denaturation at 94°C for 20 s, primer annealing at 55°C for 30 s, and extension at 72°C for 30 s, after which there was an additional cycle at 72°C for 5 min and the reaction mixture was maintained at 4°C in a soak file until fluorescence reading.
(v) Product detection.
Fluorescence reading was performed in an LS-50B luminescence spectrometer (Perkin-Elmer). The whole content of the PCR was transferred to a 96-well white microtiter plate designed for use under fluorescence. The F19T MTC probe was excited at 488 nm (maximum FAM absorption), and the emission was read at 518 nm (maximum FAM emission) and 582 nm (maximum TAMRA emission). The H19R MYC probe was excited at 535 nm (maximum HEX absorption), and the emission was read at 556 nm (maximum HEX emission) and 605 nm (maximum ROX emission). The increase in the fluorescence signal was calculated using the formula ΔRQ = RQ+ − RQ− for each probe, where RQ+ is the fluorescence of the reporter (R) divided by the fluorescence of the quencher (Q) in a sample, and RQ− is the equivalent of the negative controls. The threshold ΔRQ, used to establish a baseline for positive samples, was calculated for a 99% confidence level by using the two times the standard deviation of the results from the nontemplate control samples included in each run (TaqMan PCR reagent kit protocol; Perkin-Elmer).
Statistical analysis.
Results were expressed as proportions, and 95% confidence intervals were calculated. Statistical differences between sensitivity and specificity of different probes were tested by the McNemar exact binomial test.
RESULTS
Routine identification of samples.
Sixty-five (51.2%) out of the 127 samples analyzed by PCR were negative for auramine staining, and the remaining 62 were positive. After culture, 115 samples (90.6%) were identified as MTC, and 12 were identified as NTM (eight M. kansasii isolates, three M. avium-M. intracellulare complex isolates, and one M. xenopi isolate).
Analysis of fluorescence results.
Results from MTC and NTM samples are shown in Tables 2 and 3, respectively.
TABLE 2.
Results and sensitivities of fluorescence reading of MTC samplesd
| Staining result (n) | Sample origin (n) | No. of specimens with indicated results |
Sensitivitya (%) |
|||||
|---|---|---|---|---|---|---|---|---|
| MTC identification |
MTC−, MYC+ (n = 8)e | MTC−, MYC− (n = 26)f | ||||||
| MTC+, MYC+ (n = 78) | MTC+, MYC− (n = 3) | SO&St | St | Ov | ||||
| Negative (58) | Pulmonary (48) | 24 | 1 | 7 | 16 | 52.1 | 50.0b | 70.4 |
| Other (10) | 4 | 0 | 0 | 6 | 40.0 | 50.0 | 70.4 | |
| Positive (57) | Pulmonary (55) | 48 | 2 | 1 | 4 | 90.9 | 91.2c | 70.4 |
| Other (2) | 2 | 0 | 0 | 0 | 100.0 | 91.2 | 70.4 | |
Values refer only to identification to the MTC level. Abbreviations: SO&St, sensitivity according sample origin and staining result; St, sensitivity according to staining results; Ov, MT overall sensitivity.
MT staining, negative overall sensitivity.
MT staining, positive overall sensitivity.
For 115 samples with a final identification of MT.
MYC identification.
False negatives.
TABLE 3.
Results and sensitivities of fluorescence reading of NTM samples
| Final identification (n) | Staining result (n) | No. of specimens with indicated results |
Sensitivity (%) |
|||||
|---|---|---|---|---|---|---|---|---|
| False positive |
True positive (MTC−, MYC+ [n = 7]) | False negative (MTC−, MYC− [n = 5]) | ||||||
| MTC+, MYC+ (n = 0) | MTC+, MYC− (n = 0) | Each species | M. kansasii | Overall NTM | ||||
| M. kansasii (8) | − (4) | 0 | 0 | 1 | 3 | 25.0 | 50.0a | 58.3 |
| + (4) | 0 | 0 | 3 | 1 | 75.0 | 50.0 | 58.3b | |
| M. avium-M. intracellulare (3) | − (3) | 0 | 0 | 2 | 1 | 66.7 | 58.3 | |
| M. xenopi (1) | + (1) | 0 | 0 | 1 | 0 | 100.0 | 58.3 | |
M. kansasii overall sensitivity.
NTM overall sensitivity.
Ninety-six out of 127 patients (75.6%) were diagnosed for mycobacterial disease using the present fluorogenic approach. For the 115 MTC patients, a positive result was achieved in 77.4% (89 of 115) of the samples, although the MTC probe was positive in 70.4% (81 of 115) of the samples. Thus, the number of patients diagnosed increased 7.0% by the addition of the MYC probe, representing 9.0% (8 of 89) of the MTC patients with a positive result (Table 2).
There were no significant differences among the sensitivities and specificities for HIV-positive and HIV-negative samples.
(i) Sensitivity.
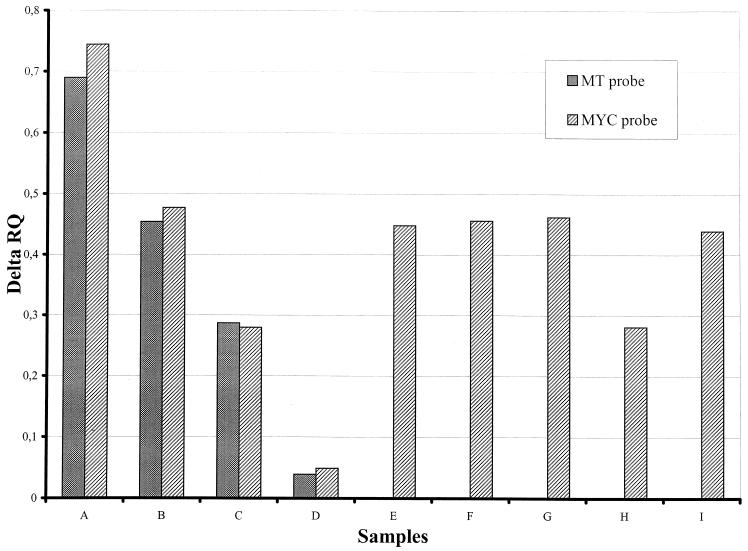
The sensitivity for fluorogenic PCR was 102 to 103 CFU/ml for mycobacterial strain dilutions (Fig. 2).
FIG. 2.
Analysis of mycobacterial strain dilutions by fluorescence. Bars A, B, C, and D, 104, 103, 102, and 10 M. tuberculosis equivalents, respectively; E, F, G, H, and I, 103 equivalents of M. kansasii, M. avium, M. intracellulare, M. xenopi, and M. fortuitum, respectively. The MTC probe was only positive in M. tuberculosis dilution samples.
In clinical specimens, the sensitivity was much higher for smear-positive samples (90.3%) than for smear-negative samples (49.2%). The sensitivity obtained was slightly higher for MTC than NTM (50.0 versus 42.9% for smear-negative samples and 91.2 versus 80.0% for smear-positive samples), although the differences in MTC and MYC probe sensitivity were not statistically significant (Tables 2 and 3).
The sensitivity for MTC pulmonary samples was 72.8 and 50.0% for samples of other origins.
The overall sensitivity of the test in this study was 69.3% (88 of 127 specimens). This value increased to 75.6% (96 of 127 specimens) when identification to any level was considered.
(ii) Specificity.
All negative controls from extraction and amplification were negative by fluorescence detection, demonstrating the absence of contamination during these steps. None of the bacterial strains and samples from the control subjects (healthy students and patients with chronic obstructive pulmonary disease) were positive by fluorescence.
There were several nonconcordant results in strain identifications by conventional methods and fluorescence results, mainly justified by the lack of sensitivity of any of the probes. Thus, in MTC samples, identification to the Mycobacterium genus level was only due to false negative results with the MTC probe in which seven out of eight samples with an MTC-negative, MYC-positive result were smear negative. In our cases, 53.3% (8 of 15) of the patient samples with only an MYC-positive result were MTC. This value fell to 20.0% (one of five) in smear-positive samples.
The three cases of MTC samples with MTC-positive, MYC-negative results had weakly positive MTC probe values but no correlation with smear status and were considered to be correctly identified.
In NTM samples, the lack of sensitivity of the MYC probe led to false-negative results. No NTM tested had an MTC-positive probe result.
DISCUSSION
The aim of this study was therefore to design a PCR-based technique for MTC identification and simultaneous screening of NTM in clinical samples in our laboratory.
Among the sequences available, the 16S rDNA gene was chosen. This target has been widely used in taxonomy since it contains both genus- and species-specific regions, it has a low mutation rate, and the patterns are well preserved (38). Kox et al. (18, 19) used this gene for the first time in the direct diagnosis of clinical samples.
The choice of a fluorogenic assay has been the most determining factor in the overall design of the experiments reported here. Conventional PCR conditions and a conventional DNA extraction method were chosen, modified, and optimized to be compatible with the 5′-exonuclease approach as described in Materials and Methods.
The selection of different quenching molecules for each probe, ROX and TAMRA instead of only TAMRA, reduces the reduction in sensitivity observed when several probes are placed together (data not shown), although other labels can certainly be used, since some fluorimeters available on the market may condition fluorescent pairs.
The sensitivities achieved here are similar to those described in the literature (1, 17). In general, the present test is more reliable in smear-positive samples of pulmonary origin. The addition of the MYC probe allows the diagnosis of an extra 7.0% of patients suffering from TB and the screening of NTM infections in 58.3% of cases. Our results show that a positive MTC probe value confirms the presence of MTC with a 100% positive predictive value. However, an MTC-negative, MYC-negative or MTC-negative, MYC-positive result does not discount the presence of MTC or NTM in the specimen, especially in smear-negative samples. This occurred in false-negative samples and in 9% of MTC patients with only an MYC-positive result. For this reason, an MTC-negative result is inconclusive. This lack of sensitivity is mainly argued by a lack of sample homogeneity (17) and the presence of a single target in the Mycobacterium genome. Some strategies are required to solve this problem. Thus, the use of 16S RNA as target, which is present in thousands of copies, instead of rDNA, could lower the detection limit severalfold (10). It was however, discarded for this initial approach, since it is less stable than DNA and requires a reverse transcription step prior to amplification (9).
Displacement of the probe instead of its breaking, leading to quenched fluorescence, may also be due to mutations in the probe targets. As described by several authors (28, 37) a single different nucleotide would be enough to produce negative results. In addition, false negatives due to degradation of the fluorogenic probes are avoided by the inclusion of positive controls in each round. Although we did not do so, tests are required to check the 5′-exonuclease activity of the polymerase (20) essential for breaking the probes.
Finally, the MTC-negative, MYC-negative values in the present study do not discount inhibition. Thus, the addition of an internal control (6, 17) may be required, as may other alternative DNA extraction methods (2, 5, 29, 35, 40) or 10-fold dilution of samples. In the present study, we have analyzed only one sample per patient. Otherwise, it would be necessary to analyze at least three samples from the same patient to increase the sensitivity (6, 17).
Although all the controls tested here were negative, it is possible that an MTC-negative, MYC-positive result may be due to environmental mycobacteria without clinical significance (A. Kolk, personal communication). Taking all this into consideration, we recommend the use of this test in conjunction with the available clinical data on the patient.
It is expected that this assay could be transferred to the LightCycler without major modifications and with nearly identical results (27). Thus, the need for larger optimal target sequences due to the need for two single-labeled probes may be avoided by the use of a single-probe approach (35).
In addition, for the identification of NTM species, an MTC probe may be substituted for by the corresponding probe.
In conclusion, with this assay patients infected with MTC may be identified and the screening of other mycobacterial species may be performed within 48 h. Moreover, this assay is easy to carry out in routine practice and is a valuable tool for the appropriate management of patients.
Acknowledgments
This work was supported by Fondo de Investigaciones Sanitarias de la Seguridad Social (FIS) grants 96/0028-01 and 98/1282 from the Ministerio de Salud, Madrid, Spain, and Sociedad Española de Neumología y Cirugía Torácica (SEPAR) grant 96/444. Albert García-Quintanilla was granted a predoctoral fellowship from the Departament de Microbiologia i Parasitologia Sanitàries, Divisió Ciències de la Salut, Universitat de Barcelona, Spain.
We thank Rosa Monté and Dolors Ricart for their assistance in routine procedures.
REFERENCES
- 1.Almeda, J., A. Garcia, J. Gonzalez, L. Quinto, P. J. Ventura, R. Vidal, G. Rufi, J. A. Martinez, M. T. Jimenez de Anta, A. Trilla, and P. L. Alonso. 2000. Clinical evaluation of an in-house IS6110 polymerase chain reaction for diagnosis of tuberculosis. Eur. J. Clin. Microbiol. Infect. Dis. 19:859-867. [DOI] [PubMed] [Google Scholar]
- 2.Böddinghaus, B., T. H. Wichelhaus, V. Brade, and T. Bittner. 2001. Removal of PCR inhibitors by silica membranes: evaluating the Amplicor Mycobacterium tuberculosis kit. J. Clin. Microbiol. 39:3750-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böddinghaus, B., T. Rogall, T. Flohr, H. Blöcker, and E. C. Böttger. 1990. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 28:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardullo, R. A., S. Agrawal, C. Flores, P. C. Zamecnik, and D. E. Wolf. 1988. Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 85:8790-8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakravorty, S., and J. S. Tyagi. 2001. Novel use of guanidinium isothiocyanate in the isolation of Mycobacterium tuberculosis DNA from clinical material. FEMS Microbiol. Lett. 205:113-117. [DOI] [PubMed] [Google Scholar]
- 6.Clarridge, J. E., III, R. M. Shawar, T. M. Shinnick, and B. B. Plikaytis. 1993. Large-scale use of polymerase chain reaction for detection of Mycobacterium tuberculosis in a routine mycobacteriology laboratory. J. Clin. Microbiol. 31:2049-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.De Beenhouwer, H., Z. Liang, P. de Rijk, C. van Eekeren, and F. Portaels. 1995. Detection and identification of Mycobacteria by DNA amplification and oligonucleotide-specific capture plate hybridization. J. Clin. Microbiol. 33:2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DesJardin, L. E., M. D. Perkins, K. Wolski, S. Haun, L. Teixeira, Y. Chen, J. L. Johnson, J. J. Ellner, et al. 1999. Measurement of sputum Mycobacterium tuberculosis messenger RNA as a surrogate for response to chemotherapy. Am. J. Repir. Crit. Care Med. 160:203-210. [DOI] [PubMed] [Google Scholar]
- 10.DesJardin, L. E., Y. Chen, M. D. Perkins, L. Teixeira, M. D. Cave, and K. D. Eisenach. 1998. Comparison of the ABI 7700 system (TaqMan) and competitive PCR for quantification of IS6110 DNA in sputum during treatment of tuberculosis. J. Clin. Microbiol. 36:1964-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falkinham, J. O., III. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Quintanilla, A., L. Garcia, G. Tudo, M. Navaro, J. Gonzalez, and M. T. Jimenez de Anta. 2000. Single-tube balanced heminested PCR for detecting Mycobacterium tuberculosis in smear-negative samples. J. Clin. Microbiol. 38:1166-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelmini, S., C. Orlando, R. Sestini, G. Vona, P. Pinzani, L. Ruocco, and M. Pazzagli. 1997. Quantitative polymerase chain reaction-based homogeneous assay with fluorogenic probes to measure c-erbB-2 oncogene amplification. Clin. Chem. 43:752-758. [PubMed] [Google Scholar]
- 14.Gonzalez, J., G. Tudo, J. Gomez, A. Garcia, M. Navarro, and M. T. Jimenez de Anta. 1998. Use of microscopic morphology in smears prepared from radiometric cultures for identification of Mycobacterium tuberculosis complex, Mycobacterium avium complex, Mycobacterium kansasii, and Mycobacterium xenopi. Eur. J. Clin. Microbiol. Infect. Dis. 17:493-500. [DOI] [PubMed] [Google Scholar]
- 15.Higgins, J. A., J. Ezzell, B. J. Hinnebusch, M. Shipley, E. A. Henchal, and M. S. Ibrahim. 1998. 5′ nuclease PCR assay to detect Yersinia pestis. J. Clin. Microbiol. 36:2284-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirschner, P., B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F.-C. Bange, and E. C. Böttger. 1993. Genotypic identification of Mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 31:2882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirschner, P., J. Rosenau, B. Springer, K. Teschner, K. Feldmann, and E. C. Böttger. 1996. Diagnosis of mycobacterial infections by nucleic acid amplification: 18-month prospective study. J. Clin. Microbiol. 34:304-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kox, L. F. F., J. van Leeuwen, S. Knijper, H. M. Jansen, and A. H. J. Kolk. 1995. PCR assay based on DNA coding for 16S rRNA for detection and identification of mycobacteria in clinical samples. J. Clin. Microbiol. 33:3225-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kox, L. F., H. M. Jansen, S. Kuijper, and A. H. Kolk. 1997. Multiplex PCR assay for immediate identification of the infecting species in patients with mycobacterial disease. J. Clin. Microbiol. 35:1492-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreuzer, K.-A., A. Bohn, U. Lass, U. R. Peters, and C. A. Schmidt. 2000. Influence of DNA polymerases on quantitative PCR results using TaqMan probe format in the LightCycler instrument. Mol. Cell. Probes 14:57-60. [DOI] [PubMed] [Google Scholar]
- 21.Kubica, G. P. W., E. Dye, M. L. Cohn, and G. Middlebrook. 1963. Sputum digestion and decontamination with N-acetyl-L-cysteine-sodium hydroxide for culture of Mycobacteria. Am. Rev. Respir. Dis. 87:775-779. [DOI] [PubMed] [Google Scholar]
- 22.Kulski, J. K., and T. Pryce. 1996. Preparation of Mycoacterial DNA from blood culture fluids by simple alkali wash and heat lysis method for PCR detection. J. Clin. Microbiol. 34:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulski, J. K., C. Khinsoe, T. Pryce, and K. Christiansen. 1995. Use of a multiplex PCR to detect and identify Mycobacterium avium and M. intracellulare in blood culture fluids of AIDS patients. J. Clin. Microbiol. 33:668-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longo, M. C., M. S. Berninger, and J. L. Hartley. 1990. DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 93:125-128. [DOI] [PubMed] [Google Scholar]
- 25.Martin Casabona, N., J. Rossello Urgell, and Grupo de Estudio sobre Micobacterias Ambientales. 2000. Micobacterias ambientales en España: aislamientos en el período 1976-1996. Med. Clin. (Barcelona) 115:663-670. [DOI] [PubMed] [Google Scholar]
- 26.Nazarenko, I. A., S. K. Bhatnagar, and R. J. Hohman. 1997. A closed tube format for amplification and detection of DNA based on energy transfer. Nucleic Acids Res. 25:2516-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nitsche, A., N. Steuer, C. A. Schmidt, O. Landt, and W. Siegert. 1999. Different real-time PCR formats compared for the quantitative detection of human cytomegalovirus DNA. Clin. Chem. 45:1932-1937. [PubMed] [Google Scholar]
- 28.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359-363. [DOI] [PubMed] [Google Scholar]
- 29.Pujol-Rique, M., F. Derouin, A. Garcia-Quintanilla, M. E. Valls, J. M. Miro, and M. T. Jiménez de Anta. 1999. Design of a one-tube hemi-nested PCR for detection of Toxoplasma gondii and comparison of three DNA purification methods. J. Med. Microbiol. 48:857-862. [DOI] [PubMed] [Google Scholar]
- 30.Roberts, G. D., E. W. Koneman, and V. K. Kim. 1991. Mycobacterium, p. 304-339, In A. Balows, W. Hausler, Jr., K. L. Herrman, H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 31.Rogall, T., T. Flohr, and E. C. Böttger. 1990. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J. Gen. Microbiol. 136:1915-1920. [DOI] [PubMed] [Google Scholar]
- 32.Shafer, R., and M. F. Sierra. 1992. Mycobacterium xenopi, Mycobacterium fortuitum, Mycobacterium kansasii, and other non-tuberculous mycobacteria in an area of endemicity for AIDS. Clin. Infect. Dis. 15:161-162. [DOI] [PubMed] [Google Scholar]
- 33.Stauffer, F., H. Haber, A. Rieger, R. Mutschlechner, P. Hasenberger, V. J. Tevere, and K. K. Y. Young. 1998. Genus level identification of mycobacteria from clinical specimens by using an easy-to-handle Mycobacterium-specific PCR assay. J. Clin. Microbiol. 36:614-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swan, D. C., R. A. Tucker, B. P. Holloway, and J. P. Icenogle. 1997. A sensitive, type-specific, fluorogenic probe assay for detection of human papillomavirus DNA. J. Clin. Microbiol. 35:886-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor, M. J., M. S. Hughes, R. A. Skuce, and S. D. Neill. 2001. Detection of Mycobacterium bovis in bovine clinical specimens using real-time fluorescence resonance energy transfer probe rapid-cycle PCR. J. Clin. Microbiol. 39:1272-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thelwell, N., S. Millington, A. Solinas, J. Booth, and T. Brown. 2000. Mode of action and application of Scorpion primers to mutation detection. Nucleic Acids Res. 28:3752-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torres, M. J., A. Criado, J. C. Palomares, and J. Aznar. 2000. Use of real-time PCR and fluorimetry for rapid detection of rifampin and isoniazid resistance-associated mutations in Mycobacterium tuberculosis. J. Clin. Microbiol. 38:3194-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rDNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 40.Witham, P. K., C. T. Yamashiro, K. J. Livak, and C. A. Batt. 1996. A PCR-based assay for the detection of Escherichia coli Shiga-like toxin genes in ground beef. Appl. Environ. Microbiol. 62:1347-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.World Health Organization. 1998. W. H. O. report on the tuberculosis epidemic 1997. World Health Organization, Geneva, Switzerland.