Abstract
The molybdenum cofactor (Moco), a highly conserved pterin compound complexing molybdenum, is required for the enzymatic activities of all molybdenum enzymes except nitrogenase. Moco is synthesized by a unique and evolutionarily old pathway that requires the activities of at least six gene products. Some of the proteins involved in bacterial, plant, and invertebrate Moco biosynthesis show striking homologies to the primary structure of gephyrin, a polypeptide required for the clustering of inhibitory glycine receptors in postsynaptic membranes in the rat central nervous system. Here, we show that gephyrin binds with high affinity to molybdopterin, the metabolic precursor of Moco. Furthermore, gephyrin expression can reconstitute Moco biosynthesis in Moco-deficient bacteria, a molybdenum-dependent mouse cell line, and a Moco-deficient plant mutant. Conversely, inhibition of gephyrin expression by antisense RNA expression in cultured murine cells reduces their Moco content significantly. These data indicate that in addition to clustering glycine receptors, gephyrin also is involved in Moco biosynthesis and illustrate the remarkable conservation of its function in Moco biosynthesis throughout phylogeny.
The molybdenum cofactor (Moco) forms the catalytically active center of all molybdoenzymes except nitrogenase (1). It consists of a molybdenum coordinated to the unique pterin compound molybdopterin (MPT). The cofactor is highly conserved in Archaea, eubacteria, and eukaryotes (2). Molybdoenzymes are essential for such diverse metabolic processes as sulfur detoxification and purine catabolism in mammals, nitrate assimilation in autotrophs, and phytohormone synthesis in plants. Therefore, the survival of the respective organisms depends on their ability to synthesize Moco.
Human Moco deficiency is characterized by the combined loss of function of three major molybdoenzymes, namely sulfite oxidase, xanthine oxidase, and aldehyde oxidase (3). The clinical signs of Moco deficiency are attributed to the accumulation of toxic metabolites caused by the reduced activity of these enzymes, mainly sulfite oxidase. Interestingly, although highest enzymatic activities are detected in visceral organs like liver, kidney, lung, and heart, the leading symptoms of Moco-deficient patients are severe neurological abnormalities such as increased muscle tone, rigid posturing, myoclonus, abnormal movements, and refractory seizures (4–6). Affected patients usually die early postnatally because no therapy is available.
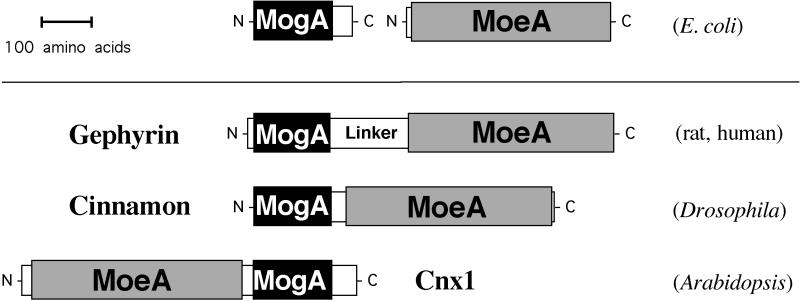
The biosynthesis of Moco requires the multistep synthesis of the MPT moiety followed by the subsequent transfer of molybdenum (2). At least six gene products involved in Moco-biosynthesis have been identified in Escherichia coli (7), plants (8), and humans (9). For the E. coli proteins MogA and MoeA and their putative homologs—cinnamon in Drososphila melanogaster (10) and Cnx1 in Arabidopsis thaliana (11)—a function in the transfer and/or activation of molybdate for incorporation into MPT has been suggested (8, 11, 12). The demonstration of high-affinity binding of MPT to Cnx1 is consistent with this idea (13). The only known mammalian homolog to MogA and MoeA is the neurotransmitter cytoskeleton linker protein gephyrin (14). Based on the homology to the E. coli proteins, the polypeptide chain can be subdivided into an N-terminal MogA-like domain and a C-terminal MoeA-like domain (Fig. 1). The domains are separated by a central region of about 160 aa without homology to any known protein. An identical domain structure also is seen in cinnamon and Cnx1; however, in the latter, the order of the domains is inverted (Fig. 1).
Figure 1.
Schematic representation of Moco biosynthetic proteins with structural homology to gephyrin. The primary structure of the mammalian polypeptide gephyrin shows striking homology to Moco synthesic proteins from E. coli (MogA and MoeA), invertebrates (cinnamon) and plants (Cnx1). MogA and MogA-like domains are shown as black bars, and MoeA and MoeA-like domains are represented as gray bars. Both domains are connected by a central region showing no significant sequence homology to other proteins. Note that the two domains are inverted in Cnx1.
In the central nervous system, gephyrin is localized at the cytoplasmic face of glycinergic and subsets of γ-aminobutyric acid (GABA)ergic postsynaptic membranes (15, 16) where it has been shown to anchor neurotransmitter receptors to the cytoskeleton (17). Gephyrin is thought to be an instructive molecule for the formation of glycinergic synapses (18), and its expression was shown to be essential for the postsynaptic aggregation of glycine receptors (19). Transcript variants of gephyrin are found not only in brain and spinal cord, but also in liver, kidney, heart, and lung (14). Because gephyrin is the only known mammalian homolog to the Moco-biosynthesis proteins MogA and MoeA, we investigated a possible additional function of gephyrin in Moco biosynthesis.
Our data show that purified gephyrin can bind MPT with high affinity and that defective Moco biosynthesis in mutant bacteria, plants, and cultured mammalian cells can be rescued by gephyrin. These data confirm that gephyrin also plays a role in Moco biosynthesis. Moreover, our data represent an example of a remarkable conservation of function throughout phylogeny underscoring the extraordinary evolutionary pressure on the biosynthetic pathway of Moco.
MATERIALS AND METHODS
Construction of Expression Vectors.
For recombinant expression of gephyrin in E. coli, the entire gephyrin P1 cDNA and parts of this cDNA encoding the MogA and MoeA domains including the linker region were cloned into the pQE30 expression vector (Qiagen, Chatsworth, CA) by insertion of KpnI restriction sites using PCR mutagenesis. Expression vectors for transfection of L929 and NR6P cell lines are based on pcDNA3 vectors (Invitrogen) by subcloning of the cnx1 cDNA into the EcoRI site, the gephyrin P1 cDNA (2.6-kb XhoI fragment from pRT101geph) in both orientations into the XhoI site, and a 1.65-kb HindIII/SpeI fragment of gephyrin P1 clone (containing the coding region of the gephyrin P1 clone up to position 1610) into the HindIII/XbaI site. The plant expression construct pRT101geph was generated by subcloning of a 2.6-kb SacI/KpnI fragment derived from the construct pBSK+geph P1 (14), containing the total coding region of the gephyrin P1 cDNA clone and the 3′-untranslated region up to position 2859, into the pRT101 (20) polylinker.
Recombinant Expression of Gephyrin, MogA Domain, MoeA Domain, and MPT Binding.
Overexpression and purification of recombinant gephyrin as well as its MogA and MoeA domain was performed on a Ni-nitrilotriacetic acid (NTA)-superflow matrix under native conditions at 4°C by using the QIAexpress kit (Qiagen) as recommended by the supplier. Pure fractions were dialyzed against 10 mM Tris⋅HCl/1 mM EDTA (pH 7.5), concentrated, sterile filtered, and stored at 4°C. Protein concentration was determined by UV absorption using the calculated extinction coefficient of 28,030 M−1⋅cm−1 for gephyrin, 7,450 M−1⋅cm−1 for MogA domain, and 20,340 M−1⋅cm−1 for MoeA domain. MPT binding experiments were performed with protein-free MPT isolated from xanthine oxidase by gel filtration as described (13). MPT was determined by quantitative conversion of MPT to its oxidized fluorescent product dephospho-Form A (13, 21).
Cell Culture.
L929 cells (ATTC CCL-1) and NR6P cells (22) were grown in DMEM medium containing 10% fetal bovine serum at 37°C in 5% CO2. For experiments involving treatments with sodium molybdate, cells were allowed to reach confluence before supplementing the growth medium to a final concentration of 1 mM sodium molybdate, and cultivation of the cells was continued for 12 h before harvesting. DNA was transfected into cells by the method of Chen and Okayama (23). Two days after transfection, selection of stable transformants was started by the addition of 300 μg/ml (NR6P) and 400 μg/ml (L929) G418 (Sigma); after 10 days, G418 concentration was increased to 500 μg/ml (NR6P) and 600 μg/ml (L929). Pools of at least 100 cells were derived from each transfection and further analyzed. RNase protection assays were performed by using the RPA II and MAXIscript in vitro transcription kit (Ambion) as recommanded by the supplier.
Stable Transformation and Regeneration of Plant Mutants.
Plants of Nicotiana plumbaginifolia cnxA mutants (24) (obtained from Christian Meyer, Institut National de la Recherche Agronomique, Versailles, France) were maintained according to Müller (25) in vitro on solid medium containing 20 mM ammonium succinate and 9.5 mM potassium nitrate at 23°C and illuminated at 25 μmol photons m−2⋅s−1 under a 16 h light/8 h dark regime. Mesophyll protoplasts were isolated from 2- to 3-month old plants after overnight digestion of leaves in 0.6% Onozuka Cellulase R10 (Serva) and 0.2% Macerocyme (Serva) dissolved in T0 (26) but omitting Tween. Transformation was performed according to ref. 27 by using the plasmids pRT101geph and pRT103neo (20). After transformation, protoplasts were cultured at a density of 8 × 104 per ml in liquid T0 supplemented with 5 mM glutamine in the dark at 25°C. Two weeks later, protoplast-derived microcolonies were plated on modified solid T8 medium (26) with macrosalts from the plant medium described above with 220 mM mannitol and 20 mM ammonium succinate containing 50 mg/l kanamycin added. Selected calli able to grow on nitrate-containing medium were regenerated into plants that were rooted and transferred to soil.
Determination of Enzyme Activities.
E. coli mogA mutant RK5206 was transformed with 200 ng of plasmid DNA and grown anaerobically at 30°C in 100 ml Luria–Bertani (LB) medium to an OD600 of 1.0. Where indicated, the medium was supplemented with 1 mM Na2MoO4. Nitrate reductase activity in protein crude extract was determined according to MacGregor et al. (28). Nitrate reductase and xanthine dehydrogenase activities in N. plumbaginifolia plants were determined in crude extracts as described (29, 30).
Determination of Moco Content.
Neurospora crassa nit-1 extract was prepared as described (31). L929 and NR6P cells obtained from a 6-cm dish were homogenized by sonication in 200 μl 50 mM potassium phosphate, 5 mM EDTA, and 5 mM reduced glutathione (pH 7.2). After clarification, 200 μl of protein extract were immediately transferred to 50 μl of nit-1 extract supplemented with 20 mM NADPH and complemented anaerobically overnight at 4°C. Reconstituted NADPH–nitrate reductase activity was assayed as described (31).
RESULTS
Gephyrin Binds Molybdopterin with High Affinity.
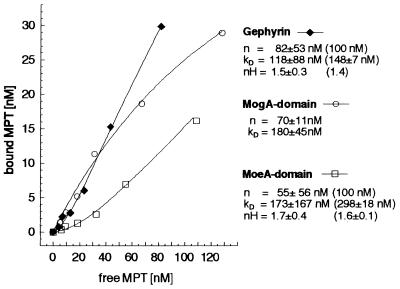
Although the precise function of the gephyrin homologs in Moco biosynthesis is not clear, it has been suggested that Cnx1 participates in the transfer of molybdate to MPT (8, 11), the last step of Moco synthesis (2, 8). Because a high-affinity binding of MPT to Cnx1 and its two separate domains has been shown (13), MPT binding of gephyrin would constitute a prerequisite for its putative implication in Moco biosynthesis. To investigate whether gephyrin can also bind MPT, we mixed purified, recombinant gephyrin (100 nM) with different amounts of MPT (5–110 nM) and determined the amount of bound and unbound MPT by gel filtration (Fig. 2). Assuming a cooperative type of binding, a Kd of 118 ± 88 nM was determined. Curve extrapolation gives 82 ± 53 nM as maximal concentration of binding sites; however, no saturation of MPT binding was reached. For equimolar binding (n = 100 nM), a Kd value of 148 ± 7 nM was calculated. The MPT binding properties (Kd and binding type) of gephyrin are consistent with values determined for Cnx1 (13).
Figure 2.
Binding of MPT to gephyrin and to its two domains. The binding of 5–157 nM MPT, isolated from denatured xanthine oxidase, to 100 nM gephyrin (♦), MogA domain (○), and MoeA-domain (□) was analyzed by using gel filtration followed by HPLC analysis of Form A, the I2/KI-oxidation product of MPT, as described (13). Data were collected from binding experiments with six different MPT concentrations. Dissociation constants (Kd), maximal concentration of binding sites (n), and Hill coefficients (nH) are shown for each curve. In the case of cooperative types of binding (gephyrin, MoeA domain), no saturation with MPT was reached. Curve regressions with n = 100 nM (equimolar binding) gave Kd values and Hill coefficients that are shown in brackets.
More detailed insights into the nature of MPT binding to gephyrin were obtained from experiments in which the MogA domain and MoeA domain of gephyrin were assayed separately for MPT binding (Fig. 2). Like Cnx1, the MogA domain binds MPT with a Michaelis–Menten type of binding (equimolar, Kd=180 ± 45 nM), whereas the MoeA domain has a cooperative binding behavior (Hill coefficient nH = 1.6–1.7) with a higher Kd value (289 ± 18 nM). These data also demonstrate that the domains of gephyrin exhibit MPT binding properties very similar to those determined for the domains of Cnx1.
Expression of Gephyrin in E. coli mogA Mutants Restores Nitrate Reductase Activity.
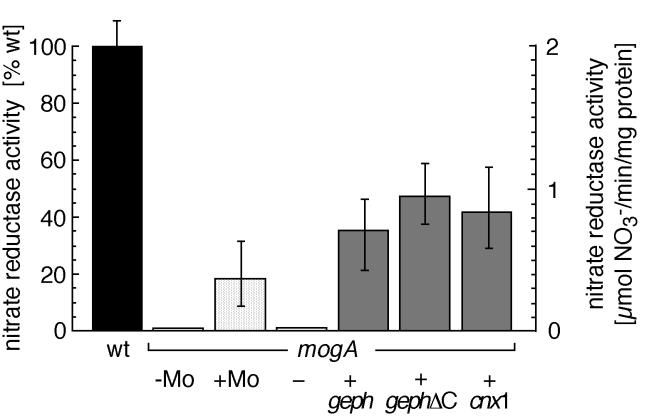
As a next step, we investigated whether the heterologous expression of gephyrin in the Moco-deficient E. coli mutant mogA can restore Moco biosynthesis (Fig. 3). In this mutant, the activity of the molybdenum enzyme nitrate reductase is strongly reduced, but the cells are able to synthesize MPT (32), which can be converted to Moco in the presence of 1 mM sodium molybdate, thus partially restoring nitrate reductase activity (33). Moreover, heterologous expression of the MogA homolog Cnx1 from A. thaliana in mogA mutant cells was shown to partially restore nitrate reductase activity (11). Although no nitrate reductase activity could be detected in mogA mutants or in mogA mutants transformed with a control vector, the expression of gephyrin (mogA+geph) or of a C-terminally truncated deletion mutant (mogA+gephΔC, retaining the entire MogA domain) restored nitrate reductase activity in E. coli mogA to 35.2 ± 8.9% and 47.8 ± 11.2%, respectively, of the activity in wild-type E. coli. In comparison, expression of Cnx1 (mogA+cnx1) leads to 42.3 ± 15.6% wild-type activity, whereas increased concentrations of molybdate induced nitrate reductase activities of only 18.5 ± 10.9%. These data demonstrate that expression of gephyrin can restore Moco synthesis in E. coli mogA mutants, suggesting a conserved function of the MogA-like domain of gephyrin.
Figure 3.
Functional complementation of the Moco-deficient E. coli mogA mutant by heterologous gephyrin expression. The columns show nitrate reductase activity of the crude protein extract derived from E. coli wild-type (wt) strain RK4353 (filled bar), untransfected E. coli mogA cells grown in the presence (+Mo) or absence (−Mo) of 1 mM sodium molybdate (stippled bars), and mogA mutant cells (RK5231, gray bars) transformed with the control plasmid pcDNA3 (−) without insert, expressing the full-length gephyrin cDNA clone P1 (+geph), the C-terminal truncated gephyrin (+gephΔC), and Cnx1 from Arabidopsis (+cnx1).
Expression of the MogA-Like Domain of Gephyrin Restores Moco Biosynthesis in the Murine Cell Line L929.
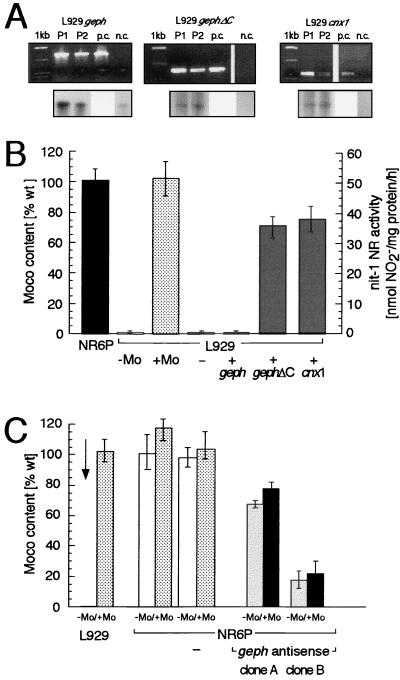
Nitrate reductase activities of Moco mutants E. coli mogA (33), Arabidopsis chl-6 (34) and N. plumbaginifolia cnxA (35), can be restored on growth on elevated concentrations of molybdate. A similar phenomenon has been described for the Moco-deficient murine fibroblast cell line L929, where Moco biosynthesis can be restored by the addition of 1 mM molybdate to the culture medium (36). To test whether gephyrin can restore Moco biosynthesis in this cell line, we generated L929 cells stably expressing (i) full-length gephyrin (L929+geph), (ii) a C-terminally truncated form of gephyrin lacking most of the MoeA domain (L929+gephΔC), or (iii) Cnx1 (L929+cnx1) by selecting pools of at least 150 independent G418-resistant clones. Incorporation of the transferred genes was monitored by using PCR amplification of genomic DNA (Fig. 4A Upper) and the expression of the transgenes was demonstrated by using RNase protection assays (Fig. 4A Lower). The Moco content of the different cell pools was determined by the sensitive nit-1 reconstitution assay and compared with that of the Moco content of murine fibroblasts NR6P or L929 cells growing in the presence or absence of 1 mM molybdate (Fig. 4B). The nitrate reductase activity reconstituted with extracts from wild-type NR6P cells was 51 ± 4.5 nmol⋅(mg protein⋅h)−1. Extracts from L929 cells expressing full-length gephyrin reconstituted only 0.8 ± 0.7% of the NR6P activity, a value comparable to that of untransfected L929 cells. In contrast, the cells expressing gephΔC restored 71.4 ± 6.2% of the NR6P activity. Similar values were determined for L929 cells expressing Cnx1 (75.9 ± 8.1% of the NR6P activity). The rescue of Moco biosynthesis in molybdate-dependent prokaryotic and eukaryotic cells indicates the functional conservation of the MogA-like domain of gephyrin. The absence of Moco biosynthesis in L929 cells expressing full-length gephyrin could be explained by the observation that recombinant gephyrin [but not the C-terminal truncated form of gephΔC (J.K., unpublished data)] forms large cytoplasmic aggregates when heterologously expressed in mammalian cells (37). This could go back to the high expression level of gephyrin—which deregulates the interaction between gephyrin molecules (18) mediated by the MoeA-domain—when using pcDNA3 vectors. Because the complementation of the E. coli mogA mutant was performed with pQE-vectors under noninducing low expression conditions, full-length gephyrin also was able to complement this mutant. Furthermore, it is remarkable that the Moco deficiency in L929 cells can be completely rescued after growth on high molybdate. In contrast, mogA mutants can be repaired only up to 15% of wild-type nitrate reductase activity, which corresponds to the reduced level of MPT (15%) in this mutant (32).
Figure 4.
Functional complementation of the molybdate-dependent murine cell line L929. L929 cells were transfected with constructs expressing the full-length gephyrin cDNA clone P1 (+geph), the C-terminal truncated gephyrin (+gephΔC) and Cnx1 (+cnx1), and pcDNA3 plasmid without insert (−) as a control. (A Upper) PCR amplification of defined parts of the gephyrin and cnx1 cDNA sequences in different transgenic cell lines. Amplifications were performed by using genomic DNA of two independent pools of transformants (P1, P2), of untransfected L929 cells in the negative control (n.c.), and plasmid DNA of the different expression constructs as positive control (p.c.). (Lower) Analysis of transgene expression by RNase protection assays in cells stably transfected. In the case of the L929 cells stably transfected with the gephyrin expression constructs (geph and gephΔC), an increase of the gephyrin mRNA level in comparison to the untransfected L929 cells can be seen, demonstrating the expression of the transgene. (B) Moco content of transfected L929 cells. The columns show Moco activity obtained from multiple nit-1 complementation assays of NR6P cells (black bar), L929 cells (stippled bars) grown in the presence (+Mo) or absence (−Mo) of 1 mM sodium molybdate and pools of at least 100 stable transformants (gray bars). 100% of wild-type Moco content represent a NADPH–nitrate reductase activity of 51 ± 4.5 nmol/mg protein⋅h in the nit-1 complementation assay. (C) Antisense expression of gephyrin cDNA in murine cells. For gephyrin depletion, cells of the murine wildtype cell line NR6P were transfected with the pcDNA3 gephyrin antisense expression construct and as a negative control with pcDNA3 without insert (indicated by −), and stable transformants were selected. The colums show Moco contents of L929 cells, untransfected NR6P cells, NR6P cells transfected with control vector (NR6P−), and cell clones derived from the transfection with the antisense construct (NR6Pgeph antisense clone A and clone B) grown on normal medium (−Mo, white and gray bars) and on medium supplemented with 1 mM sodium molybdate for 12 h (+Mo, stippled and black bars). Values are expressed in % of the Moco content of the untransformed NR6P wild-type cells (51 ± 4.5 nmol/mg protein⋅h).
Inhibition of Gephyrin Expression Reduces the Moco Content of Murine NR6P Cells.
To further corroborate the role of gephyrin in Moco biosynthesis, it was investigated whether the inhibition of gephyrin expression by antisense RNA can reduce the Moco content in NR6P cells. Therefore NR6P cells were stably transfected with a full-length gephyrin cDNA in antisense orientation. Incorporation of the transduced gene was confirmed by using PCR analysis of genomic DNA from 5 independent clones as well as from a pool consisting of at least 150 individual transformants. The expression of the antisense RNA was demonstrated by using the RNase protection assays (data not shown). The Moco content, determined by nit-1 nitrate reductase reconstitution assays in protein extracts derived from antisense expressing clonal cell lines was significantly reduced compared with that of extracts derived from untransfected NR6P cells or cells transfected with control vector (Fig. 4C). The Moco content in individual clonal lines was decreased to 67.4 ± 2.2% (clone A) and 17.7 ± 5.8% (clone B) of the activity of NR6P cells. The variation may be explained by different efficiencies in the inhibition of gephyrin expresssion caused by dosis and positional variations.
We also investigated whether inhibition of gephyrin expression in NR6P cells results in a molybdate-repairable phenotype comparable to that of L929 cells. Surprisingly, the nitrate reductase activities reconstituted from protein extracts of gephyrin-depleted NR6P cells were not significantly increased when the cells were cultured in the presence of 1 mM molybdate, whereas protein extracts from L929 cells cultured under identical conditions exhibit a dramatic increase in nitrate reductase activity [from background activity to values comparable to that of untransfected NR6P cells (Fig. 4C)]. These results confirm the previously postulated role of the MogA-like domain of gephyrin in Moco biosynthesis and indicate that the combined loss of MogA and MoeA functions cannot be corrected for by elevated molybdate.
Gephyrin Rescues the Phenotype of the N. plumbaginifolia Moco Mutant cnxA.
Our data support the view that the MogA-like domain of gephyrin is indeed the mammalian homolog of the E. coli protein MogA, and gephyrin is the direct homolog of the plant protein Cnx1. To prove this assumption, we investigated whether the phenotype of the Moco-deficient and molybdate-dependent plant N. plumbaginifolia mutant cnxA (35) can be rescued by gephyrin expression. Therefore, the full-length gephyrin cDNA under the control of the cauliflower mosaic virus 35S promoter was transfected into cells of N. plumbaginifolia mutant cnxA D70. In eight independent transformation experiments, we obtained 289 kanamycin-resistant colonies. After exposure to a medium containing potassium nitrate, 21 colonies continued to grow because of their ability to assimilate nitrogen and regenerated plants. The successful incorporation of the transgene into the plant genome was confirmed with Southern blot analysis of 11 randomly chosen colonies (data not shown). The nitrate reductase activity of 9 transformants was determined, and all of them displayed activities ranging from 67.5 ± 31.7 to 133.3 ± 38.2% of N. plumbaginifolia wild-type cells (Fig. 5A), whereas the untransfected cnxA mutant D70 had 12.5 ± 8.9%. Also, the activity of another Moco-containing enzyme, xanthine dehydrogenase, was restored after gephyrin expression (Fig. 5A). Furthermore, the phenotype of the cnxA mutant plant, whose habitus is characterized by chlorotic, small, and crinkled leaves, was completely reversed to wild type-like green plants in the gephyrin transformants (Fig. 5B). These experiments demonstrate unequivocally that gephyrin can substitute for plant gene products involved in the biosynthetic pathway of Moco.
Figure 5.
Expression of the gephyrin P1 cDNA in the N. plumbaginifolia cnxA Moco mutant. (A) Restoration of molybdoenzyme activities. Protoplasts of in vitro-grown plants of the N. plumbaginifolia mutant line D70 defective in the locus cnxA were transformed with the gephyrin P1 cDNA driven by the Cauliflower Mosaic Virus 35S promoter. Transgenic selected colonies were grown as callus cultures and assayed for nitrate reductase and xanthine dehydrogenase activities. Because of the defect in Moco synthesis, the cnxA mutant has only 12.5 ± 8.9% of the wild-type level of nitrate reductase activity. Colonies transformed with the gephyrin cDNA show nitrate reductase activities ranging from 67.5 ± 31.7% (D70/585, presented here) up to 133.3 ± 38.2% of the wild-type level. Wild-type activity levels of 100% represent a NADH–nitrate reductase activity of 1.23 μmol of nitrite formed per gram of fresh weight per hour. Mean values ± SD of nitrate reductase activity determined in three independent subculturings of the cells are shown with duplicate measurements for each subculture. The same callus colonies as described above were tested for xanthine dehydrogenase activity by using an in situ gel assay (30). Because of the defect in Moco synthesis, the cnxA mutant has only trace amounts of xanthine dehydrogenase activity, whereas the plants (D70/585) transformed with the gephyrin cDNA show strongly increased enzyme activities. (B) Normalization of the mutant phenotype. All plants shown were grown in vitro and transferred on the same day into soil fertilized with potassium nitrate as the nitrogen source. The Moco-defective cnxA mutant line D70 (Left) is unable to grow in soil and dies. (Center) The N. plumbaginifolia wild type is shown. (Right) A representative gephyrin-expressing cnxA mutant plant (line D70/585) is shown. The wild type-like phenotype of this plant is representative of 10 gephyrin-transformed and regenerated cnxA-mutant plants.
DISCUSSION
The striking homology of the neurotransmitter anchoring protein gephyrin to the prokaryotic Moco proteins MogA and MoeA and to their plant and invertebrate homologs Cnx1 (11) and cinnamon (10), respectively, suggested an additional function of gephyrin in Moco biosynthesis. Here, we demonstrate that (i) gephyrin binds with high affinity to MPT; (ii) gephyrin can efficiently restore the activity of the Moco-dependent enzyme nitrate reductase in E. coli mogA mutants; (iii) Moco biosynthesis in murine L929 fibroblasts can be restored by expression of the MogA-like domain of gephyrin; (iv) inhibition of gephyrin expression in murine NR6P fibroblasts leads to a reduction of Moco, which cannot be compensated for by increased concentrations of molybdate; and (v) the phenotype of a Moco-deficient plant mutant can be rescued by gephyrin expression.
Moco synthesis is believed to be a multistep biochemical pathway (2, 8) converting GTP via precursor Z (7, 38) to MPT (39) followed by the transfer of molybdate to the MPT moiety (13). One could argue that those Moco deficiencies that can be partially restored by increased concentrations of molybdate (32, 35, 36) should be affected in the last step in Moco biosynthesis. In line with this hypothesis, the results presented here indicate that the MogA-like domain of gephyrin and its homologs is indeed capable of catalyzing the transfer of molybdate to MPT.
The MoeA-like domain of gephyrin seems to be essential for the conversion of precursor Z to Moco, because E. coli moeA mutants accumulate precursor Z (40). Furthermore, a role of the E. coli MoeA in molybdate utilization has been suggested (12) and would explain the independent fusion of MoeA and MogA domains in gephyrin and other homologs to facilitate a coordinated and efficient transfer of molybdate to MPT. Our finding that gephyrin depletion in NR6P cells could not be overcome by elevated molybdate in the culture medium is consistent with this idea, arguing for an additional and independent role of the MoeA-like domain of gephyrin in molybdate utilization. It may be that both the MogA- and the MoeA-like protein domains must act coordinately for efficient transfer of molybdate to MPT, thus forming Moco. The evolutionary advantage of domain fusion in gephyrin and Cnx1 could be the compartmentalization of the Moco synthetic machinery to defined regions of the cytoplasm.
The conservation of a protein function bridging an evolutionary distance estimated at one billion years has been described in very few cases (41). This remarkable conservation underscores not only the ancient origin of Moco during phylogeny but also the extraordinarily high evolutionary pressure on the gene products involved in its synthesis. One could speculate that mogA and moeA genes were fused during evolution, resulting in putatively bifunctional gene products like cinnamon and Cnx1. The central region, which is already rudimentarily present in Cnx1 but most prominent in the phylogenetically youngest member of the family, namely gephyrin, might have acquired additional functions independent from Moco biosynthesis. Thus, it is possible that the property of gephyrin that is best characterized—clustering of inhibitory neurotransmitter receptors—resides in this relatively young part of the polypeptide chain. At present it is completely unclear, however, why such highly specialized fusion proteins could have acquired a completely independent function like receptor clustering or vice versa and why the last step of Moco biosynthesis in neurons should be compartmentalized to the subsynaptic membrane of inhibitory synapses. The topographical connection of Moco biosynthesis with receptor clustering remains enigmatic, but it is tempting to speculate that there may be a functional link between synaptic inhibition and Moco biosynthesis.
Gephyrin’s necessity for Moco biosynthesis renders it a candidate gene for Moco deficiency in humans. However, the recently described mocs1 gene (9) has been shown to be responsible for the majority of cases (42), and most of the remaining cases can be attributed to mutations in mocs2 encoding molybdopterin synthase (unpublished observations). Whether a few patients bear mutations in mocs3 (GenBank accession no. G3851719) or gephyrin remains to be determined. Patients with deficiencies in mocs1 and mocs2 show neurological symptoms (3) that are also seen on impaired synaptic inhibition (43); however, patients with isolated sulfite oxidase deficiency experience the same symptoms. The only difference between the isolated form of sulfite oxidase and the combined loss of molybdoenzyme activity in Moco deficiency is the activity of xanthine dehydrogenase (3). The neurological complications of Moco deficiency therefore may be attributable to sulfite toxicity and/or sulfate deficiency even in the case of a gephyrin deficiency.
Acknowledgments
We thank Heinrich Betz for providing the gephyrin P1 clone. The financial support by the Fritz Thyssen Stiftung (R.R.M.) and the Deutsche Forschungsgemeinschaft (J.K., J.R., and R.R.M.) is gratefully acknowledged. J.K. holds an endowed professorship of the Stifterverband für die Deutsche Wissenschaft.
ABBREVIATIONS
- Moco
molybdenum cofactor
- MPT
molybdopterin
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Hille R. Chem Rev. 1996;96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- 2.Rajagopalan K V, Johnson J L. J Biol Chem. 1992;267:10199–10202. [PubMed] [Google Scholar]
- 3.Johnson J L, Wadman S K. In: The Metabolic Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly E S, Valle D L, editors. New York: McGraw–Hill; 1995. pp. 2271–2283. [Google Scholar]
- 4.Beemer F A, Duran M, Wadman S K, Cats B P. Ophthalmic Paediatr Genet. 1985;5:191–195. doi: 10.3109/13816818509006133. [DOI] [PubMed] [Google Scholar]
- 5.Bamforth F J, Johnson J L, Davidson A G F, Wong L T K, Lockitsch G, Applegrath D A. Clin Biochem. 1990;23:537–542. doi: 10.1016/0009-9120(90)80046-l. [DOI] [PubMed] [Google Scholar]
- 6.Johnson J L, Rajagopalan K V, Wadman S K. In: Chemistry and Biology of Pteridines and Folates. Ayling J E, Nair M G, Baugh C M, editors. New York: Plenum; 1993. pp. 373–378. [Google Scholar]
- 7.Rajagopalan K V. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 674–679. [Google Scholar]
- 8.Mendel R R. Planta. 1997;203:399–405. doi: 10.1007/s004250050206. [DOI] [PubMed] [Google Scholar]
- 9.Reiss J, Cohen N, Dorche C, Mandel H, Mendel R R, Stallmeyer B, Zabot M T, Dierks T. Nat Genet. 1998;20:51–53. doi: 10.1038/1706. [DOI] [PubMed] [Google Scholar]
- 10.Kamdar K P, Primus J P, Shelton M E, Archangeli L L, Wittle A E, Finnerty V. Biochem Soc Trans. 1997;25:778–783. doi: 10.1042/bst0250778. [DOI] [PubMed] [Google Scholar]
- 11.Stallmeyer B, Nerlich A, Schiemann J, Brinkmann H, Mendel R R. Plant J. 1995;8:751–762. doi: 10.1046/j.1365-313x.1995.08050751.x. [DOI] [PubMed] [Google Scholar]
- 12.Hasona A, Ray R M, Shanmugam K T. J Bacteriol. 1998;180:1466–1472. doi: 10.1128/jb.180.6.1466-1472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarz G, Boxer D H, Mendel R R. J Biol Chem. 1997;272:26811–26814. doi: 10.1074/jbc.272.43.26811. [DOI] [PubMed] [Google Scholar]
- 14.Prior P, Schmitt B, Grenningloh G, Pribilla I, Multhaup G, Beyreuther K, Maulet Y, Werner P, Langosch D, Kirsch J, et al. Neuron. 1992;8:1161–1170. doi: 10.1016/0896-6273(92)90136-2. [DOI] [PubMed] [Google Scholar]
- 15.Triller M, Triller C. Rev Odonto-Stomatol. 1987;16:327–336. [PubMed] [Google Scholar]
- 16.Sassoe-Pognetto M, Kirsch J, Grunert U, Greferath U, Fritschy J M, Mohler H, Betz H, Wassle H. J Comp Neurol. 1995;357:1–14. doi: 10.1002/cne.903570102. [DOI] [PubMed] [Google Scholar]
- 17.Kirsch J, Betz H. J Neurosci. 1995;15:4148–4156. doi: 10.1523/JNEUROSCI.15-06-04148.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirsch J, Betz H. Nature (London) 1998;392:717–720. doi: 10.1038/33694. [DOI] [PubMed] [Google Scholar]
- 19.Kirsch J, Wolters I, Triller A, Betz H. Nature (London) 1993;366:745–748. doi: 10.1038/366745a0. [DOI] [PubMed] [Google Scholar]
- 20.Töpfer R, Schell J, Steinbiss H H. Nucleic Acids Res. 1988;16:8725. doi: 10.1093/nar/16.17.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson J L, Hainline B E, Rajagopalan K V, Arison B H. J Biol Chem. 1984;259:5414–5422. [PubMed] [Google Scholar]
- 22.Sorscher S M, Green M R, Feramisco J R. Biochem Biophys Res Commun. 1995;206:518–524. doi: 10.1006/bbrc.1995.1074. [DOI] [PubMed] [Google Scholar]
- 23.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabard J, Pelsy F, Marion-Poll A, Caboche M, Saalbach I, Grafe R, Müller A J. Mol Gen Genet. 1988;213:275–281. [Google Scholar]
- 25.Müller A. Mol Gen Genet. 1983;192:275–281. [Google Scholar]
- 26.Crepy, L., Chupeau, M.-C. & Chupeau, Y. (1982) Z. Pflanzenphysiol. 123–131.
- 27.Negrutiu I, Shillito R, Potrykus I, Biasini G, Sala F. Plant Mol Biol. 1987;8:363–373. doi: 10.1007/BF00015814. [DOI] [PubMed] [Google Scholar]
- 28.MacGregor C H, Schnaitman C A, Normansell D E. J Biol Chem. 1974;249:5321–5327. [PubMed] [Google Scholar]
- 29.Scheible W R, Gonzalez-Fontes A, Morcuende R, Lauerer M, Geiger M, Glaab J, Gojon A, Schulze E D, Stitt M. Planta. 1997;203:304–319. doi: 10.1007/s004250050196. [DOI] [PubMed] [Google Scholar]
- 30.Mendel R R, Müller A J. Biochem Physiol Pflanzen. 1976;170:538–541. [Google Scholar]
- 31.Nason A, Lee K Y, Pan S S, Ketchum P A, Lamberti A, DeVries J. Proc Natl Acad Sci USA. 1971;68:3242–3246. doi: 10.1073/pnas.68.12.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joshi M S, Johnson J L, Rajagopalan K V. J Bacteriol. 1996;178:4310–4312. doi: 10.1128/jb.178.14.4310-4312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart V, MacGregor C H. J Bacteriol. 1982;151:788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braaksma A, Haaker H, Grande H J, Veeger C. Eur J Biochem. 1982;121:483–491. doi: 10.1111/j.1432-1033.1982.tb05813.x. [DOI] [PubMed] [Google Scholar]
- 35.Müller A J, Mendel R R. In: Molecular and Genetic Aspects of Nitrate Assimilation. Wray J L, Kinghorn J R, editors. Oxford: Oxford Univ. Press; 1989. pp. 166–185. [Google Scholar]
- 36.Falciani F, Terao M, Goldwurm S, Ronchi A, Gatti A, Minoia C, Li Calzi M, Salmona M, Cazzaniga G, Garattini E. Biochem J. 1994;298:69–77. doi: 10.1042/bj2980069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirsch J, Kuhse J, Betz H. Mol Cell Neurosci. 1995;6:450–461. doi: 10.1006/mcne.1995.1033. [DOI] [PubMed] [Google Scholar]
- 38.Wuebbens M M, Rajagopalan K V. J Biol Chem. 1995;270:1082–1087. doi: 10.1074/jbc.270.3.1082. [DOI] [PubMed] [Google Scholar]
- 39.Pitterle D M, Rajagopalan K V. J Biol Chem. 1993;268:13499–13505. [PubMed] [Google Scholar]
- 40.Johnson M E, Rajagopalan K V. J Bacteriol. 1987;169:117–125. doi: 10.1128/jb.169.1.117-125.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallois P, Makishima T, Hecht V, Despres B, Laudie M, Nishimoto T, Cooke R. Plant J. 1997;11:1325–1331. doi: 10.1046/j.1365-313x.1997.11061325.x. [DOI] [PubMed] [Google Scholar]
- 42.Reiss, J., Christensen, E., Kurlemann, G., Zabot, M.-T. & Dorche, C. (1998) Hum. Genet. [DOI] [PubMed]
- 43.Becker C M. FASEB J. 1990;4:2767–2774. doi: 10.1096/fasebj.4.10.2165011. [DOI] [PubMed] [Google Scholar]