Abstract
We analyzed the possible causes of imipenem (IPM) resistance in multidrug-resistant isolates of Acinetobacter baumannii. Comparison of the outer membrane protein (OMP) profiles of two genomically related strains (Ab288 [IPM sensitive] and Ab242 [IPM resistant]) indicated the conspicuous loss of a 29-kDa polypeptide in the Ab242 strain. No carbapenemase activity was detected in any of these strains. The treatment of Ab288 with sodium salicylate resulted in IPM resistance and the loss of the 29-kDa OMP. In addition, IPM-resistant clones of Ab288 which were selected by repetitive culturing in increasing concentrations of this antibiotic also showed the absence of this 29-kDa OMP.
In the last few years, the number of infections caused by Acinetobacter baumannii has increased significantly, particularly among patients confined to hospital intensive care units, where the widespread use of antibiotics has resulted in the selection of strains with resistance to a wide range of antimicrobial agents (2, 3). Often, imipenem (IPM) remains one of the few therapeutic alternatives, a situation aggravated in recent years by the worldwide emergence of carbapenem-resistant isolates of this bacterium.
In this report, we characterized multidrug-resistant strains of A. baumannii which were isolated in Hospital Municipal Clemente Alvarez (HECA), Rosario City, Argentina. HECA is a public, 150-bed tertiary-care teaching hospital with an average attendance of 35,200 patients per year (mostly members of the low-income population of Rosario City and neighboring areas). The intensive care unit of this hospital is constituted by a 10-bed facility, to which around 730 patients are admitted per year. IPM therapy was introduced in 1996 and since that time has been extensively used in HECA for the treatment of multidrug-resistant bacteria.
Identification of A. baumannii was done by using the API 20NE identification system (bioMérieux, Lyon, France) and by accessing the ability of the bacteria to grow at 37, 41, and 44°C (4). MIC determinations were performed by using the agar dilution method on Mueller-Hinton agar (MHA; Oxoid, Basingstoke, United Kingdom) according to National Committee for Clinical Laboratory Standards guidelines (15). Carbapenemase activity was measured spectrophotometrically in crude extracts of bacteria disrupted with sonic treatment by employing 0.1 mM IPM as substrate (13). Outer membrane protein (OMP) fractions were prepared by the N-lauroyl sarcosine method from envelopes isolated from cells in the logarithmic phase of growth, following essentially the procedure described previously for Pseudomonas aeruginosa (10). OMP profiles were analyzed by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), employing a 10 to 18% polyacrylamide gradient (11). N-terminal sequence determination was done by Edman degradation of the electroblotted 29-kDa polypeptide onto polyvinylidene difluoride membranes (14) at the ICBR Protein Chemistry Core Facility, University of Florida, Gainesville, Fla.
Multidrug-resistant A. baumannii clinical strains isolated in HECA since 1997 were genotypically characterized by random PCR amplification (12), and two highly related isolates (Ab242 and Ab288) were selected for further study. The MICs obtained for both strains showed similar resistance to piperacillin (256 μg/ml), piperacillin-tazobactam (128 μg/ml), cefotaxime (32 μg/ml), gentamicin (16 μg/ml), and ciprofloxacin (16 μg/ml). Ab242 showed additional resistance to IPM (MIC of 16 μg/ml), while Ab288 was sensitive to this antibiotic (MIC of 0.5 μg/ml).
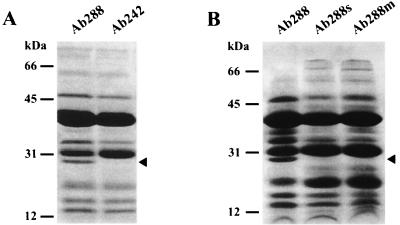
Imipenem resistance in A. baumannii isolates has been attributed to diverse causes, including alterations in the affinity of penicillin-binding proteins (9), the presence of carbapenemases (2, 8, 17, 19), a reduced expression of OMPs in the 33- to 36-kDa range (a situation that also conferred resistance to other antimicrobial agents [5]), and both synergistic effects among β-lactamases capable of hydrolyzing IPM and decreased expression of certain OMPs (3, 7). Since no carbapenemase activity could be detected by spectrophotometric analysis of either Ab242 or Ab288 (data not shown), we speculated whether IPM resistance could be ascribed to differences in the composition of OMPs between these strains. The corresponding patterns in SDS-PAGE analysis are shown in Fig. 1A. As seen in the figure, the OMP patterns of Ab288 and Ab242 were very similar except for the conspicuous absence of a 29-kDa OMP in the IPM-resistant Ab242 strain. A reduced content of a 36-kDa OMP was also observed in this strain (Fig. 1A), which could be related to phenomena reported by other authors (5, 7).
FIG. 1.
IPM resistance is linked to the loss of a 29-kDa OMP in A. baumannii. The corresponding purified outer membrane fractions (∼30 μg of protein) were incubated with 2% SDS for 5 min in a boiling water bath, subjected to SDS-PAGE as described in the text, and stained with Coomassie blue. (A) OMP profiles of IPM-sensitive (Ab288) and IPM-resistant (Ab242) clinical strains. (B) OMP profiles of Ab288, salicylate-treated Ab288 (Ab288s), and an IPM-resistant mutant clone of Ab288 (Ab288m). The closed arrowheads at the right margins indicate the position of the 29-kDa OMP, and the locations and molecular masses of the protein markers are indicated at the left margins. Corel DRAW 9.0 software was used.
We further analyzed whether IPM resistance could be linked to the loss of the above-mentioned 29-kDa OMP by using two different approaches. First, salicylate has been shown to repress expression of certain OMPs and to induce multiple antibiotic resistance in a number of gram-negative bacteria, including Escherichia coli and P. aeruginosa (6, 18). Thus, by following essentially the procedures described previously for P. aeruginosa (18), we examined whether growth of the IPM-sensitive Ab288 strain in the presence of this compound could reduce expression of the 29-kDa OMP and result in IPM resistance. This was found to be the case; the MICs of IPM for Ab288, as determined in MHA as described above, increased from 0.5 μg/ml (no salicylate) to 16 μg/ml in the presence of 16 mM sodium salicylate. In turn, growth of this strain in Luria-Bertani broth in the presence of 16 mM sodium salicylate resulted in the loss of the 29-kDa OMP, as determined by using SDS-PAGE analysis (Fig. 1B, lane Ab288s).
Second, an IPM-resistant mutant was obtained from Ab288 by successive selection in liquid media containing increasing concentrations of this antibiotic. For this purpose, Ab288 was first grown overnight in Luria-Bertani broth containing 0.5 μg of IPM/ml and then 0.1-ml aliquots of this culture were spread on MHA plates containing the same antibiotic concentration. Clones of resistant bacteria were reisolated in a second agar plate at the same IPM concentration, and the procedure was subsequently repeated employing 1, 2, 4, and 8 μg of IPM/ml, respectively. A clone resistant to 8 μg of IPM/ml after repeated isolation in agar plates was finally chosen for further study. The OMP pattern of this mutant (Fig. 1B, lane Ab288m) again indicated a disappearance of the 29-kDa polypeptide, reinforcing the idea that the loss of this protein is closely linked to IPM resistance.
The determination of the N-terminal region of this 29-kDa OMP revealed the following sequence for the first 24 aminoacid residues: Asp-Glu-Ala-Val-Val-His-Asp-Ser-Tyr-Ala-Phe-Asp-Lys-Asn-Gln-Leu-Ile-Pro-Val-Gly-Ala-Arg-Ala-Glu-. Employing the BLAST algorithm (1), a comparison of this sequence with those present in the GenBank database indicated that a region spanning residues 5 to 23 shares 36% identity and 78% overall homology with an internal region (residues 126 to 163) of a putative P. aeruginosa PAO1 OMP (accession no. AAG07977), a species closely related to A. baumannii in 16S rRNA phylogenetic trees (16). A similar search using the preliminary release of the Acinetobacter ADP1 genome (http://www.genoscope.cns.fr) indicated that this sequence also shares >80% identity with the N-terminal region of a predicted protein of 277 amino acid residues.
It is tempting to speculate that this 29-kDa OMP plays an important role in the transport of IPM across the outer bacterial membrane. This study and previous work by other authors (2, 3, 5, 7-9, 17, 19) provide evidence for the diversity of carbapenem resistance mechanisms evolved among Acinetobacter isolates. It still remains possible that a different interplay of these various factors operates in geographically separated strains. Further studies are being conducted to clarify both the role of the 29-kDa polypeptide and its putative interactions with other OMPs in conferring carbapenem resistance in A. baumannii.
Nucleotide sequence accession number.
The protein sequence information reported in this study is to appear in the SWISS-PROT and TrEMBL knowledge database under accession number P8346.
Acknowledgments
This work was supported by grants from the Subsecretaria de Investigación y Tecnología, Ministerio de Salud de la Nación Argentina; Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT); and Municipalidad de Rosario.
A.M.V. is a Career Investigator of the Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina, and M.A.M. is a Fellow of this institution. A.S.L. is an Adjunct Professor and Researcher of the National University of Rosario.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amyes, S. G. B., and H.-K. Young. 1996. Mechanisms of antibiotic resistance in Acinetobacter spp.—genetics of resistance, p. 185-223. In E. Bergogne-Berezin, M. L. Jolly-Guillou, and K. J. Towner (ed.), Acinetobacter: microbiology, epidemiology, infections, management. CRC Press, New York, N.Y.
- 3.Bou, G., G. Cerveró, M. A. Domínguez, C. Quereda, and J. Martínez-Beltrán. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of β-lactamases. J. Clin. Microbiol. 38:3299-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouvet, P. J. M., and P. A. Grimont. 1987. Identification and biotyping of clinical isolates of Acinetobacter. Ann. Inst. Pasteur/Microbiol. 138:569-578. [DOI] [PubMed] [Google Scholar]
- 5.Clark, R. B. 1996. Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33-36-kDa outer membrane protein. J. Antimicrob. Chemother. 38:245-251. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, S. P., S. B. Levy, J. Foulds, and J. L. Rosner. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 175:7856-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa, S. F., J. Woodcock, M. Gill, R. Wise, A. A. Barone, H. Caiafa, and A. S. S. Levin. 2000. Outer-membrane proteins pattern and detection of β-lactamases in clinical isolates of imipenem-resistant Acinetobacter baumannii from Brazil. Int. J. Antimicrob. Agents 13:175-182. [DOI] [PubMed] [Google Scholar]
- 8.Da Silva, G. J., G. J. Leitao, and L. Peixe. 1999. Emergence of carbapenem-hydrolyzing enzymes in Acinetobacter baumannii clinical isolates. J. Clin. Microbiol. 37:2109-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehrlein, M., H. Leying, W. Cullmann, S. Wendt, and W. Opferkuch. 1991. Imipenem resistance in Acinetobacter baumannii is due to altered penicillin-binding proteins. Chemotherapy 37:405-412. [DOI] [PubMed] [Google Scholar]
- 10.Hamzehpour, M. M., J.-C. Pecher, P. Plesiat, and T. Kohler. 1995. OprK and OprM define two genetically distinct multidrug efflux systems in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:2392-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 12.Limansky, A. S., and A. M. Viale. 2002. Can composition and structural features of oligonucleotides contribute to their wide-scale applicability as random PCR primers in mapping bacterial genome diversity? J. Microbiol. Methods 50:291-297. [DOI] [PubMed] [Google Scholar]
- 13.Matagne, A., A.-M. Misselyn-Baudin, B. Joris, T. Erpicum, B. Granier, and J.-M. Frere. 1990. The diversity of the catalytic properties of class A beta-lactamases. Biochem. J. 265:131-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsudaira, P. 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262:10035-10038. [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 16.Olsen, G. J., C. R. Woese, and R. Overbeek. 1994. The winds of (evolutionary) change: breathing new life into microbiology. J. Bacteriol. 176:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sumita, Y., and M. Fukasawa. 1993. Transient carbapenem resistance induced by salicylate in Pseudomonas aeruginosa associated with suppression of outer membrane protein D2 synthesis. Antimicrob. Agents Chemother. 37:2743-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi, A., S. Yomoda, I. Kobayashi, T. Okubo, M. Tsunoda, and S. Iyobe. 2000. Detection of carbapenemase-producing Acinetobacter baumannii in a hospital. J. Clin. Microbiol. 38:526-529. [DOI] [PMC free article] [PubMed] [Google Scholar]