Abstract
A closed-tube, real-time PCR assay was developed for sensitive and specific detection and differentiation of the two closely related intestinal protozoan parasites Entamoeba histolytica and Entamoeba dispar directly from human feces. The assay is performed with the LightCycler system using fluorescence-labeled detection probes and primers amplifying a 310-bp fragment from the high-copy-number, ribosomal DNA-containing ameba episome. The assay was able to detect as little as 0.1 parasite per g of feces. The two pairs of primers used were specific for the respective ameba species, and results were not influenced by the presence of other Entamoeba species even when present in exceeding amounts. PCR was evaluated using several hundred stool samples from areas of amebiasis endemicity in Vietnam and South Africa, and results were compared with those of microscopy and ameba culture. PCR was found to be significantly more sensitive than microscopy or culture, as all samples positive by microscopy and 22 out of 25 (88%) samples positive by culture were also positive by PCR, but PCR revealed a considerable number of additional E. histolytica- or E. dispar-positive samples. Compared to culture and subsequent ameba differentiation by isoenzyme analysis, PCR was 100% specific for each of the two Entamoeba species. Interestingly, the comparison with PCR revealed that culture, in particular, underestimates E. histolytica infections. Given the high sensitivity and specificity of the developed PCR assay, the inability of microscopy to distinguish between the two ameba species, and the time it takes to culture and subsequently differentiate entamoebae by isoenzyme analysis, this assay is more suitable than microscopy or culture to correctly diagnose intestinal E. histolytica or E. dispar infection.
The intestinal protozoan parasite Entamoeba histolytica is endemic in large parts of the world and is considered responsible for millions of cases of dysentery and liver abscess each year (32). Prior to invasive disease, E. histolytica may reside in the human gut for months or even years as asymptomatic infection (15, 37). To interrupt transmission of the parasite and to avoid progression of infected individuals to invasive disease, treatment of E. histolytica carriers has been recommended (36).
Traditionally, the laboratory detection of E. histolytica in human feces has relied upon the microscopic examination of fresh or fixed stool samples. However, the recent identification of Entamoeba dispar as a separate but nonpathogenic species which is morphologically indistinguishable from E. histolytica and does not require treatment has indicated the need of alternative detection methods which are able to differentiate between the two organisms (8). A number of assays have been developed during recent years, such as protein and DNA detection systems, which are able to distinguish E. histolytica from E. dispar (1, 2, 5, 7, 10, 14, 18, 21, 22, 24, 28, 29, 30). However, most of these assays are not suitable for a rapid diagnosis directly from stool samples, in particular when large numbers of samples have to be processed.
Amplification of ameba DNA fragments by PCR has been proven to constitute a sensitive and specific method to detect E. histolytica or E. dispar from human feces (1, 5, 14, 21, 30). The PCR protocols reported so far, however, require further processing of the amplicon, which is time-consuming and prone to false-positive results due to possible cross-contamination. Recently developed closed-tube, real-time PCR methods can circumvent these problems (35). These methods allow specific detection of the amplicon by binding to one or two fluorescence-labeled probes during PCR. Thus, further downstream analysis is not required, which reduces the time needed to obtain results. In addition, the closed reaction tube minimizes the potential for cross-contamination, and the assay output is numerical rather than qualitative, allowing appropriate diagnostic statistics to be applied.
Here we report the application of closed-tube, real-time PCR for the detection and differentiation of E. histolytica and E. dispar directly from fecal samples.
MATERIALS AND METHODS
Entamoeba strains and culture conditions.
The two E. histolytica isolates HK-9 and HM-1:IMSS (obtained from the American Type Culture Collection) and the two E. dispar isolates SAW142 and SAW760 (kindly provided by Peter Sargeaunt, London School of Hygiene and Tropical Medicine) were used to spike fecal samples. All isolates were cultured in TY-S-33 medium (9). The E. histolytica isolates were grown axenically, whereas the E. dispar isolates were grown monoxenically in the presence of Crithidia fasciculata (6).
Spiking of fecal samples.
Cultured ameba trophozoites were sedimented by centrifugation and resuspended in phosphate-buffered saline, pH 7.4. Cell density was measured by counting the amebae in a hemocytometer (Neubauer chamber) under the microscope. Serial dilutions of cells were mixed with 10 g of human feces prior to DNA extraction. Human feces microscopically free of any parasite were obtained from German residents who had never been in an area where amebiasis is endemic.
Real-time PCR.
DNA was extracted from human feces using the QIAamp DNA stool mini kit (Qiagen, Hilden, Germany), according to the manufacturer's protocol. Real-time PCR was performed using the LightCycler (34). Basic reagents for the LightCycler PCR were purchased from Roche Diagnostics, Mannheim, Germany. Oligonucleotide primers and probes (TibMolBiol, Berlin, Germany) were selected by comparing the available ribosomal DNA (rDNA) sequences from public databases using ClustalW (www2.ebi.ac.uk/clustalw/) and designed using the Oligo software (version 5.0; National Biosciences Inc., Plymouth, Minn.) to minimize primer dimer and other primer secondary structures. The 10-μl reaction mixture volume in a glass capillary tube contained 1 μl of FastStart reaction mix hybridization probes (a component of the FastStart DNA master hybridization probes kit; Roche Diagnostics), 1.2 μl of MgCl2 (25 mM), 3.8 μl of H2O, 1 μl each of sense and antisense primer (10 pmol/μl), 0.5 μl each of LC-Red 640- and fluorescein-labeled probe (4 pmol/μl), and 1 μl of DNA extract. Primers and probes for E. histolytica-specific PCR were Eh-S26C (5′-GTA CAA AAT GGC CAA TTC ATT CAA CG), Eh/Ed-AS25 (5′-GAA TTG ATT TTA CTC AAC TCT AGA G), Eh/Ed-24-LC-Red 640 (LC-Red 640-TCG AAC CCC AAT TCC TCG TTA TCC p), and Eh/Ed-25-fluorescein (fluorescein-GCC ATC TGT AAA GCT CCC TCT CCG A X). For E. dispar-specific PCR the same primers and probes were used except that Eh-S26C was replaced by Ed-27C, 5′-GTA CAA AGT GGC CAA TTT ATG TAA GCA. Reaction conditions were chosen according to a standard LightCycler protocol in our laboratory and were 5 min at 95°C, followed by 50 cycles of 10 s at 58°C and 20 s at 72°C. Temperature change rates were 20°C/s for the first two steps and 3°C/s for the last step. In addition, a touch-down PCR mode was incorporated in that the annealing temperature was stepwise decreased from 62 to 58°C by steps of 0.5°C within the first 8 of the 50 cycles. Readout was performed in channel F2/Back-F1. A sample was regarded as positive when the LightCycler software, version 3.5, determined a crossing point in the quantification analysis screen.
Stool microscopy, culture, and serology.
Stool samples were examined microscopically for the presence of protozoan parasites using the Formol-ether concentration technique and subsequent staining with Lugol's iodine solution (19, 20). Amebae were cultured from fecal samples and differentiated by isoenzyme analysis essentially as described by Sargeaunt and coworkers (24). Serum samples were investigated for the presence of antiameba antibodies using an enzyme-linked immunosorbent assay based on a recombinant E. histolytica surface antigen as previously described (17). This assay has been proven to be highly specific even when applied in countries where amebiasis is endemic, and the test is able to detect anti-E. histolytica antibodies for about 6 to 12 months after successful antiamebic treatment (16).
Statistical analysis.
All data collected were computer coded and analyzed by use of Sigma-Stat (SAS Software; Jandel Scientific, Erkrath, Germany). Binomial or χ2 tests were used for comparisons between two groups. A P value of <0.05 was considered to be significant.
RESULTS
Sensitivity and specificity of PCR for the detection of E. histolytica or E. dispar in fecal samples.
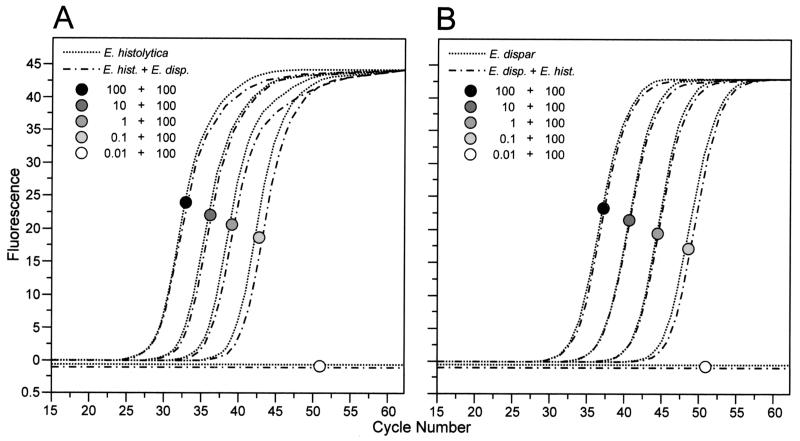
Sensitivity of the developed real-time PCR assay was evaluated using Entamoeba-negative human feces, which were spiked with various numbers of cultured E. histolytica or E. dispar trophozoites prior to DNA extraction and subjected to LightCycler PCR. Depending on the number of cells introduced, positive results were obtained after 20 to 45 cycles. For both ameba species, detection limits were 0.1 cell per g of feces (Fig. 1). Each of the two sets of primers used for PCR was specific and did not amplify DNA from the opposite ameba species. In addition, PCR was negative with both sets of primers when DNA was introduced which had been isolated from stool samples containing other Entamoeba species such as Entamoeba coli, Entamoeba hartmanni, or Entamoeba chattoni. Moreover, double infections with E. histolytica and E. dispar did not influence PCR results, as cross-contamination of E. histolytica-spiked fecal samples with E. dispar trophozoites or vice versa in a ratio between 1:1 and 1:10,000 produced virtually identical results as did cases without contamination (Fig. 1).
FIG. 1.
Detection of E. histolytica or E. dispar from human feces by LightCycler PCR. An Entamoeba-negative stool sample was spiked with various numbers of cultured E. histolytica or E. dispar trophozoites prior to DNA extraction and subjected to PCR. One microliter of DNA extract was introduced into the PCR representing 0.01 to 100 cells per g of feces (dotted line). In parallel experiments, E. histolytica-spiked samples were contaminated with E. dispar and vice versa in a ratio between 1:1 and 1:10,000 as indicated (dashed line). Shown are the LightCycler PCR results obtained by using template DNA of the respective ameba species as indicated. (A) PCR with primers specific for E. histolytica; (B) PCR with primers specific for E. dispar.
Comparison of PCR with microscopy and serology.
A total of 491 fecal samples collected from residents of an area with a high incidence of amebic liver abscess (ALA) in Hué, Vietnam (4), were subjected to PCR analysis. Outcome was compared with the results of microscopy and antiamebic antibody status (Table 1). Prevalence of E. histolytica and E. dispar was found to be 9.2% (45 of 491) based on microscopy versus 14.3% (70 of 491) according to the results of PCR (P = 0.02). Remarkably, all samples positive by microscopy were also positive by PCR. There was a good correlation between the number of PCR cycles required in order to reach the crossing point and the relative parasite burden. In general, PCR became positive within 20 to 30 cycles when relatively high numbers of amebae were seen under the microscope, whereas more than 40 cycles were required with samples containing very low parasite numbers. There was a strong association between PCR and microscopy, as none of the samples negative by PCR were positive by microscopy and 64.3% (45 of 70) of PCR-positive samples were also positive by microscopy (P < 0.001). Differentiation into E. histolytica and E. dispar by PCR revealed 9.4% (46 of 491) of samples to be positive for E. histolytica versus 4.9% (24 of 491) positive for E. dispar (P < 0.01). The association between positive PCR and microscopy was slightly but not significantly higher for E. dispar than for E. histolytica (66.7 versus 63.0%; P = 0.93). Interestingly, PCR revealed no double infection with E. histolytica and E. dispar.
TABLE 1.
Comparison of fecal PCR with microscopy and serologya
| Specimen group (n) | No. (%) of specimens with result |
|
|---|---|---|
| Positive microscopy for E. histolytica and E. dispar | Positive serology for E. histolytica | |
| All samples (491) | 45 (9.2) | 120 (24.4) |
| E. histolytica-positive PCR (46) | 29 (63.0)* | 38 (82.6)* |
| E. dispar-positive PCR (24) | 16 (66.7)* | 5 (20.8)** |
| E. histolytica- and E. dispar- negative PCR (421) | 0* | 77 (18.3)*** |
Symbols for significant difference compared to all samples: *, P < 0.001; ***, P < 0.03; **, not significantly different(P = 0.63).
In addition to microscopy, a strong association was found between E. histolytica-positive PCR and serology. A significant antiameba antibody titer was present in 24.4% (120 of 491) of all individuals tested but in 82.6% (38 of 46) of individuals with a positive E. histolytica fecal PCR result (P < 0.001). In contrast, individuals with fecal PCR results positive for E. dispar or negative for both organisms revealed positive serology in only 20.8% (5 of 24) and 18.3% (77 of 421) of cases, respectively (P = 0.79).
Comparison of PCR with Entamoeba culture.
Ameba culture and subsequent isoenzyme analysis is considered to be the “gold standard” for the differentiation between E. histolytica and E. dispar from fecal samples. Thus, results of ameba culture were compared with those of PCR (Table 2). DNA was extracted from 181 fecal samples, which were collected during a study in Durban, South Africa. Parallel samples were taken into culture, and those cultures positive for E. histolytica and/or E. dispar were subjected to isoenzyme analysis for further differentiation. Of the 181 samples, 81 were obtained from patients with a recent history of ALA. These samples were collected 3 to 9 months after successful ALA treatment with metronidazole. The remaining 100 samples were collected from family members of the various ALA patients living under similar conditions (controls). Culture and isoenzyme analysis revealed a prevalence of 7.2% (13 of 181) for E. histolytica and of 6.6% (12 of 181) for E. dispar. Eleven of the 13 positive E. histolytica cultures were from recent ALA patients, whereas the majority of positive E. dispar cultures (10 out of 12) were from the group of controls. Comparison with PCR revealed a good concordance between culture and fecal PCR, as 12 out of 13 (92.3%) samples with a positive E. histolytica culture and 10 out of 12 (83.3%) samples with a positive E. dispar culture were also positive by the respective PCR. None of the samples with an E. histolytica-positive culture was positive by E. dispar-specific PCR and vice versa. However, PCR identified an additional 21 E. histolytica-positive and two E. dispar-positive samples, which were negative by culture. All of the additional E. dispar-positive samples were from the group of controls, whereas the majority of additional E. histolytica-positive samples (14 of 21, P < 0.02) were from the group of recent ALA patients.
TABLE 2.
Comparison of fecal PCR with culture and isoenzyme analysisa
| Specimen group (n) | No. (%) of specimens with result(s) |
|||||
|---|---|---|---|---|---|---|
|
E. histolytica |
E. dispar |
|||||
| Culture positive | PCR positive | Culture and PCR positive | Culture positive | PCR positive | Culture and PCR positive | |
| All samples (181) | 13 (7.2) | 33 (18.2)* | 12 (6.6) | 12 (6.6) | 12 (6.6) | 10 (5.5) |
| ALA patients (81) | 11 (13.6) | 24 (29.6)** | 10 (12.3) | 2 (2.5) | 2 (2.5) | 2 (2.5) |
| Controls (100) | 2 (2.0) | 9 (9.0) | 2 (2.0) | 10 (10.0) | 10 (10.0) | 8 (8.0) |
Symbols for significant difference compared to culture: *, P < 0.001; **, P < 0.03.
DISCUSSION
The recent identification of E. dispar as a separate but nonpathogenic species which cannot be distinguished by microscopy from pathogenic E. histolytica has prompted the World Health Organization to recommend the development of improved methods for the specific identification of E. histolytica from human feces (36). In this respect, we have evaluated the application of closed-tube, real-time PCR technology for the detection and differentiation of E. histolytica and E. dispar directly from stool samples. The LightCycler PCR method employed here is fast, because it takes only 1 h to process 30 samples, and it is specific, as it requires two primers as well as two fluorescence-labeled probes for DNA detection. DNA was extracted from human feces by a commercially available kit, which has been proven to allow reproducible preparations of PCR-amplifiable DNA, as >99% of all extracted samples do not contain inhibitory activity (30). This was confirmed in this study, as 99.1% of all samples negative for E. histolytica or E. dispar by PCR were PCR positive with primers amplifying a fragment of the human HLA-DRB gene (27). The rDNA sequence was selected as the target DNA for amebic PCR because it is well conserved between different E. histolytica isolates as well as between different E. dispar isolates (7) and because it is located on an episomal plasmid, which is present in about 200 copies per cell (3). Compared to chromosomal DNA the episomal plasmid is less sensitive to DNA degradation, and the high copy number should allow sensitive detection even if only a few cells are present. Sensitivity was considered further improved by selecting a relatively small fragment (310 bp only) for PCR amplification. However, rDNA sequences of E. histolytica and E. dispar share 98.4% nucleotide identity (25). Thus, only very few sites are suitable for the construction of appropriate primers for PCR differentiation. We took advantage of a considerable number of nucleotide polymorphisms around position 190 of ameba rDNA sequences, which represents one of the two sites where, compared to E. histolytica, the E. dispar sequence contains a nucleotide insertion. In addition, a point mutation was introduced into each of the specific sense primers, in that a thymidine was replaced by a cytidine residue. This created three consecutive mismatches between the 3′ ends of each primer and the respective rDNA sequences of the opposite Entamoeba species, which in the case of “cross priming” will strongly prevent DNA elongation by the polymerase (33). Our results with spiked fecal samples indicate that the two sets of primers are indeed species specific and that PCR is not influenced by the presence of considerable amounts of other Entamoeba species. In addition, PCR is highly sensitive, as as little as 0.1 cell per g of feces could be detected. The high sensitivity and specificity of PCR was further confirmed by comparison with stool microscopy and ameba culture. All 45 samples positive by microscopy and 22 out of 25 (88%) samples positive by culture were also positive by PCR. According to isoenzyme analysis, PCR classified all samples correctly into E. histolytica and E. dispar, respectively. Moreover, positive PCR for E. histolytica was strongly associated with ameba serology, which is consistent with previous findings indicating that in contrast to E. dispar, E. histolytica intestinal infection, even if asymptomatic, usually induces a significant systemic antiamebic antibody response (13, 23, 29, 31). However, compared to microscopy or culture, PCR identified a considerably larger number of additional positive samples, suggesting that PCR is more sensitive. Sensitivity of microscopy for the detection of Entamoeba by examination of a single fecal sample is considered to be about 70% (11). This value is in good agreement with the results of PCR, as regardless of whether E. histolytica or E. dispar was identified, about 65% of PCR-positive samples were positive by microscopy. The sensitivity of ameba culture has not been established unambiguously, but is considered to be higher than that of microscopy (19). Interestingly, sensitivity for the detection of E. dispar was identical between PCR and culture, but detection of E. histolytica was significantly higher by PCR. As for both ameba species the associations of PCR with microscopy and the PCR detection limits were identical, the results strongly suggest that culture in particular underestimates E. histolytica infections.
According to PCR about 30% of previous ALA patients were found to be positive for E. histolytica. Whether this relatively high number is the result of a high rate of reinfection in these individuals or whether this is due to ineffective treatment remains to be determined. However, the latter is more likely, as all patients were only treated with metronidazole and none of them received a luminal antiamebic agent such as diloxanide furoate or paromomycin. A 30% E. histolytica prevalence in previous ALA patients 3 to 9 months after metronidazole therapy would be fully consistent with previous findings, which indicated a cure rate of intestinal infection by metronidazole of about 50% (12, 26).
Another interesting result that came out of this study was the absence of any double infection with E. histolytica and E. dispar. This was obviously not due to technical problems, as the two specific sets of primers used for PCR were suitable to detect double infections, even if one of the two species were greatly underrepresented. Thus, it is intriguing to speculate whether intestinal infection with one of the two species is able to prevent colonization with the other one. However, to reach a definite conclusion a larger number of samples has to be analyzed.
Taken together, our results indicate that the PCR protocol presented here is suitable for the detection and differentiation of E. histolytica and E. dispar directly from human feces. Given the many advantages of closed-tube, real-time PCR technology, the high sensitivity and specificity of the developed PCR assay, the inability of microscopy to distinguish between the two ameba species, and the time it takes to culture and subsequently differentiate Entamoeba species by isoenzyme analysis, it is obvious that this protocol or similar protocols are substantially more appropriate than microscopy or culture to correctly diagnose intestinal E. histolytica or E. dispar infections. On the other hand, although the costs for reagents to extract DNA from human feces and to perform PCR are less than $7 per sample, at present, the application of this method is limited to specifically equipped laboratories running the LightCycler.
Acknowledgments
This study was supported by the Volkswagen Foundation.
REFERENCES
- 1.Acuna-Soto, R., J. Samuelson, P. De Girolami, L. Zarate, F. Millan-Velasco, G. Schoolnick, and D. Wirth. 1993. Application of the polymerase chain reaction to the epidemiology of pathogenic and nonpathogenic Entamoeba histolytica. Am. J. Trop. Med. Hyg. 48:58-70. [DOI] [PubMed] [Google Scholar]
- 2.Aguirre, A., D. C. Warhurst, F. Guhl, and I. A. Frame. 1995. Polymerase chain reaction-solution hybridization enzyme-linked immunoassay (PCR-SHELA) for the differential diagnosis of pathogenic and non-pathogenic Entamoeba histolytica. Trans. R. Soc. Trop. Med. Hyg. 89:187-188. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya, S., I. Som, and A. Bhattacharya. 1994. The ribosomal DNA plasmids of Entamoeba. Parasitol. Today 14:181-185. [DOI] [PubMed] [Google Scholar]
- 4.Blessmann, J., P. Van Linh, P. A. Nu, H. D. Thi, B. Muller-Myhsok, H. Buss, and E. Tannich. 2002. Epidemiology of amebiasis in a region of high incidence of amebic liver abscess in central Vietnam. Am. J. Trop. Med. Hyg. 66:578-583. [DOI] [PubMed] [Google Scholar]
- 5.Britten, D., S. M. Wilson, R. McNerney, A. H. Moody, P. L. Chiodini, and J. P. Ackers. 1997. An improved colorimetric PCR-based method for detection and differentiation of Entamoeba histolytica and Entamoeba dispar in feces. J. Clin. Microbiol. 35:1108-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, C. G., C. C. Cunnick, and L. S. Diamond. 1992. Entamoeba histolytica: is conversion of nonpathogenic amebae to the pathogenic form a real phenomenon? Exp. Parasitol. 74:307-314. [DOI] [PubMed] [Google Scholar]
- 7.Clark, C. G., and L. S. Diamond. 1991. Ribosomal RNA genes of ‘pathogenic’ and ‘nonpathogenic’ Entamoeba histolytica are distinct. Mol. Biochem. Parasitol. 49:297-302. [DOI] [PubMed] [Google Scholar]
- 8.Diamond, L. S., and R. D. Clark. 1993. A rediscription of Entamoeba histolytica Schaudinn, 1903 (emended Walker, 1911) separating it from Entamoeba dispar Brumpt, 1925. J. Eukaryot. Microbiol. 40:340-344. [DOI] [PubMed] [Google Scholar]
- 9.Diamond, L. S., D. R. Harlow, and C. C. Cunnick. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431-432. [DOI] [PubMed] [Google Scholar]
- 10.Haque, R., L. M. Neville, P. Hahn, and W. A. Petri, Jr. 1995. Rapid diagnosis of Entamoeba infection by using Entamoeba and Entamoeba histolytica stool antigen detection kits. J. Clin. Microbiol. 33:2558-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiatt, R. A., E. K. Markell, and E. Ng. 1995. How many stool examinations are necessary to detect pathogenic intestinal protozoa. Am. J. Trop. Med. Hyg. 53:36-39. [PubMed] [Google Scholar]
- 12.Irusen, E. M., T. F. Jackson, and A. E. Simjee. 1992. Asymptomatic intestinal colonization by pathogenic Entamoeba histolytica in amebic liver abscess: prevalence, response to therapy, and pathogenic potential. Clin. Infect. Dis. 14:889-893. [DOI] [PubMed] [Google Scholar]
- 13.Jackson, T. F., V. Gathiram, and A. E. Simjee. 1985. Seroepidemiological study of antibody responses to the zymodemes of Entamoeba histolytica. Lancet 30:716-719. [DOI] [PubMed] [Google Scholar]
- 14.Katzwinkel-Wladarsch, S., T. Loscher, and H. Rinder. 1994. Direct amplification and differentiation of pathogenic and nonpathogenic Entamoeba histolytica DNA from stool specimens. Am. J. Trop. Med. Hyg. 51:115-118. [DOI] [PubMed] [Google Scholar]
- 15.Knobloch, J., and E. Mannweiler. 1983. Development and persistence of antibodies to Entamoeba histolytica in patients with amebic liver abscess: analysis of 216 cases. Am. J. Trop. Med. Hyg. 32:727-732. [DOI] [PubMed] [Google Scholar]
- 16.Lotter, H., T. F. G. H. Jackson, and E. Tannich. 1995. Evaluation of three serological tests for the detection of antiamebic antibodies applied to sera of patients from an area endemic for amebiasis. Trop. Med. Parasitol. 71:401-407. [PubMed] [Google Scholar]
- 17.Lotter, H., E. Mannweiler, M. Schreiber, and E. Tannich. 1992. Sensitive and specific serodiagnosis of invasive amoebiasis by using a recombinant surface protein of pathogenic Entamoeba histolytica. J. Clin. Microbiol. 30:3163-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackenstedt, U., and A. M. Johnson. 1995. Genetic differentiation of pathogenic and nonpathogenic strains of Entamoeba histolytica by random amplified polymorphic DNA polymerase chain reaction. Parasitol. Res. 81:217-221. [DOI] [PubMed] [Google Scholar]
- 19.McMillan, A., and G. J. McNeillage. 1984. Comparison of the sensitivity of microscopy and culture in the laboratory diagnosis of intestinal protozoal infection. J. Clin. Pathol. 37:809-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moody, A. H. 1996. Laboratory diagnosis, p. 1738-1742. In G. C. Cook (ed.), Manson's tropical diseases, 20th ed. W. B. Saunders Company, Philadelphia, Pa.
- 21.Nunez, Y. O., M. A. Fernandez, D. Torres-Nunez, J. A. Silva, I. Montano, J. L. Maestre, and L. Fonte. 2001. Multiplex polymerase chain reaction amplification and differentiation of Entamoeba histolytica and Entamoeba dispar DNA from stool samples. Am. J. Trop. Med. Hyg. 64:293-297. [DOI] [PubMed] [Google Scholar]
- 22.Pillai, D. R., and K. C. Kain. 1999. Immunochromatographic strip-based detection of Entamoeba histolytica-E. dispar and Giardia lamblia coproantigen. J. Clin. Microbiol. 37:3017-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravdin, J. I., T. F. H. G. Jackson, W. A. Petri, Jr., C. F. Murphy, B. L. P. Ungar, V. Gathiram, J. Skilogiannis, and A. E. Simjee. 1990. Association of serum antibodies to adherence lectin with invasive amebiasis and asymptomatic infection with pathogenic Entamoeba histolytica. J. Infect. Dis. 162:768-772. [DOI] [PubMed] [Google Scholar]
- 24.Sargeaunt, P. G., J. E. Williams, and J. D. Grene. 1978. The differentiation of invasive and non-invasive Entamoeba histolytica by isoenzyme electrophoresis. Trans. R. Soc. Trop. Med. Hyg. 72:519-521. [DOI] [PubMed] [Google Scholar]
- 25.Silberman, J. D., C. G. Clark, L. S. Diamond, and M. L. Sogin. 1999. Phylogeny of the genera Entamoeba and Endolimax as deduced from small-subunit ribosomal RNA sequences. Mol. Biol. Evol. 16:1740-1751. [DOI] [PubMed] [Google Scholar]
- 26.Spillmann, R., S. C. Ayala, and C. E. Sanchez. 1978. Double-blind test of metronidazole and tinidazole in the treatment of asymptomatic Entamoeba histolytica and Entamoeba hartmanni carriers. Am. J. Trop. Med. Hyg. 25:549-551. [DOI] [PubMed] [Google Scholar]
- 27.Spurkland, A., I. Knutsen, G. Markussen, F. Vartdal, T. Egeland, and E. Thorsby. 1993. HLA matching of unrelated bone marrow transplant pairs: direct sequencing of in vitro amplified HLA-DRB1 and -DQB1 genes using magnetic beads as solid support. Tissue Antigens 41:155-164. [DOI] [PubMed] [Google Scholar]
- 28.Tachibana, H., S. Ihara, S. Kobayashi, Y. Kaneda, T. Takeuchi, and Y. Watanabe. 1991. Differences in genomic DNA sequences between pathogenic and nonpathogenic isolates of Entamoeba histolytica identified by polymerase chain reaction. J. Clin. Microbiol. 29:2234-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tannich, E., and G. D. Burchard. 1991. Differentiation of pathogenic from nonpathogenic Entamoeba histolytica by restriction fragment analysis of a single gene amplified in vitro. J. Clin. Microbiol. 29:250-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verweij, J. J., J. Blotkamp, E. A. Brienen, A. Aguirre, and A. M. Polderman. 2000. Differentiation of Entamoeba histolytica and Entamoeba dispar cysts using polymerase chain reaction on DNA isolated from faeces with spin columns. Eur. J. Clin. Microbiol. Infect. Dis. 19:358-361. [DOI] [PubMed] [Google Scholar]
- 31.Verweij, J. J., L. Van Lieshout, C. Blotkamp, E. A. T. Brienen, S. Van Duivenvoorden, M. Van Esbroek, and A. M. Polderman. 2000. Differentiation of Entamoeba histolytica and Entamoeba dispar using PCR-SHELA and comparison with antibody response. Arch. Med. Res. 31(Suppl. 4):44-46. [DOI] [PubMed] [Google Scholar]
- 32.Walsh, J. A. 1986. Problems in recognition and diagnosis of amebiasis. Estimation of the global magnitude of morbidity and mortality. Rev. Infect. Dis. 66:228-238. [DOI] [PubMed] [Google Scholar]
- 33.Whitcombe, D., J. Theaker, S. P. Guy, T. Brown, and S. Little. 1999. Detection of PCR products using self-probing amplicons and fluorescence. Nat. Biotechnol. 17:804-807. [DOI] [PubMed] [Google Scholar]
- 34.Wittwer, C. T., K. M. Ririe, R. V. Andrew, D. A. David, R. A. Gundry, and U. J. Balis. 1997. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques 22:176-181. [DOI] [PubMed] [Google Scholar]
- 35.Wittwer, C. T., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22:130-138. [DOI] [PubMed] [Google Scholar]
- 36.World Health Organization. 1997. Amebiasis. W. H. O. Wkly. Epidemiol. Rec. 72:97-100. [Google Scholar]
- 37.Wynants, H., J. Van den Ende, J. Randria, A. Van Gompel, E. Van den Enden, C. Brands, P. Coremans, P. Michielsen, L. Verbist, and R. Colebunders. 1995. Diagnosis of amoebic infection of the liver: report of 36 cases. Ann. Soc. Belg. Med. Trop. 75:297-303. [PubMed] [Google Scholar]