Abstract
In this study, we determined the sequences of four intimin variant genes detected in attaching and effacing Escherichia coli isolates of human origin. Three of them were novel and were designated eae-η (eta), eae-ι (iota), and eae-κ (kappa). The fourth was identical to the recently described eae-ζ (zeta), isolated from a bovine E. coli O84:NM isolate. We compared these sequences with those of published intimin-α, intimin-β, intimin-γ1, intimin-γ2, intimin-ɛ, and intimin-θ alleles. Sequence analysis of these 10 intimin alleles confirmed extensive genetic diversity within the intimin gene family in E. coli. The genetic diversity was more prominent in the 3′ region (starting at bp 2112), which encodes the binding domain of intimin. Phylogenetic analyses revealed four groups of closely related intimin genes: α and ζ; β and κ; γ1 and γ2/θ; and ɛ and η. Calculation of homoplasy ratios of sequences of the 5′ region of eae (positions 1 to 2111) revealed evidence for intragenic recombination. Split decomposition analysis also indicates that recombination events have played a role in the evolutionary history of eae. In conclusion, we recommend an eae nomenclature system based on the Greek alphabet and provide an updated PCR scheme for amplification and typing of E. coli eae.
Attaching and effacing Escherichia coli (AEEC) strains have a similar mode of interaction to epithelial cells: following initial adherence to epithelial cell surfaces, they exert their pathological effects by causing histopathological alterations termed attaching and effacing (A/E) lesions (13). A/E lesions result from the destruction of the microvillus brush border through restructuring of the underlying cytoskeleton by signal transduction between bacterial and host cells. The ability to cause A/E lesions is encoded on a large pathogenicity island, the locus of enterocyte effacement (LEE) (13, 23). LEE is usually integrated adjacent to either the selC, pheU, or pheV tRNA loci and consists of three functionally different modules (17, 23, 29, 35). A type III secretion system exports effector molecules and is encoded at one end of the island. The secreted proteins EspA, EspB, and EspD, which function as part of the type III secretion apparatus, are encoded at the opposite end of LEE. The central portion of LEE encodes intimin (Eae), which mediates intimate attachment to the host cell, and Tir, the intimin receptor, which is chaperoned by CesT and translocated into the host cell plasma membrane by the type III system (6, 13).
The intimin-encoding eae gene was sequenced initially from enteropathogenic E. coli (EPEC) strain E2348/69 (16) and later from enterohemorrhagic E. coli (EHEC) strain EDL933 (43). The 5′ regions of both genes are conserved, whereas the 3′ regions are heterogeneous. This observation led to the construction of universal PCR primers (SK1 and SK2) and allele-specific PCR primers (LP1 and LP2, and LP1 and LP3), which made it possible to differentiate between the two intimin genes (32, 34).
The C-terminal end of intimin is responsible for receptor binding, and it has been suggested that different intimins may be responsible for different host tissue cell tropism (30). In a recent work, Fitzhenry et al. (11) showed that intimin gamma appears to restrict colonization of EHEC to human follicle-associated epithelium.
Molecular analyses by Adu-Bobie et al. (2) yielded additional intimin types; these workers defined the groups α, β, γ, and δ. In another molecular study, nucleotide sequence analysis revealed the novel eae type ɛ, which was found predominantly in E. coli strains of serogroup O103 (28). All intimin alleles currently known demonstrate more similarity in their 5′ regions than in their 3′ regions. Intimin alleles α, β, and γ have been subgrouped by restriction analysis into α1-α2, β1-β2 and γ1-γ2 groups (28). The β2 allele was also termed δ by Adu-Bobie et al. (2). Recently, Tarr and Whittam (39) presented a paper on the molecular evolution of intimin genes in E. coli O111 clones and defined the new group eae-θ. They suggested that amino acid substitutions that alter the residue charge occur more frequently than would be expected under random substitution in the C-terminal domain. As well as these, some further intimin genes, such as eae from Citrobacter rodentium, have been characterized (4).
In this paper, we report three new eae variants from human AEEC isolates, recommend PCR protocols and reference strains for detection of these types of genes and perform phylogenetic analyses on the basic eae types currently known. Only published E. coli eae sequences were included in our analyses.
MATERIALS AND METHODS
Bacterial strains.
AEEC strains 4795/97 (stx1+, O84:H4), 7476/96 (stx, O145:H4), and 6044/95 (stx, O118:H5) were isolated from patient stools in Würzburg, Germany, during routine diagnostic work. EPEC O125:H− strain CF11201 (stx) was isolated from the stools of a child with diarrhea in Burundi, Africa (12). EPEC O127:H6 strain E2348/69 (20) was used as a reference strain for eae-α, rabbit-pathogenic E. coli O15:H− strain RDEC-1 (44) was used as a reference strain for eae-β, EHEC O157:H7 strain EDL933 (27) was used as a reference strain for eae-γ1, E. coli O111:H− strain 95NR1 (42) was used as a reference strain for eae-γ2, EHEC O103:H2 strain PMK5 (22) was used as a reference strain for ɛ- eae, and EHEC O111:H8 strain CL-37 was used as a reference for eae-θ (39). As a reference for eae-ζ, Stx-producing O84:H4 strain 4795/97 was used (Table 1). Bacterial strains were routinely grown in Luria broth at 37°C with vigorous shaking at 37°C.
TABLE 1.
Designations, sequence characteristics, and origin of the intimin alleles used in this study
| Designation of intimin allele | ORF length (bp) | G+C content (%) | Reference strain (serotype) | Accession no. |
|---|---|---|---|---|
| α (alpha) | 2,820 | 42.09 | E2348/69 (O127:H6) | M58154 |
| β (beta) | 2,820 | 42.98 | RDEC-1 (O15:H−) | AF200363 |
| γ 1 (gamma 1) | 2,805 | 42.67 | EDL933 (O157:H7) | Z11541.1 |
| γ 2 (gamma 2) | 2,808 | 42.95 | 95NR1 (O111:H−) | AF025311 |
| ɛ (epsilon) | 2,847 | 42.78 | PMK5 (O103:H2) | AF116899 |
| ζ (zeta) | 2,817 | 42.60 | 537/89 (O84:NM) | AJ298279 |
| 4795/95 (O84:H4) | AJ271407 | |||
| η (eta) | 2,847 | 42.50 | CF11201 (O125:H−) | AJ308550 |
| θ (theta) | 2,808 | 43.07 | CL37 (O111:H8) | AF449418 |
| ι (iota) | 2,814 | 41.70 | 7476/96 (O145:H4) | AJ308551 |
| κ (kappa) | 2,820 | 42.06 | 6044/95 (O118:H5) | AJ308552 |
DNA techniques.
DNA fragments carrying novel intimin genes were analyzed by amplification of a portion of the LEE using primers orfU-upper (5′-TAT GAT GAT CTA TGG CGT CTG T-3′) and escD-lower (5′-TAT TTT CAA AAA GAA TGA TGT C-3′) as described previously (28). The PCR conditions for amplification of intimin alleles from AEEC were as described in Table 2 except for SK1/LP3: here we used 2.0 mM MgCl2 and 2.5 U of Taq polymerase and increased the number of cycles to 31. Three cycles were performed with a low annealing temperature and 28 cycles were performed with a higher one, as described in Table 2.
TABLE 2.
PCR primers and conditions for amplification of intimin alleles
| Primerc | Sequence of primer (5′ → 3′) | Target sequence | Reference strain(s) | PCR program | Size of product (bp) | Reference |
|---|---|---|---|---|---|---|
| SK1 | CCCGAATTCGGCACAAGCATAAGC | Conserved re- gion of eae | 94°C, 30 s; 52°C, 60 s; 72°C, 60 sa,b | 863 | 32 | |
| SK2 | CCCGGATCCGTCTCGCCAGTATTCG | EDL933, E2348/69 | ||||
| LP2 | CCCGAATTCTTATTTTACACAAGTGGC | eae-α | E2348/69 | 94°C, 30 s; 55°C, 60 s; 72°C, 120 sa,b | 2,807 | 33 |
| LP3 | CCCGAATTCTTATTCTACACAAACCGC | eae-γ | EDL933 | 94°C, 30 s; 48°C, 60 s; 72°C, 90 sa,b | 2,792 | 33 |
| 94°C, 30 s; 55°C, 60 s; 72°C, 90 sb,e | ||||||
| LP4 | CCCGTGATACCAGTACCAATTACGGTC | eae-β | RDEC-1 | 94°C, 30 s; 55°C, 60 s; 72°C, 120 sa,b | 2,287 | 28 |
| LP5 | AGCTCACTCGTAGATGACGGCAAGCG | eae-ɛ | PMK5 | 94°C, 30 s; 55°C, 60 s; 72°C, 120 sa,b | 2,608 | 28 |
| LP6B | TAGTTGTACTCCCCTTATCC | eae-ζ | 4795/95 | 94°C, 30 s; 53°C, 60 s; 72°C, 150 sa,b | 2,430 | This study |
| LP7 | TTTATCCTGCTCCGTTTGCT | eae-ι | 7476/97 | 94°C, 30 s; 52°C, 60 s; 72°C, 150 sa,b | 2,685 | This study |
| LP8 | TAGATGACGGTAAGCGAC | eae-η | CF11201 | 94°C, 30 s; 52°C, 60 s; 72°C, 150 sa,b | 2,590 | This study |
| LP10 | GGCATTGTTATCTGTTGTCT | eae-κ | 6044/95 | 94°C, 30 s; 52°C, 60 s; 72°C, 150 sa,b | 2,769 | This study |
| LP11B | GTTGATAACTCCTGATATTTTA | eae-θ | CL-37 | 94°C, 30 s; 50°C, 60 s; 72°C, 150 sa | 2,686 | This study |
| 94°C, 30 s; 50°C, 60 s; 72°C, 150 sb |
Before the first cycle the sample was denatured for 2 min at 94°C.
After the last cycle, the sample was extended for 5 min at 72°C.
Primer SK1 was used as forward primer in all PCR approaches in combination with SK2, LP2, LP3, LP4, LP5, LP6B, LP7, LP8, LP10, and LP11B.
Three cycles
Twenty-eight cycles.
Phylogenetic analyses.
DNA sequences were edited with BioEdit, version 4.8.10 (http://jwbrown.mbio.ncsu.edu/BioEdit/bioedit.html) (14) and converted into Mega and Nexus files with START (Sequence Type Analysis and Recombinational Tests; http://outbreak.ceid.ox.ac.uk/software.htm). Phylogenetic trees were compiled with Mega 2.1 (http://www.megasoftware.net/) using the unweighted pair group method with arithmetic mean UPGMA (19). Assessment of natural selection was also performed with Mega 2.1 by determining proportions of synonymous differences per synonymous site (pS) and proportions of nonsynonymous differences per nonsynonymous site (pN) by the implemented Nei- Gojobori method. These quantities were used previously to detect adaptive evolution (26). Evolution rates per site were expected to be equal for neutral mutation (pN = pS), whereas pN exceeds pS for purifying (negative) selection (pN > pS), and pS exceeds pN for diversifying (positive) selection (pS > pN). Aligning softwares were BioEdit version 4.8.10, Seqverter (http://www.gene-studio.com/seqverter.htm), and Clustal (HUSAR software package; http://genome.dkfz-heidelberg.de/). Genetic distances were calculated with ClustalW included in the BioEdit software (40). To detect and quantify intragenic recombination of intimin alleles, homoplasy tests were performed with Homoplasy software (38). As a second method of analyzing the gene and population structure for evidence of recombination, split decomposition analysis (3, 15) was performed with Splitstree 2.0 (http://bibiservchfak.uni-bielefeld.de/splits/). Numerical analysis of polymorphic sites was performed with DnaSP (31), and graphical analysis was performed with Happlot (http://www.shigatox.net/stec/programs/happlot/).
Nucleotide sequence accession numbers.
The sequences of eae-ζ (4795/97), eae-η, eae-ι, and eae-κ were submitted to the European Bioinformatics Institute (EBI) and have been assigned accession numbers AJ271407, AJ308550, AJ308551, and AJ308552, respectively.
RESULTS
Detection of novel members of the intimin gene family.
All intimin genes of E. coli isolates from patients with diarrhea which were positive with universal eae primers SK1 and SK2 were routinely subtyped with primer SK1 in combination with LP2, LP3, LP4, or LP5 as described earlier (28). E. coli strains which were PCR negative with these four specific primer combinations were considered to carry novel intimin variant genes. Four of these, O84:H4 strain 4795/97, O145:H4 strain 7476/96, O125:H− strain CF11201, and O118:H5 strain 6044/95, were chosen for further analysis. An approximately 3.8-kb region of the LEE including eae (orfU-escD region) was amplified and sequenced from each of these strains as described previously (28). By sequence analysis, the respective intimin genes could be identified. Three of the eae variants were new and have been designated eae-η (eta), eae-ι (iota) and eae-κ (kappa), in order to follow the Greek alphabet. The fourth one was identical to eae-ζ (zeta) as described by Jores et al. (accession number AJ298279; mentioned in reference 39), which was sequenced from the bovine E. coli O84:NM isolate 537/98 and therefore also termed eae-ζ. In Table 1, we describe some basic features of the nucleotide sequences of these intimin variant genes along with features of the most important, already published, intimin genes. Interestingly, eae variants display different sequence lengths ranging from 2,805 to 2,847 bp (Table 1). The G+C contents of the various intimin genes are similar and range from 41.7 to 43.07%. The intimin alleles α2 and δ/β2 are not considered in this study because no nucleotide sequences are available.
Sequence comparison and evolutionary analysis of E. coli intimin genes.
We started our genetic analysis by determining the genetic relationship of eae-α, eae-β, eae-γ1, eae-γ2, eae-ɛ, eae-ζ, eae-η, eae-θ, eae-ι, and eae-κ (Table 1). Since the nucleotide sequences were of different lengths, we used ClustaIW (40) for optimal sequence alignment. Genetic distances were calculated by constructing a sequence identity matrix of the nucleotide sequences (Table 3). The values represent ratios of identities to the length of the longer sequence of a given sequence pair.
TABLE 3.
Sequence pair distances of intimin genes on the basis of a ClustalW alignment using sequence distance matrix from Bioedit
| Gene | Distance with respect toa: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| α | β | γ1 | γ2 | ɛ | ζ | η | θ | ι | κ | |
| α | 1.000 | 0.833 | 0.857 | 0.845 | 0.824 | 0.916 | 0.847 | 0.845 | 0.870 | 0.879 |
| β | 1.000 | 0.830 | 0.840 | 0.863 | 0.827 | 0.818 | 0.838 | 0.828 | 0.886 | |
| γ1 | 1.000 | 0.913 | 0.832 | 0.846 | 0.841 | 0.909 | 0.870 | 0.856 | ||
| γ2 | 1.000 | 0.840 | 0.829 | 0.828 | 0.992 | 0.859 | 0.843 | |||
| ɛ | 1.000 | 0.816 | 0.921 | 0.839 | 0.830 | 0.819 | ||||
| ζ | 1.000 | 0.835 | 0.828 | 0.850 | 0.858 | |||||
| η | 1.000 | 0.828 | 0.846 | 0.853 | ||||||
| θ | 1.000 | 0.858 | 0.841 | |||||||
| ι | 1.000 | 0.868 | ||||||||
| κ | 1.000 | |||||||||
Values represent ratios of identities to the length of the longer sequence of a given pair. Values in bold type are mentioned in the text.
The sequences of eae-γ2 and eae-θ were almost identical (0.992 or 99.2%), and these two sequences should be considered one eae variant (γ2/θ). Identities of 0.909 were calculated between eae-γ1 and eae-θ. The two genes are of similar lengths of 2,805 and 2,808 bp. The third-greatest identity, with a value of 0.921, is shared between eae-ɛ and eae-η. Both genes are of the same length, 2,847 bp. This is an interesting finding, since E. coli host strains CF11201 and PMK3 are from different geographic locations (Burundi and France, respectively) and belong to EPEC (stx) and EHEC (stx+) pathovars, respectively. Genes eae-α and eae-ζ are also closely related (0.916). Interestingly, eae-α originates from an EPEC strain and eae-ζ was detected in bovine and human Stx1-producing E. coli strains 537/89 and 4795/95. The genetic distance of eae-γ1 and eae-γ2 is 0.913. Therefore, eae-γ1 is more closely related to eae-θ than to eae- γ2. Finally, eae-β and eae-κ (0.886) are also closely related, although eae-β originates from a rabbit diarrheagenic E. coli isolate and eae-κ originates from a human Stx-negative O145:H− isolate.
Due to these results, we propose a cutoff value of less than 95% (0.95) sequence identity to define a new intimin allele. Above this value, subgroup names (e.g., ζ1, ζ2, and ζ3,) should be used.
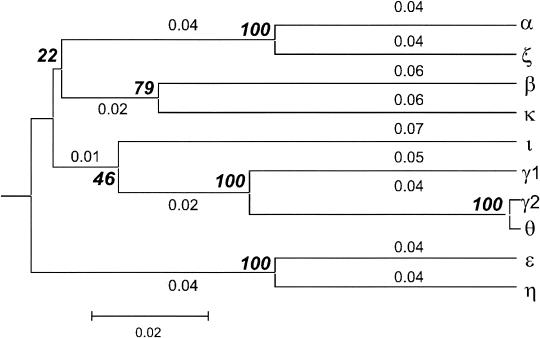
A phylogenetic tree of entire eae sequences was constructed with Mega 2.1 by UPGMA. The phylogenetic tree supports the data of the sequence identity values (Table 3; Fig. 1). Only groups β-κ and ɛ-θ are exchanged in the general arrangement of the tree. From this analysis, it appears to be appropriate to define four groups of closely related intimin genes: (i) eae-α and eae-ζ, (ii) eae-γ1 and eae-θ/γ2, (iii) eae-ɛ and eae-η, and (iv) eae-β and eae-κ. eae-ι is loosely related to the alleles of the γ group. The relationship is closest between group 1 and group 3.
FIG. 1.
Phylogenetic tree of intimin variant genes constructed by the UPGMA method of Mega 2.1. Numbers on the branches represent the branch length and denote the genetic distance (e.g., number of substitutions per unit time) between the two taxa they connect. Bold numbers in italics represent bootstrap values.
Bootstrap values of the phylogenetic tree support the existence of the 4 groups eae- α/eae-ζ; eae-γ1/eae-γ2/θ; eae-ɛ/eae-η with values of 100 and eae-β/eae-κ, with a value of 79 at the widest points of the tree (Fig. 1). The bootstrap value for the relation of ι to the γ/θ group is 46. The relation of the latter groups with each other cannot correctly be demonstrated by a phylogenetic tree, since bootstrap values are too low.
One of the basic challenges in population biology is the determination of the evolutionary process underlying genetic variation, i.e., point mutation or recombination. The relationship between these forces is complex and may vary between species and even between subpopulations. Recombination between multiple commensal lineages of E. coli in the gut may be one of the driving forces in generating novel pathotypes (8). Recombination processes produce networks of sequences rather than strictly bifurcating evolutionary trees (10). Such networks can be detected by split decomposition analysis, a method which depicts parallel edges between sequences if there are conflicting phylogenetic signals in the data. When the sequence data do not support a phylogenetic tree, i.e., such as one evolved by successive point mutations, networks appear that suggest horizontal exchange of DNA by recombination.
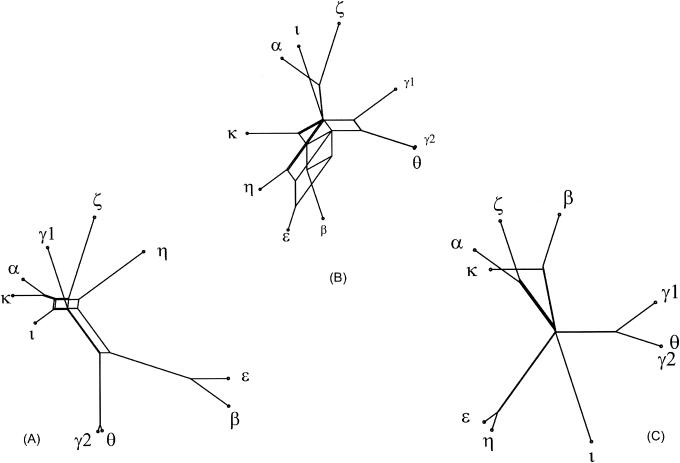
By this method, we detected conflicting signals in the phylogeny of eae (Fig. 2). Split decomposition analysis of the eae sequences (Fig. 2) showed an interconnected network with several parallel edges, indicating the occurrence of recombination events during evolution of eae.
FIG. 2.
Split decomposition analysis of intimin variant genes with Splitstree. By this method, evolutionary data are canonically decomposed into a sum of weakly compatible splits and represented by a splits graph. For ideal data, this is a tree; less ideal data will give rise to a tree-like network, which can be interpreted as evidence for different and conflicting phylogenies, such as recombination in the evolutional history. Splitstree demonstrates network-like structures in the 5′ region (positions 1 to 2112) (A), the entire gene (B), and the 3′ region (positions 2112 to end) (C) of intimin alleles.
Similar results were obtained when the 5′ region (positions 1 to 2112) and the 3′ region (position 2112 to end) were analyzed separately. The split decomposition analyses provided strong evidence for networked evolution of current intimin alleles (Fig. 2). Slight differences in the graphs displaying the 5′ and 3′ ends may be interpreted as differences in the evolution of the two ends and hence as evidence that those ends have a different evolutionary history. Although the descent of the major eae groups indicates recombination events, the Splitstree graphs support the existences of these groups, as indicated above.
We further analyzed the number of polymorphic sites by using DnaSP software. The alignment consists of 10 sequences of 2,869 bp, including alignment gaps. The analysis included 2,782 sites (excluding 69 gaps). There are 1,981 monomorphic sites where no mutations occurred and 801 polymorphic sites including a total of 1,128 mutations. Analysis of singleton polymorphic and parsimony-informative polymorphic sites revealed an interesting result when extrapolated on the respective 5′ and 3′ regions of eae. Whereas the distribution of polymorphic sites with two variants is similar in the 5′ and 3′ regions, sites with three or four variants are more frequent in the 3′ region. This irregular distribution of sites may be an indicator of a greater heterogeneity of the 3′ region (Table 4).
TABLE 4.
Number and distribution of singleton polymorphic sites and parsimony-informative polymorphic sites with two, three, and four different nucleotides in the 5′ and 3′ regions of 10 aligned intimin genes
| Sitea (no. of nucleotides) | No. of sites in region: |
|
|---|---|---|
| 5′ | 3′ | |
| SPM (2) | 108 | 45 |
| PIPM (2) | 200 | 185 |
| SPM (3) | 6 | 9 |
| PIPM (3) | 27 | 157 |
| SPM (4) | 0 | 0 |
| PIPM (4) | 7 | 57 |
SPM, singleton polymorphic sites; PIPM, parsimony-informative polymorphic sites.
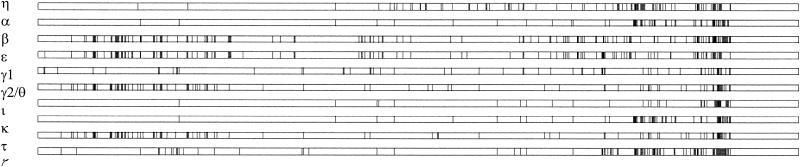
To better illustrate recombination events, we used Happlot analysis to display polymorphic sites (Fig. 3). Since we could use only sequences without gaps for this analysis, we present a Happlot graph of the 5′ eae regions. Interestingly, two clusters of polymorphic sites were detected in the conserved region from positions 1 to 2112. These clusters span from ca. bp 142 to 575 (encoding parts of PP and TM domain) and bp 1613 to 2112 (encoding parts of the D0 domain) and also indicate increased recombination activity in the 5′ region, although this region was originally considered to be conserved.
FIG. 3.
Illustration of polymorphic sites in the 5′ regions of intimin alleles by Happlot. Vertical lines indicate polymorphic sites.
To assess the type of natural selection, we determined the number of synonymous differences per synonymous site (pS) and the proportion of nonsynonymous differences per nonsynonymous site (pN) by using Mega2 (the Nei-Gojobori method). The pS and pN values per 100 sites for the entire intimin alleles were 23.6 and 9.36, respectively. Calculation of the pS and pN values per 100 sites for the 5′ and 3′ ends of intimin alleles revealed values of 15.5, 3.58, 50.0, and 29.8 respectively. Analyses of synonymous and nonsynonymous substitutions for both parts and the entire intimin sequence shows that pS > pN, indicating that the molecule is generally under purifying selection rather than neutral mutation or diversifying selection.
Homoplasy tests (38) were performed in the 2,112-bp 5′ region of eae. Since expression of eae under laboratory conditions is thought to be low (18), we applied the low-gene-expression mode. The mean homoplasy ratio, H, was 0.27. For four E. coli housekeeping genes, the homoplasy ratio was calculated earlier to be 0.26 (38). That indicates that the 5′ regions of eae variants demonstrate a similar extent of recombination to E. coli housekeeping genes.
PCR technique for analysis of intimin alleles.
Differentiation of intimin alleles represents an important tool for strain typing in routine diagnostics as well as in epidemiological studies. Therefore, we elaborated a PCR scheme for amplification and typing of the E. coli intimin genes currently known (Table 2). The PCR scheme was based on a single PCR start primer for all intimin alleles and variant-specific end primers and was able to amplify and subtype all currently known intimin variant genes of E. coli. The PCR scheme was tested with at least two strains from each of these eae groups, and PCR products of the expected sizes were achieved in all cases. Differentiation of eae-γ2 and eae-θ was not possible under the PCR conditions described. This supports our efforts to handle these alleles as a single one.
By investigating a sample of 111 eae-positive, stx-negative strains from patients with diarrhea isolated in Würzburg and Berlin, Germany, we could detect these new intimin gene variants in different numbers (Table 5). However, five strains could not be typed with the primer pairs applied, suggesting the existence of further intimin alleles.
TABLE 5.
Frequency of eae types in 111 stx-negative E. coli isolates from patients and carriers
| eae type | No. of strains | Serotype (no. of strains) |
|---|---|---|
| α | 3 | ONT:HNT, O63:H29; O132:H4 |
| β | 38 | O26:H− (12), O26:H11 (9), O14:H−, O81:H−, O90:H−, O108:H−, O103:H2, O5:H4, O121:H−, O20:H−, O33:H6, O33:H2, ONT:H6, Orough:H26, ONT:H32, Orough:HNT, Orough:H11, O145:HNT (2) |
| γ | 35 | O157:H− (11), O145:H− (7), O145:HNT, O145:H19, O25:H28, O70:H11, O76:H8 (2); O76:H−, O2:H37, O8:H− (2), Orough:H19, ONT:H33, O111:H2, O118:HNT, O125:HNT, O156:H25, O156:H35 |
| ɛ | 14 | O103:H3, O103:H2 (3), O103:H− (3), O96:H19, ONT:HNT, O121:H−, O157:H−, O8:H−, Orough:H− (2) |
| ζ | 5 | O156:H25, O104:H−, O85:H− (2), O105:H31 |
| η | 3 | O125:H−; O2:H49,a O12:H19a |
| θ | 0 | |
| ι | 6 | O145:H4 (3), ONT:H8, Orough:HNT, O98:H2 |
| κ | 2 | O118:H5; O157:H42a |
| NDb | 5 | ONT:H−/HNT (2), O5:H−, O132:H4, O54:H− |
Isolates from the RKI, Berlin, Germany. The O2:H49 strain originates from a healthy carrier.
ND, not determined.
DISCUSSION
AEEC strains represent a large and heterogeneous group of E. coli strains sharing the capability of producing A/E lesions at the microvillus brush border of enterocytes of animals and humans (7, 16, 25). These lesions are caused by the function of the LEE, a pathogenicity island best characterized in EPEC and EHEC strains (9, 13). The intimin gene is localized in the central region of LEE and is involved in the interaction with the Tir receptor. The sequence diversity of eae has been the subject of several investigations, and it has been hypothesized that the heterologous sequences of eae of EPEC and EHEC explains the different host cell tropism (small bowel/large bowel) of these two pathogroups. Studies with infected in vitro organ cultures obtained from different regions of the gut mucosa have demonstrated that EPEC may colonize almost all regions of the small bowel whereas EHEC binding is restricted to the follicle-associated epithelium of Peyer's patches (11). It has also been shown that exchanging intimin-α of EPEC with intimin-γ of EHEC without exchanging Tir resulted in a recombinant EPEC strain showing enhanced tropism to Peyer's patches. Conversely recombinant EHEC strains expressing α-intimin instead of γ-intimin were able to spread to all small- intestine regions (11). Such a tropism could also be shown in animal models (5, 41). Crystallization of the intimin receptor complex demonstrated that intimin is composed of five functional domains: an N-terminal anchor region which includes the periplasmic (PP) and transmembrane (TM) domain, and four extracellular domains labeled D0 to D3 (21). Further work by Tarr and Whittam (39) showed that particular domains of the C-terminal region are under positive selection pressure.
The LEE represent a mosaic of genes that probably evolved by micro- and macrorecombination events. This is reflected in the mosaic structure of intimin, which has been investigated by McGraw et al. (24). It has been concluded that the protein divergence of restricted parts of intimin, e.g., the extracellular domain, has been accelerated by recombination and diversifying selection (39).
In this study, we have shown that the intimin family is increasing and confirmed that recombination has played a role in the history of eae. A homoplasy ratio of 0.27 could be calculated for the 5′ region consisting of 2,112 nucleotides and indicates the participation of recombination in generating allelic diversity of eae. The homoplasy ratio for free recombination is defined as 1.0, and the value for a clonal descent is 0.0. Homoplasy ratios have been determined for housekeeping genes of a variety of microorganisms and range from 0.06 (Borrelia burgdorferi, 1 gene), 0.26 (E. coli, 4 genes) (38), 0.2 (Streptococcus pneumoniae, 2 genes) (36), 0.34 (Neisseria meningitidis, 11 genes) (38), 0.42 (Campylobacter jejuni, 5 genes) (37) to 0.65 (Helicobacter pylori, 7 genes) (1). Recombination between intimin alleles thus apparently occurs with similar frequency to that in E. coli housekeeping genes. The increasing number of intimin gene variants described raises the question of a universal nomenclature that can easily accommodate novel variants. We recommend that new eae variants, which cannot be amplified with the current primer pairs may be considered to be new. When they can be amplified but demonstrate a different restriction pattern or if their sequences are more than 95% homologous, they may be designated an eae subgroup such as eae-γ1 and eae-γ2. eae-θ may also be considered a subgroup of eae-γ, since it is possible to amplify the gene with primers SK1 and LP3.
The use of our typing scheme on a sample of eae-positive strains obtained from patients with diarrhea suggests that the variants θ, η, ι, and κ do not occur frequently in strains isolated from human stool samples. These intimin alleles are also rare among strains associated with severe human disease (20). However, they could play a role as a reservoir for recombination of intimin genes. Further work is in progress to evaluate the occurrence of such variants in AEEC strains isolated from animals. Epidemiological investigations and pathogenetic studies are needed to clarify the mechanism and the role of eae variability in human disease.
Acknowledgments
We thank Gad Frankel, London, United Kingdom, James Paton, Adelaide, Australia, Thomas Whittam, East Lansing, Mich., and Christine Forestier, Clermont-Ferrand, France, for generously providing strains. We also thank Beatrix Henkel and Stefanie Müksch for skillful technical assistance.
This work was supported by grants from the European Union (E.U. project QLK2-2000-0060) and by grant SU133/3-3 from the Deutsche Forschungsgemeinschaft to S.S.
We are solely responsible for the work described in this paper, and our opinions are not necessarily those of the E.U.
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Adu-Bobie, J., G. Frankel, C. Bain, A. G. Goncalves, L. R. Trabulsi, G. Douce, S. Knutton, and G. Dougan. 1998. Detection of intimins alpha, beta, gamma, and delta, four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol. 36:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandelt, H. J., and A. W. Dress. 1992. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol. Phylogenet. Evol. 1:242-252. [DOI] [PubMed] [Google Scholar]
- 4.Deng, W., Y. Li, B. A. Vallance, and B. B. Finlay. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 69:6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnenberg, M. S., S. Tzipori, M. L. McKee, A. D. O'Brien, J. Alroy, and J. B. Kaper. 1993. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J. Clin. Investig. 92:1418-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott, S. J., S. W. Hutcheson, M. S. Dubois, J. L. Mellies, L. A. Wainwright, M. Batchelor, G. Frankel, S. Knutton, and J. B. Kaper. 1999. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol. Microbiol. 33:1176-1189. [DOI] [PubMed] [Google Scholar]
- 7.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 8.Feil, E. J., and B. G. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55:561-590. [DOI] [PubMed] [Google Scholar]
- 9.Finlay, B. B., I. Rosenshine, M. S. Donnenberg, and J. B. Kaper. 1992. Cytoskeletal composition of attaching and effacing lesions associated with enteropathogenic Escherichia coli adherence to HeLa cells. Infect. Immun. 60:2541-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitch, W. M. 1997. Networks and viral evolution. J. Mol. Evol. 44:65-75. [DOI] [PubMed] [Google Scholar]
- 11.Fitzhenry, R. J., D. J. Pickard, E. L. Hartland, S. Reece, G. Dougan, A. D. Phillips, and G. Frankel. 2002. Intimin type influences the site of human intestinal mucosal colonisation by enterohaemorrhagic Escherichia coli O157:H7. Gut 50:180-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forestier, C., A. Darfeuille-Michaud, E. Wasch, C. Rich, E. Petat, F. Denis, and B. Joly. 1989. Adhesive properties of enteropathogenic Escherichia coli isolated from infants with acute diarrhea in Africa. Eur. J. Clin. Microbiol. Infect. Dis. 8:979-983. [DOI] [PubMed] [Google Scholar]
- 13.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 14.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 15.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 16.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jores, J., L. Rumer, S. Kiessling, J. B. Kaper, and L. H. Wieler. 2001. A novel locus of enterocyte effacement (LEE) pathogenicity island inserted at pheV in bovine Shiga toxin-producing Escherichia coli strain O103:H2. FEMS Microbiol. Lett. 204:75-79. [DOI] [PubMed] [Google Scholar]
- 18.Knutton, S., J. Adu-Bobie, C. Bain, A. D. Phillips, G. Dougan, and G. Frankel. 1997. Down regulation of intimin expression during attaching and effacing enteropathogenic Escherichia coli adhesion. Infect. Immun. 65:1644-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 10:189-191. [DOI] [PubMed] [Google Scholar]
- 20.Levine, M. M., J. P. Nataro, H. Karch, M. M. Baldini, J. B. Kaper, R. E. Black, M. L. Clements, and A. D. O'Brien. 1985. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J. Infect. Dis. 152:550-559. [DOI] [PubMed] [Google Scholar]
- 21.Luo, Y., E. A. Frey, R. A. Pfuetzner, A. L. Creagh, D. G. Knoechel, C. A. Haynes, B. B. Finlay, and N. C. Strynadka. 2000. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405:1073-1077. [DOI] [PubMed] [Google Scholar]
- 22.Mariani-Kurkdjian, P., E. Denamur, A. Milon, B. Picard, H. Cave, N. Lambert-Zechovsky, C. Loirat, P. Goullet, P. J. Sansonetti, and J. Elion. 1993. Identification of a clone of Escherichia coli O103:H2 as a potential agent of hemolytic-uremic syndrome in France. J. Clin. Microbiol. 31:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGraw, E. A., J. Li, R. K. Selander, and T. S. Whittam. 1999. Molecular evolution and mosaic structure of alpha, beta, and gamma intimins of pathogenic Escherichia coli. Mol. Biol. Evol. 16:12-22. [DOI] [PubMed] [Google Scholar]
- 25.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nei, M. K. S. 2000. Molecular evolution and phylogenetics. Oxford University Press, Oxford, United Kingdom.
- 27.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 28.Oswald, E., H. Schmidt, S. Morabito, H. Karch, O. Marches, and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perna, N. T., G. F. Mayhew, G. Posfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips, A. D., and G. Frankel. 2000. Intimin-mediated tissue specificity in enteropathogenic Escherichia coli interaction with human intestinal organ cultures. J. Infect. Dis. 181:1496-1500. [DOI] [PubMed] [Google Scholar]
- 31.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt, H., B. Plaschke, S. Franke, H. Rüssmann, A. Schwarzkopf, J. Heesemann, and H. Karch. 1994. Differentiation in virulence patterns of Escherichia coli possessing eae genes. Med. Microbiol. Immunol. Berl. 183:23-31. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt, H., H. Rüssmann, and H. Karch. 1993. Virulence determinants in nontoxinogenic Escherichia coli O157 strains that cause infantile diarrhea. Infect. Immun. 61:4894-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt, H., H. Rüssmann, A. Schwarzkopf, S. Aleksic, J. Heesemann, and H. Karch. 1994. Prevalence of attaching and effacing Escherichia coli in stool samples from patients and controls. Int. J. Med. Microbiol. Virol. Parasitol. Infect. Dis. 281:201-213. [DOI] [PubMed] [Google Scholar]
- 35.Sperandio, V., J. B. Kaper, M. R. Bortolini, B. C. Neves, R. Keller, and L. R. Trabulsi. 1998. Characterization of the locus of enterocyte effacement (LEE) in different enteropathogenic Escherichia coli (EPEC) and Shiga-toxin producing Escherichia coli (STEC) serotypes. FEMS Microbiol. Lett. 164:133-139. [DOI] [PubMed] [Google Scholar]
- 36.Suerbaum, S. 2000. Genetic variability within Helicobacter pylori. Int. J. Med. Microbiol. 290:175-181. [DOI] [PubMed] [Google Scholar]
- 37.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarr, C. L., and T. S. Whittam. 2002. Molecular evolution of the intimin gene in O111 clones of pathogenic Escherichia coli. J. Bacteriol. 184:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tzipori, S., F. Gunzer, M. S. Donnenberg, L. de-Montigny, J. B. Kaper, and A. Donohue-Rolfe. 1995. The role of the eaeA gene in diarrhea and neurological complications in a gnotobiotic piglet model of enterohemorrhagic Escherichia coli infection. Infect. Immun. 63:3621-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voss, E., A. W. Paton, P. A. Manning, and J. C. Paton. 1998. Molecular analysis of Shiga toxigenic Escherichia coli O111:H− proteins which react with sera from patients with hemolytic-uremic syndrome. Infect. Immun. 66:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, J., and J. B. Kaper. 1992. Cloning and characterization of the eae gene of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 6:411-417. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, C., T. S. Agin, S. J. Elliott, L. A. Johnson, T. E. Thate, J. B. Kaper, and E. C. Boedeker. 2001. Complete nucleotide sequence and analysis of the locus of enterocyte effacement from rabbit diarrheagenic Escherichia coli RDEC-1. Infect. Immun. 69:2107-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]