Abstract
Numerous eukaryotic mRNAs contain sequences complementary to segments of the 18S and 28S rRNAs. Previous studies raised the possibility that these complementarities might allow mRNA–rRNA interactions and affect rates of translation. In the present study, we investigated the mRNA encoding the mouse Gtx homeodomain protein. This mRNA contains within its 5′ untranslated region (UTR) a segment that is complementary to two regions of the 18S rRNA, located at nucleotides 701–741 and 1104–1136. A Gtx RNA probe containing this complementarity could be photochemically cross-linked to ribosomal subunits through a linkage to 18S rRNA but not to 28S rRNA. Oligonucleotide-directed RNase H digestion of the rRNA and a reverse transcription analysis localized the cross-linked probe to the complementary segment of 18S rRNA at nucleotides 1104–1136 but not at nucleotides 701–741. To determine whether complementarity in the Gtx mRNA affected translation, a mutational analysis was performed with a Gtx-luciferase fusion construct and four related constructs with altered complementarity to the 18S rRNA. These constructs were examined for their ability to be translated in cell-free lysates prepared from P19 embryonal carcinoma and C6 glioma cell lines and after cellular transfection into these same cell lines. In both cell-free translation and transfection studies, the rate of translation decreased more than 9-fold as the degree of complementarity to nucleotides 1104–1136 of the 18S rRNA increased. We hypothesize that segments complementary to rRNA, such as those contained within the Gtx mRNA, form a category of cis-acting regulatory elements in mRNAs that affect translation by base pairing to rRNA within ribosomes.
The translation of eukaryotic mRNAs is subject to regulation at multiple levels. One well-characterized regulatory mechanism involves the binding of specific mRNA sequences to proteins (for reviews, see refs. 1 and 2). Recent studies from our laboratory suggest that translation may also be controlled by sequence motifs that bind directly to rRNA at sites of complementarity (3, 4). This so-called riboregulation was proposed on the basis of database analyses, cross-linking studies, and functional experiments (3–5). These investigations revealed large numbers of mRNAs containing segments resembling those in the 18S and 28S rRNAs (3). These rRNA-like sequences appear both as similarities or “sense” matches and as complementarities or “antisense” matches with 50–100% identity over lengths of 10 to several hundred nucleotides. The accessibility of rRNA within ribosomes to complementary mRNA sequences was shown in cross-linking studies (4) and is consistent with the results of studies that have used short DNA oligonucleotides to map such accessibility (e.g., see refs. 6 and 7).
In prokaryotes, the presence of rRNA-like sequences within individual mRNAs has been shown to affect the translation of a number of mRNAs (see refs. 8–10). In eukaryotes, the occurrence of rRNA-like sequences had previously been observed in some gene sequences, and it has been noted that particular mRNA probes cross-hybridize to rRNAs on Northern blots (e.g., refs. 11–13). rRNA-complementarity has been reported in the internal ribosome entry sites of several viral mRNAs (for review, see ref. 14) and has also been shown to be essential in the function of small nucleolar RNAs (for reviews, see refs. 15 and 16).
Evidence that a complementary sequence could function as a cis-regulatory element in a eukaryotic mRNA was obtained by analyzing the mRNA encoding mouse ribosomal protein S15 (4); this mRNA contains a sequence within its coding region that is complementary to the 18S rRNA. When RNA molecules containing the complementary segment of the S15 gene or variants of this sequence were fused to a luciferase reporter gene, the degree of complementarity was strongly correlated to the inhibition of translation. The S15 mRNA was complementary to a stem–loop structure at the 3′ end of the 18S rRNA, a region that had previously been investigated for its role in translation initiation (17, 18). Inasmuch as most of the messages identified in our database searches contained sequences with complementarity to other regions of the 18S and 28S rRNAs (3) and because some of these regions were shown to be accessible to complementary mRNA sequences (4), we sought to examine messages with complementarity to some of these other regions.
In the present study, we therefore examined the mRNA that encodes the mouse Gtx homeodomain protein (19). This mRNA encodes a transcriptional repressor expressed in oligodendrocytes of the central nervous system, as well as in germ cells of the testis (19, 20). The 5′ untranslated region (UTR) of the Gtx mRNA contains a sequence complementary to two segments of the 18S rRNA (nucleotides 701–741 and nucleotides 1104–1136) that are distinct from the region of the 18S rRNA complementary to the previously studied S15 mRNA. Our analysis of cross-linking and effects on translation of the complementary sequence contained within the Gtx mRNA is consistent with the notion that it acts by base pairing to one of the two regions of complementarity identified within the 18S rRNA. We discuss the possibility that rRNA-complementarity such as that contained within the Gtx and S15 mRNAs may reflect the occurrence of a class of cis-regulatory elements in eukaryotic mRNAs that affect translation by base pairing to rRNA within ribosomes.
MATERIALS AND METHODS
Cross-Linking.
Ribosomes were prepared from the mouse P19 cell line (21). Subunits were dissociated in the presence of puromycin, recovered by centrifugation, distributed into aliquots, and stored at −80°C before use (22). Subunit dissociation was confirmed by sucrose gradient profiles (23). Cross-linking to ribosomes or to purified rRNA was performed as described (4, 24) with RNA probes containing the cross-linking reagent 4-thiouridine [s4U (25)]. All probes were 32P-labeled except for those used in the reverse transcription analysis. Probes were transcribed by using T7 RNA polymerase (4) from DNA templates that contained the T7 promoter sequence (26) at the 5′ end of the probe sequence.
RNase H Localization of Cross-Linked Probes.
The position within the rRNA at which the GTX probes were cross-linked within intact ribosomal subunits was localized by annealing short DNA oligonucleotides to the rRNA at various locations flanking the regions of complementarity, cleaving the rRNA at these DNA⋅RNA hybrids with RNase H, and analyzing the fragments by electrophoresis and autoradiography as described (4, 27).
Inhibition of Primer Extension (Toeprinting).
Ribosome subunits were cross-linked to nonradioactive s4U-containing probe, and RNA was purified (4). Two pmoles of 32P-labeled primer complementary to the 18S rRNA at nucleotides 1174–1193 was annealed to cross-linked or control rRNA and this template was reverse-transcribed (28). The RNA templates were then hydrolyzed with 0.16 M NaOH, and samples were electrophoresed on 6% polyacrylamide/urea gels. M13 sequencing reaction products were used as size markers.
Cell-Free Translation and Cellular Transfection.
Transient transfection experiments were performed with reporter constructs in which nucleotides 1–196 of the Gtx cDNA sequence, encompassing the entire 5′ UTR of the Gtx mRNA, were cloned immediately upstream of the initiation codon, in the 5′ UTR of the luciferase gene in the pGL3-Control vector (Promega). The wild-type sequence, as well as various mutations, were synthesized as oligonucleotides, made double-stranded, and cloned into the vector at HindIII and NcoI restriction sites. The Gtx-luciferase reporter constructs were transfected into cells by using Lipofectamine (GIBCO/BRL). Transfection efficiencies were normalized by cotransfection with the pCMVβ vector (CLONTECH). Cells were harvested after 48 h and luciferase activity was determined (4). β-Galactosidase activity was assayed by using the Fluoreporter lacZ kit (Molecular Probes), and fluorescence was measured with the Millipore Cytofluor 2450 system.
For cell-free translation studies, the T7 promoter sequence (26) was cloned upstream of the Gtx-luciferase gene at the BlnI and XhoI restriction sites present within the vector. Plasmids were linearized with BamHI, and RNA was transcribed with T7 RNA polymerase and then purified (4). Lysates for cell-free translation and cell-free protein synthesis were performed as described (4). The integrity of the RNA templates was confirmed by electrophoresis on 1.2% agarose/formaldehyde gels (3).
RESULTS
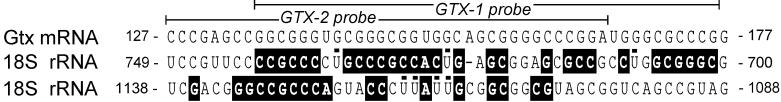
The Gtx cDNA was identified with the fasta program (29) in a search of the GenBank and EMBL nucleic acid databases for sequences with complementarity to segments of the mouse 18S rRNA (3). The fasta analysis identified a segment within the 5′ UTR of the Gtx mRNA complementary to the 18S rRNA at nucleotides 701–741, within helix 21.1of the 18S rRNA (ref. 30; see Fig. 1). This complementarity is 71% over 41 nucleotides and contains a segment that is 100% complementary over 9 nucleotides (outlined in black in Fig. 1). Further analysis of this same segment revealed a second site of complementarity at nucleotides 1104–1136 within helix 27 of the 18S rRNA (30). This second match was 55% over 33 nucleotides and also contains a stretch that is 100% complementary over 9 nucleotides. Four potential G⋅U base-pairs increase the overall complementarity of this second match to 67%.
Figure 1.
Complementarity between 5′ UTR of Gtx mRNA and 18S rRNA. The top sequence is that of the mouse Gtx 5′ UTR at nucleotides 127–177 (19). The lower two sequences are segments of the mouse 18S rRNA at nucleotides 700–749 and at 1088–1138 (30) with complementary nucleotides outlined in black. Potential G⋅U base pairing within these two segments is indicated by black bars positioned over uracil (U) nucleotides. The sequence of the GTX-1 and GTX-2 probes are indicated above the Gtx mRNA sequence.
Cross-Linking of Gtx Probes to Ribosomes.
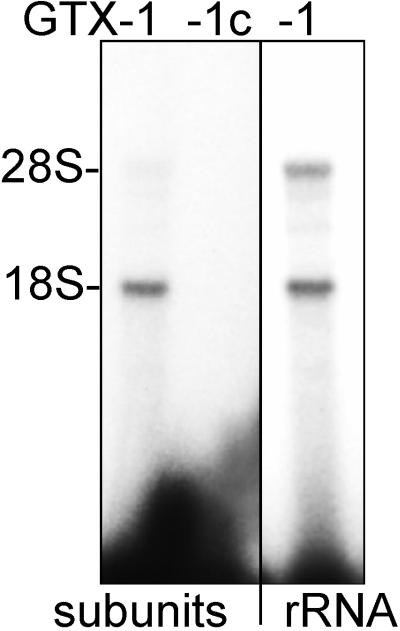
To determine whether the sequence complementarities identified within the 5′ UTR of the Gtx mRNA would enable the mRNA to interact with ribosomes by base pairing to the 18S rRNA, photochemical cross-linking experiments were performed with 32P-labeled RNA probes containing the modified nucleotide s4U. The probes included GTX-1 (see Fig. 1) and GTX-1c. GTX-1 is based on the Gtx mRNA sequence at nucleotides 135–176, and GTX-1c is the complement of GTX-1 and was used as a control. Probes were incubated with puromycin-dissociated 60S and 40S ribosomal subunits and were cross-linked by exposure to 365-nm UV radiation. The GTX-1 probe cross-linked preferentially to the 18S rRNA but not to the 28S rRNA, whereas GTX-1c failed to cross-link to either rRNA (Fig. 2).
Figure 2.
Cross-linking of GTX probes to dissociated ribosomal subunits or to purified rRNA. The Gtx mRNA probe GTX-1 and the control probe (GTX-1c) were cross-linked to 80S ribosomal subunits or to rRNA purified from these subunits. The positions of the rRNAs were determined by ethidium bromide staining of the agarose gels.
Binding of the Gtx Probe to 18S rRNA Does Not Appear to Require Proteins.
To determine whether the GTX-1 probe cross-linked to the 18S rRNA because of interactions with ribosomal proteins, cross-linking experiments were performed with rRNA purified from the ribosomal proteins, and the results were compared with those obtained with intact ribosomes (Fig. 2). These experiments showed that the extent of cross-linking of the GTX-1 probe to the 18S rRNA is similar with intact ribosomes and with purified rRNA. Interestingly, however, there was cross-linking to purified 28S rRNA that did not occur when ribosomal subunits were used. This result suggests that the 28S rRNA contains a segment of complementarity that is inaccessible within the intact 60S subunit. A computer analysis of the 28S rRNA identified sites with potential complementarity (up to 76% in 29 nucleotides, with perfect stretches of 5 nucleotides) that might account for the cross-linking. Overall, these results indicate that cross-linking of the GTX probe to the 18S rRNA was a consequence of interactions with the rRNA and thus could occur in the absence of ribosomal proteins.
Localizing the Position of Cross-Linked Gtx Probe.
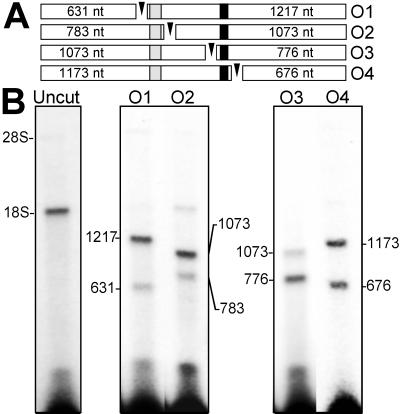
To localize the position of the cross-linked GTX probe within the 18S rRNA, the cross-linked rRNA was purified from protein and cleaved at locations flanking the two regions of complementarity. This was accomplished by hybridizing short DNA oligonucleotides to these flanking regions and digesting rRNA within these DNA⋅RNA hybrids with RNase H (Fig. 3A). The result of RNase H digestions flanking the first region of complementarity (nucleotides 701–741) indicated that there was no detectable cross-linking of the GTX-1 probe to this segment of the 18S rRNA and that most of the probe was cross-linked to a region located 3′ of this sequence (Fig. 3B). Ribosomes were then cross-linked to the GTX-2 probe (Figs. 1 and 3B). This probe overlaps the GTX-1 probe and is a better match to the second region of complementarity (nucleotides 1104–1136) than GTX-1. The result of RNase H digestions flanking this second region of complementarity indicated that 50–60% of the GTX-2 probe cross-linked to this segment of the 18S rRNA. Cross-linking also occurred to the region of rRNA located 3′ of the second region of complementarity. This cross-linking might indicate the presence of other binding sites or may reflect technical limitations of the cross-linking technique (25).
Figure 3.
Mapping the binding site of the GTX RNA probes by RNase H digestion of the rRNA (A). The open bars represent the 18S rRNA fragments after RNase H digestion, the sizes of which are indicated within each bar. The shaded and solid segments indicate the positions of GTX complementarity at nucleotides 701–741 and 1104–1136, respectively. The oligonucleotides (O1–O4) used to direct digestion of rRNA by RNase H are represented as arrowheads. O1 is complementary to the 18S rRNA at nucleotides 632–652, O2 is complementary at nucleotides 784–796, O3 is complementary at nucleotides 1074–1093, and O4 is complementary at nucleotides 1174–1193. (B) Results of the RNase H digestions. After cross-linking the GTX-1 or GTX-2 probe to dissociated ribosomes, the RNAs were purified away from protein and annealed to the O1 or O2 oligonucleotides (for the GTX-1 probe) or to the O3 or O4 oligonucleotides (for the GTX-2 probe), and the rRNA at these positions was digested with RNase H. The positions of the rRNAs were determined by ethidium bromide staining of the agarose gels.
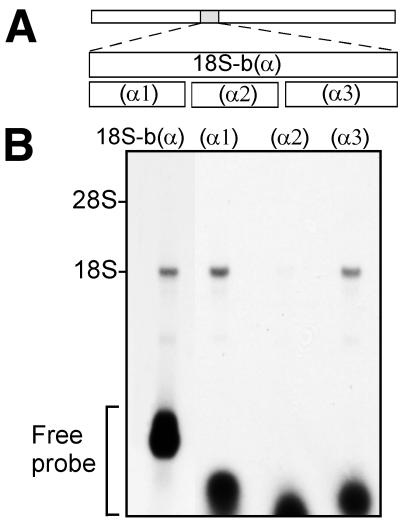
The results of these experiments indicated that the GTX probes did not cross-link to the first region of complementarity at nucleotides 701–741 of the 18S rRNA. However, in a earlier study (4) in which probes constructed to be exactly complementary to the rRNA sequences themselves were used to map the accessibility of various segments of the 18S and 28S rRNAs, this region of the 18S rRNA appeared to be accessible to a probe previously designated 18S-b(α) (Fig. 4A). This probe was 100% complementary to the 18S rRNA at nucleotides 645–783, which includes the region complementary to the GTX probes. To examine the accessibility of this region of the rRNA further, we divided the 18S-b(α) probe (4) into three smaller probes (α1), (α2), and (α3). The 18S-b(α) probe contains nine uracil bases, and each of the smaller probes contains three uracil residues. These probes were cross-linked to ribosomal subunits, and the rRNA was purified and examined. The results of this analysis showed that probes (α1) and (α3) cross-linked to the 18S rRNA but that (α2) did not (Fig. 4B). (α2) is antisense to the 18S rRNA at nucleotides 715–743, the segment containing the greatest degree of complementarity to the GTX probes. These results suggest that the 18S rRNA at nucleotides 715–743 is inaccessible for cross-linking to a probe with perfect complementarity (α2) or to GTX probes with complementary mRNA sequences.
Figure 4.
Fragment analysis of 18S-b(α). (A) Schematic representation of probes that are antisense to the 18S rRNA. Top bar represents 18S rRNA. The shaded region indicates the segment of 18S rRNA, at nucleotides 680–784, complementary to probe 18S-b(α). Probe (α1) is complementary to the 18S rRNA at nucleotides 680–713, (α2) is complementary at nucleotides 715–743, and (α3) is complementary at nucleotides 746–784. (B) Gel electrophoresis after cross-linking of 32P-labeled RNA probes to ribosomal subunits. The positions of the rRNAs were determined by ethidium bromide staining of the agarose gels.
Toeprinting Analysis of 18S rRNA: Evidence for a GTX Cross-Linked Complex.
To further characterize the binding of the GTX-2 probe to the 18S rRNA at nucleotides 1104–1136, a reverse transcription analysis was performed on rRNA purified from ribosomes cross-linked to the GTX-2 probe or to a probe we synthesized to be 100% complementary to the 18S rRNA at nucleotides 1108–1136 designated 18Sα(1108). As a control, rRNA was purified from ribosomes that were similarly treated and processed but in the absence of probe.
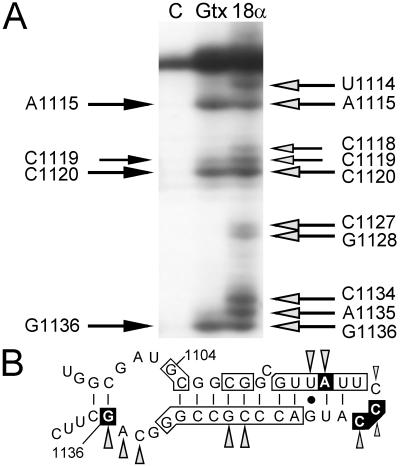
As shown in Fig. 5A, in the presence of the GTX-2 or 18Sα(1108) probes, extension of reverse transcriptase was blocked at particular sites that were not blocked in the control or that differed dramatically in band intensity. In the presence of GTX-2 probe, extension of reverse transcriptase was blocked at nucleotides A1115, C1119, C1120, and G1136. The stop at nucleotide A1115 of the 18S rRNA corresponds to the location of a complementary s4U residue within the GTX-2 probe (nucleotide U203 of the Gtx 5′ UTR) and nucleotide G1136 of the 18S rRNA where a stop occurs corresponds to the first base-paired nucleotide encountered by the reverse transcriptase at the 5′ end of the GTX-2 probe. In the presence of 18Sα(1108), a probe with perfect complementarity to this same segment of the 18S rRNA, extension was blocked at the same nucleotides as with the GTX probe, and there were additional stops that may have resulted from the presence of two additional s4U residues within this probe, one of which corresponds to the stop observed at nucleotide A1134 (summarized in Fig. 5B). These reverse transcription results provide additional evidence that the GTX-2 probe is base-paired to the 18S rRNA at nucleotides 1104–1136 and that complementarity plays a role in base-pairing interactions to this segment of the 18S rRNA.
Figure 5.
Reverse transcription analysis of one region of 18S rRNA after cross-linking of RNA probes to ribosomes. (A) Lanes contain primer extension products from control ribosomes without probe (lane C), from ribosomes cross-linked to the GTX-2 probe (lane Gtx), and from ribosomes cross-linked to the 18Sα(1108) probe (lane 18α). The black arrows to the left of the autoradiogram indicate the nucleotide positions where reverse transcriptase stopped when ribosomes were cross-linked to the GTX-2 probe. Similarly, the shaded arrows to the right of the gel indicate stop sites in the presence of the 18Sα(1108) probe. All of these sites have been seen in at least four experiments. (B) Schematic representation of 18S rRNA at nucleotides 1097–1140 indicating complementarity to Gtx mRNA and summary of toeprinting results. The secondary structure of this segment of the 18S rRNA is based on Holmberg et al. (30). Vertical lines indicate base pairing and the solid dot represents G⋅U base pairing. Complementarity between the Gtx mRNA and the 18S rRNA is indicated by the boxed nucleotides. The nucleotide positions at which reverse transcriptase stopped in the presence of the GTX-2 probe are boxed in black. Stop sites in the presence of the 18Sα(1108) probe are indicated as shaded arrowheads.
18S rRNA Sequence Complementarity Inhibits Expression in Vitro.
The results of the localization studies support the notion that a segment of the 5′ UTR of the Gtx mRNA base- pairs to the 18S rRNA at nucleotides 1104–1136. To test whether this complementary mRNA sequence has any effect on the translation of a full-length message, we performed cell-free translation and transfection experiments following an approach that has been used successfully to define the effects of rRNA-complementarity within some prokaryotic mRNAs (e.g., refs. 10 and 31) and within eukaryotic S15 mRNA (4). Both cell-free translation and transfection studies used the constructs shown in Fig. 6 and the data from both studies are in Table 1.
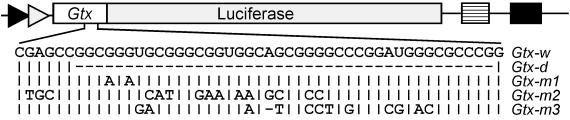
Figure 6.
Schematic representation of the Gtx-luciferase reporter plasmids. The Gtx sequences are indicated as a open bar, the luciferase sequences as a shaded bar, the poly(A) signal as a hatched bar, the simian virus 40 promoter as a solid arrow, the simian virus 40 enhancer as a solid bar, and the T7 promoter as a shaded arrow. The Gtx mRNA sequence at nucleotides 129–177 is written below the schematic. Below this sequence are indicated the modifications present in the other constructs. A vertical line indicates nucleotides identical to those in Gtx-w, and a dash indicates a deleted nucleotide.
Table 1.
Cell-free translation and transfection analysis of Gtx-luciferase fusion constructs
| Construct | %
complementarity to 18S rRNA
|
Luciferase activity assayed
|
||||
|---|---|---|---|---|---|---|
| By cell-free translation
|
By
transfaction
|
|||||
| nt. 701–741 | nt. 1104–1136 | P19 | C6 | P19 | C6 | |
| Gtx-w | 78 | 67 | 100 | 100 | 100 ± 9 | 100 ± 2 |
| Gtx-d | (38)* | (43)* | 477 ± 46 | 300 ± 57 | 398 ± 9 | 252 ± 22 |
| Gtx-m1 | 73 | 61 | 208 ± 36 | 230 ± 50 | 196 ± 1 | 370 ± 12 |
| Gtx-m2 | 66 | 100 | 53 ± 7 | 41 ± 10 | 40 ± 18 | 55 ± 33 |
| Gtx-m3 | 100 | 61 | 204 ± 27 | 148 ± 37 | 448 ± 16 | 354 ± 2 |
For the various constructs, the percentage complementarity to two sites within the 18S rRNA are indicated, including GU base-pairs, along with the activity as measured by luciferase activity. Constructs were assayed in cell-free lysates prepared from P19 or C6 cells and after transfection into P19 or C6 cells. nt., Nucleotides. Luciferase activity was measured as raw light units, and the values were normalized to the activity of construct Gtx-w. The luciferase activity for each construct was calculated from at least three experiments, performed in duplicate. Data are the mean ± SEM.
Construct Gtx-d contains a 42-nucleotide deletion of the Gtx 5′UTR. The numbers in parentheses are the best complementary matches to the new junction, comparing up to 50 nucleotides to either side of the junction.
Cell-free translation experiments were performed with the full-length 5′ UTR of the Gtx mRNA fused to the coding sequence of the luciferase gene (Gtx-w; Fig. 6). To evaluate whether complementarity to the 18S rRNA affects expression, the complementary nucleotides within this construct were deleted (Gtx-d), or mutated (Gtx-m1, -m2, and -m3) to alter complementarity.
mRNA transcribed from the Gtx-luciferase reporter constructs were translated in cell-free lysates prepared from the C6 and P19 cell lines under conditions in which the translation machinery was not saturated by message. Translation efficiency was determined by measuring luciferase activity. Similar results were obtained in both C6 and P19 lysates (Table 1). A 41-nucleotide deletion of the rRNA-like sequence in construct Gtx-d increased luciferase activity up to 4.8-fold as compared with that of construct Gtx-w, which contains the complementary sequence. Two point mutations in the region of complementarity (Gtx-m1) that slightly lowered the degree of complementarity to both regions of the 18S rRNA resulted in an increase in luciferase activity of up to 2.5-fold.
We predicted that if complementarity were the basis for the inhibition of translation, the efficiency of translation should decrease as complementarity increased. Inasmuch as the localization studies indicated that the GTX probes were cross-linking to the 18S rRNA at nucleotides 1104–1136, we mutated the Gtx sequence to increase complementarity to this segment of the 18S rRNA (construct Gtx-m2). The result of this mutation was a further inhibition of translation: the Gtx-m2 RNA was translated at approximately 50% of the rate the wild-type Gtx-w RNA, consistent with a postulated base-pairing interaction with this segment of the 18S rRNA. In increasing the complementarity to nucleotides 701–741 of the 18S rRNA with construct Gtx-m3, the degree of complementarity to region 1104–1136 was decreased (see Fig. 1) and, consistent with this, the rate of translation also increased.
Expression of Gtx-Luciferase Fusion RNAs in Cells.
To determine whether complementarity between the Gtx 5′ UTR and the 18S rRNA also affects translation in cells, transient transfection experiments were performed. The same Gtx-luciferase reporter constructs used in the cell-free studies, but lacking the T7 promoter sequences, were transfected into P19 and C6 cell lines (Table 1). The results of these experiments were similar to those obtained in the cell-free translation experiments. A Northern blot analysis of RNA prepared from the transfected cells (data not shown) indicated that mutations in the complementary sequence did not affect the Gtx-luciferase mRNA levels, confirming that the effects of complementarity on translation were primarily translational. The results of the cell-free translation and transfection studies, as well as those of the RNase H localization and toeprinting analyses, are all in accord with the notion that base pairing occurs between nucleotides 129–161 of the Gtx 5′ UTR and nucleotides 1104–1136 of the 18S rRNA.
DISCUSSION
The 5′ UTR of the Gtx mRNA contains a sequence with complementarity to two segments of the 18S rRNA. Photochemical cross-linking and localization techniques demonstrated that a probe containing this sequence could bind to one of these two segments, located at nucleotides 1104–1136, a ribosomal region known as helix 27, the structure of which is conserved both in eukaryotic 18S rRNAs and in prokaryotic 16S rRNAs (32). The presence of the complementary segment of the Gtx mRNA was also shown to diminish translation both in vitro and in cells when assayed with a luciferase fusion construct. In both cases, the extent of inhibition increased with the degree of complementarity to this segment of the 18S rRNA.
The short sequence complementarity contained within the Gtx mRNA corresponded to a different segment of the 18S rRNA than the segment interacting with the S15 mRNA (4). The location of the complementary sequences within each of these mRNAs also differed: the sequence present within the S15 mRNA was located in the coding region whereas the sequence present within the Gtx mRNA was located in the 5′ UTR. Despite these differences, in both cases the presence of the complementary sequence reduced the translation efficiency of the mRNA in cell-free translation assays. Moreover, translation efficiency was reduced when the degree of complementarity to the 18S rRNA was increased.
In addition to the cross-linking that occurred between Gtx mRNA probes and the 18S rRNA at nucleotides 1104–1136, cross-linking also took place 3′ of this segment of the 18S rRNA. This additional cross-linking might indicate binding to other, shorter, regions of complementarity or reflect interactions of the probes with the mRNA binding tract (33). Alternatively, the binding to the 3′ sequences might simply reflect limitations of the technique: it is known that s4U-containing probes can cross-link to RNA that is adjacent to and in close contact to the base paired sequence (25).
Although the role of rRNA-complementarity in eukaryotic messages has not been extensively studied, the role of complementary sequences in prokaryotic mRNAs has received more attention. In addition to base pairing that occurs during initiation at the Shine–Dalgarno sequence (34–36), base pairing also appears to be the basis for several distinct mechanisms of translational control. Sequences complementary to different segments of the 16S rRNA have been identified in untranslated and coding regions of messages and have been shown to function as cis- and trans-acting regulatory elements that can either enhance or repress translation (see refs. 8–10). Our recent identification of a significant number of eukaryotic mRNAs that contain sequences complementary to rRNA in untranslated and coding regions (3) suggests that, as in prokaryotes, such sequences may be involved in various translational processes and this may be one basis for mechanisms of translational control.
While our studies of the S15 and Gtx mRNAs have focused on how primary nucleotide sequences might affect translation by binding to complementary rRNA sequences, they do not discount the potential roles of RNA structures or of RNA-binding proteins in modulating the efficiency of translation. It is likely that structural motifs and protein interactions will directly affect the accessibility of these complementary sequences, both within the mRNA and rRNA. As was observed in the present study, the presence of a complementary sequence match between a segment of the Gtx 5′ UTR and nucleotides 701–741 of the 18S rRNA did not prove to be sufficient for binding to occur. This inaccessibility might be due to the structure of the rRNA in this region or because this segment of rRNA was blocked by ribosomal proteins. The potential role of the protein kinase PKR in these phenomena deserves additional consideration. PKR is activated by double-stranded RNA (for review, see refs. 37 and 38) and phosphorylates eukaryotic initiation factor 2α, inhibiting translation. PKR appears to be activated by perfect RNA duplexes of 30–85 nucleotides and to be inactivated by shorter double-stranded RNAs (39). The complementary sequences identified within the S15 and Gtx mRNAs are not likely to activate PKR because the longest perfect duplex possible within this segment itself or after base pairing to the 18S rRNA is only 10 nucleotides long. Nonetheless, other mRNAs with longer or more perfect complementary sequence matches (see ref. 3) might activate PKR after base pairing to the rRNA and thus deserve further study.
The question arises: how might complementarity to rRNA reduce the translation efficiency of the Gtx and S15 mRNAs? Base pairing between mRNAs and rRNA might occur dynamically during the process of translation or could occur as a static event independent of translation. If base pairing occurs during the process of translation, the location of the complementary sequence within the mRNA is likely to be significant for function and several possibilities arise: (i) sequences in 5′ UTRs and coding regions might make the initial contacts with ribosomes during initiation, (ii) sequences located in coding regions might affect elongation, and (iii) sequences located in coding regions and 3′ UTRs might affect termination. If, however, base pairing occurs as a static event independent of translation, the location of complementary sequences within the mRNA may not affect function. In this case, messages might simply be sequestered and translation inhibited by an antisense mechanism (40). An alternative is that binding of an mRNA sequence to a segment of rRNA may cause structural changes by disrupting base-paired stems or by displacing ribosomal protein interactions. Such changes might alter the accessibility of particular sites in the rRNA, perhaps affecting the binding and translation of other mRNAs. It may therefore be informative to explore the effects of complementarity in a given mRNA on the translation of other mRNAs.
The ability of rRNA-like sequences in messages to affect translation by binding to complementary segments of rRNA within ribosomes thus raises the possibility that complementary sequence matches can function as cis-acting regulatory elements. Complementary sequences might be expected to affect expression indirectly by altering the ability of the mRNAs to compete against other mRNAs for ribosomes or, in some cases, by affecting mRNA stability (41) by altering ribosome associations. More extensive characterization of complementary sequences may establish whether there are specific rules related to the location of these sequences within mRNAs and the segments of rRNA to which they are complementary. Such rules may be difficult to establish if different complementary sequences or combinations of sequences can have similar effects on the rate of translation. These effects will depend upon the location of the complementary sequence within the mRNA, the region of 18S or 28S rRNA to which the message is complementary, and the extent of complementarity. In any event, the immediate implications of the present study are that mRNAs might interact with ribosomes by complementary sequence matches that are not restricted to the 3′ end of the 18S rRNA and that such interactions may serve to regulate translation.
Acknowledgments
We thank Kristine Flores and Erin Powrie for excellent technical assistance and Drs. Kathyrn Crossin, Bruce Cunningham, Joseph Gally, and Frederick Jones for critical reading of the manuscript. This work was supported by a grant from the G. Harold and Leila Y. Mathers Charitable Foundation (to G.M.E.). G.M.E. is a consultant to Becton Dickinson and Company.
ABBREVIATIONS
- UTR
untranslated region
- s4U
4-thiouridine
References
- 1.Day D A, Tuite M F. J Endocrnol. 1998;157:361–371. doi: 10.1677/joe.0.1570361. [DOI] [PubMed] [Google Scholar]
- 2.Morris D R. In: mRNA Metabolism & Post-Transcriptional Gene Regulation. Harford J B, Morris D R, editors. Vol. 17. New York: Wiley; 1997. pp. 165–180. [Google Scholar]
- 3.Mauro V P, Edelman G M. Proc Natl Acad Sci USA. 1997;94:422–427. doi: 10.1073/pnas.94.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tranque P, Hu M C-Y, Edelman G M, Mauro V P. Proc Natl Acad Sci USA. 1998;95:12238–12243. doi: 10.1073/pnas.95.21.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rastinejad F, Conboy M J, Rando T A, Blau H M. Cell. 1993;75:1107–1117. doi: 10.1016/0092-8674(93)90320-p. [DOI] [PubMed] [Google Scholar]
- 6.Hill W E, Tassanakajohn A. Biochimie. 1987;69:1071–1080. doi: 10.1016/0300-9084(87)90007-1. [DOI] [PubMed] [Google Scholar]
- 7.Graifer D M, Malygin A A, Matasova N B, Mundus D A, Zenkova M A, Karpova G G. Biochim Biophys Acta. 1997;1350:335–344. doi: 10.1016/s0167-4781(96)00176-5. [DOI] [PubMed] [Google Scholar]
- 8.Sprengart M L, Porter A G. Mol Microbiol. 1997;24:19–28. doi: 10.1046/j.1365-2958.1997.3161684.x. [DOI] [PubMed] [Google Scholar]
- 9.Jackson R J. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 71–112. [Google Scholar]
- 10.Bensing B A, Dunny G M. Mol Microbiol. 1997;24:295–308. doi: 10.1046/j.1365-2958.1997.3311709.x. [DOI] [PubMed] [Google Scholar]
- 11.Dooley S, Farber U, Welter C, Theisinger B, Blin N. Gene. 1992;110:263–264. doi: 10.1016/0378-1119(92)90659-d. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida T, Schneider E L, Mori N. Gene. 1994;151:253–255. doi: 10.1016/0378-1119(94)90666-1. [DOI] [PubMed] [Google Scholar]
- 13.Matveeva O V, Shabalina S A. Nucleic Acids Res. 1993;21:1007–1011. doi: 10.1093/nar/21.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheper G C, Voorma H O, Thomas A A M. FEBS Lett. 1994;352:271–275. doi: 10.1016/0014-5793(94)00975-9. [DOI] [PubMed] [Google Scholar]
- 15.Bachellerie J-P, Cavaillé J. Trends Biochem Sci. 1997;22:257–261. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- 16.Maxwell E S. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 17.Nakashima K, Darzynkiewicz E, Shatkin A J. Nature (London) 1980;286:226–230. doi: 10.1038/286226a0. [DOI] [PubMed] [Google Scholar]
- 18.Ganoza M C, Farrow N A, An G. Mol Biol Rep. 1992;16:277–284. doi: 10.1007/BF00419668. [DOI] [PubMed] [Google Scholar]
- 19.Komuro I, Schalling M, Jahn L, Bodmer R, Jenkins N A, Copeland N G, Izumo S. EMBO J. 1993;12:1387–1401. doi: 10.1002/j.1460-2075.1993.tb05783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Awatramani R, Scherer S, Grinspan J, Collarini E, Skoff R, O’Hagan D, Garbern J, Kamholz J. J Neurosci. 1997;17:6657–6668. doi: 10.1523/JNEUROSCI.17-17-06657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin T E, Rolleston F S, Low R B, Wool I G. J Mol Biol. 1969;43:135–149. doi: 10.1016/0022-2836(69)90084-9. [DOI] [PubMed] [Google Scholar]
- 22.Sherton C C, Wool I G. J Biol Chem. 1972;247:4460–4467. [PubMed] [Google Scholar]
- 23.Davies E, Abe S. Methods Cell Biol. 1995;50:209–222. doi: 10.1016/s0091-679x(08)61032-8. [DOI] [PubMed] [Google Scholar]
- 24.Bhangu R, Wollenzien P. Biochemistry. 1992;31:5937–5944. doi: 10.1021/bi00140a033. [DOI] [PubMed] [Google Scholar]
- 25.Dubreuil Y L, Expert-Bezançon A, Favre A. Nucleic Acids Res. 1991;19:3653–3660. doi: 10.1093/nar/19.13.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milligan J F, Uhlenbeck O C. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 27.Hill W E, Camp D G, Tapprich W E, Tassanakajohn A. Methods Enzymol. 1988;164:401–419. doi: 10.1016/s0076-6879(88)64057-2. [DOI] [PubMed] [Google Scholar]
- 28.Wollenzien P L. Methods Enzymol. 1988;164:319–329. doi: 10.1016/s0076-6879(88)64052-3. [DOI] [PubMed] [Google Scholar]
- 29.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmberg L, Melander Y, Nygard O. Nucleic Acids Res. 1994;22:1374–1382. doi: 10.1093/nar/22.8.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sprengart M L, Fuchs E, Porter A G. EMBO J. 1996;15:665–674. [PMC free article] [PubMed] [Google Scholar]
- 32.Raue H A, Klootwijk J, Musters W. Prog Biophys Mol Biol. 1988;51:77–129. doi: 10.1016/0079-6107(88)90011-9. [DOI] [PubMed] [Google Scholar]
- 33.Graifer D M, Juzumiene D I, Karpova G G, Wollenzien P. Biochemistry. 1994;33:6201–6206. doi: 10.1021/bi00186a020. [DOI] [PubMed] [Google Scholar]
- 34.Shine J, Dalgarno L. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hui A, De Boer H A. Proc Natl Acad Sci USA. 1987;84:4762–4766. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacob W F, Santer M, Dahlberg A E. Proc Natl Acad Sci USA. 1987;84:4757–4761. doi: 10.1073/pnas.84.14.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson H D, Mathews M B. Biochimie. 1996;78:909–914. doi: 10.1016/s0300-9084(97)86712-0. [DOI] [PubMed] [Google Scholar]
- 38.Clemens M J. Int J Biochem Cell Biol. 1997;29:945–949. doi: 10.1016/s1357-2725(96)00169-0. [DOI] [PubMed] [Google Scholar]
- 39.Manche L, Green S R, Schmedt C, Mathews M B. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sweeney R, Fan Q, Yao M-C. Proc Natl Acad Sci USA. 1996;93:8518–8523. doi: 10.1073/pnas.93.16.8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobson A. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]