Abstract
We investigated the diversity of the primary sequences of 16S rRNA genes among Neisseria meningitidis strains (Men) and evaluated the use of this approach as a molecular subtyping tool. We aligned and compared a 1,417-bp fragment of the 16S rRNA gene from 264 Men strains of serogroups A, B, C, and Y (MenA, MenB, MenC, and MenY, respectively) isolated throughout the world over a 30-year period. Thirty-one positions of difference were found among 49 16S types: differences between types ranged from 1 to 14 positions (0.07 to 0.95%). 16S types and serogroups were highly associated; only 3 out 49 16S types were shared by two or more serogroups. We have identified 16S types that are exclusively associated with strains of certain hypervirulent clones: 16S type 5 with MenA subgroup III, 16S type 4 with the MenB electrophoretic type 5 (ET-5) complex, and 16S types 12 and 13 with MenC of the ET-37 complex. For MenC strains, 16S sequencing provided the highest sensitivity and specificity and the best overall association with the outbreak-related versus sporadic isolates when compared with pulsed-field gel electrophoresis, multilocus enzyme electrophoresis, and multilocus sequence typing. We demonstrated for the first time an unexpected diversity among 16S rRNA genes of Men strains, identified 16S types associated with well-defined hypervirulent clones, and showed the potential of this approach to rapidly identify virulent strains associated with outbreaks and/or an increased incidence of sporadic disease.
rRNA is essential for the survival of all cells, and the genes encoding the rRNA are highly conserved in the bacteria and other kingdoms. The sequences of the rRNA and proteins comprising the ribosome are highly conserved throughout evolution, because they require complex inter- and intramolecular interactions to maintain the protein-synthesizing machinery (13, 38). Consequently, determination of differences in the sequence of the 16S rRNA gene is well established as a standard method for the identification and phylogenetic classification of prokaryotic species, genera, and families (3, 38) and infers the evolution of the organisms. The sequences of rRNA genes in species with multiple rRNA operons are identical or nearly identical. A few studies have clearly shown the presence of sequence heterogeneity between rRNA operons within single genomes (16, 19). In most organisms, 16S rRNA gene sequences from only a few strains of any particular species have been studied; therefore, variation among operons and its significance have not been fully evaluated (2, 16, 19, 31, 35; http://rdp.cme.msu.edu/).
Differences in 16S rRNA gene sequences and the use of 16S rRNA gene diversity to monitor and identify epidemiologically related isolates became quite apparent in an investigation of the meningococcal disease outbreak associated with the 2000 Hajj pilgrimage in Saudi Arabia and caused by Neisseria meningitidis serogroup W135 (MenW135) strains of the electrophoretic type 37 (ET-37) complex (22). Among the 12 molecular markers used, 16S typing was the most sensitive (100%) and specific (98%) in identifying strains epidemiologically associated with the 2000 Hajj outbreak, while only a combination of other molecular markers allowed for such differentiation (18).
Few N. meningitidis 16S rRNA gene sequences were available in the GenBank database at the time of this study. In the present work, nearly the entire 16S rRNA gene sequences of 264 N. meningitidis strains of various serogroups were determined, and the aligned sequences were analyzed. This study was designed to (i) determine the diversity of 16S rRNA genes among representative N. meningitidis strains of different serogroups, (ii) evaluate the association of particular 16S rRNA gene sequences with hypervirulent clonal groups as defined by multilocus enzyme electrophoresis (MEE) and multilocus sequence typing (MLST), and (iii) assess the potential of 16S typing to rapidly identify virulent strains associated with outbreaks and/or an increased incidence of sporadic disease.
MATERIALS AND METHODS
Epidemiologic definitions. (i) Outbreak.
A meningococcal disease outbreak is defined by the Centers for Disease Control and Prevention's (CDC) Advisory Committee on Immunization Practices as an occurrence of three or more confirmed or probable cases of meningococcal disease during a period of <3 months in persons who have a common affiliation, but no close contact with one another (an organizational-based outbreak), or in two or more persons residing in the same area who are not close contacts and who do not share a common affiliation (a community-based outbreak), resulting in a primary disease attack rate of at least 10 cases per 100,000 persons (4, 23).
(ii) Sporadic case.
A sporadic case of meningococcal disease is defined as the isolation of N. meningitidis from a normally sterile site, such as blood or cerebrospinal fluid (CSF), in a resident of a surveillance area and not part of an outbreak. Active laboratory-based, population-based surveillance for sporadic invasive disease caused by N. meningitidis is part of an ongoing multistate Active Bacterial Core Surveillance (ABCS) coordinated by the CDC. Between 1989 and 1996, the CDC collaborated with investigators at state or local health departments or universities in seven geographically dispersed areas of the United States with an aggregate population of 22 million.
Bacterial strains.
Strains of serogroups A, B, C, and Y were selected to represent the diversity in previously defined hypervirulent clonal groups: subgroup III and the ET-5 and ET-37 complexes. Additional strains were selected based on their association with well-defined and epidemiologically investigated outbreaks or sporadic cases of meningococcal disease (strains of serogroups B, C, and Y) (7, 21, 23, 37). The 264 N. meningitidis strains included in this study belong to the CDC strain collection and were isolated throughout the world over a 30-year period (Table 1).
TABLE 1.
Origins and 16S rRNA types of 264 N. meningitidis strains collected worldwide from 1963 to 2000
| Sero- group | No. of strains | Country of origin | 16S type(s) |
||
|---|---|---|---|---|---|
| Total no. identifieda | Outbreak or epidemic associated | Association with hypervirulent clones | |||
| A | 23 | Burkina Faso, Chad, Djibouti, Ethiopia, Ghana, Iran, Kenya, Niger, Tanzania, Uganda, United States, South Africa, Egypt, Saudi Arabia, and Nigeria | 4 | Type 5 | Clone III-1 |
| B | 58 | Brazil, Cuba, Chile, Norway, New Zealand, and United States | 18 | Type 4 | ET-5 complex |
| C | 119 | United States | 18 | Types 12 and 13 | ET-37 complex |
| Y | 64 | United States | 6 | Types 13 and 19 | Mostly sporadic cases |
| Total | 264 | ||||
The remaining three 16S types were seen in strains of more than one serogroup; 16S type 1 was present in strains of serogroups B and C; 16S type 13 was present in strains of serogroups B, C, and Y; and 16S type 35 was present in strains of serogroups C and Y (Table 2).
(i) Serogroup A strains.
Twenty-three N. meningitidis serogroup A (MenA) strains analyzed in this study were previously characterized at CDC by MEE as belonging to subgroups I (seven strains), II (one strain), and III (15 strains) (data not published). They were isolated in 15 different countries: one strain each from Burkina Faso, Chad, Djibouti, Ethiopia, Ghana, Iran, Kenya, Niger, Tanzania, Uganda, and the United States; two strains from South Africa; three strains each from Egypt and Saudi Arabia; and four strains from Nigeria.
(ii) Serogroup B strains.
We selected 58 N. meningitidis serogroup B (MenB) strains isolated in six different countries: 37 strains from the United States, 12 from Brazil, 4 from Norway, 2 from Chile, 2 from New Zealand, and 1 from Cuba. Of the 37 MenB U.S. strains, 24 were from sporadic cases and were selected to provide statistical representation of isolates collected through the ABCS conducted in the United States between 1989 and 1996. Ten strains were associated with an outbreak of meningococcal B disease in Oregon (1996 to 1998), and 3 strains were from sporadic cases from Oregon and California. No epidemiological information was available to distinguish epidemic from sporadic MenB strains from the other five countries (21 strains). Thirteen of these 21 strains (6 from Brazil, 4 from Norway, 2 from Chile, and 1 from Cuba) belong to the ET-5 complex, the hypervirulent clone responsible for meningococcal disease epidemics in Cuba, Brazil, Chile, and United States (7-10, 26). The remaining eight strains were isolated in New Zealand and Brazil (26), but are not members of the ET-5 complex.
(iii) Serogroup C strains.
We selected 119 N. meningitidis serogroup C (MenC) strains for this study. Ninety-three were collected during the investigations of four well-defined MenC outbreaks in the United States: 35 strains from Texas (24), 12 strains from New Mexico (20), 23 strains from Arizona, and 23 strains from California (including three carrier isolates epidemiologically linked to that outbreak) (32). Of the 93 strains, only 66 met the epidemiological definition as outbreak associated; the remaining 27 strains were isolated at the same time as outbreak investigations were conducted from sporadic cases. Twenty-six additional MenC isolates from sporadic cases were selected to represent isolates collected through the ABCS between 1992 and 1996 (25).
(iv) Serogroup Y strains.
We selected 64 N. meningitidis serogroup Y (MenY) strains isolated in 19 different U.S. states. Fifteen were collected during the investigations of MenY outbreaks in Massachusetts (5 outbreak-associated and 6 sporadic cases) and Los Angeles (4 outbreak-associated cases). Thirty-seven additional MenY isolates were selected to represent geographic distribution of all isolates collected during the periods 1972 to 1975 (16 strains) and 1992 to 1998 (21 strains). The remaining 12 MenY strains were isolated from U.S. Army personnel between 1970 and 1974.
Molecular and epidemiological data.
MEE, MLST, PFGE, and 16S rRNA gene typing data obtained on 93 MenC isolates collected from four well-defined outbreaks of MenC infection in the United States were used to evaluate the sensitivity, specificity, and positive and negative predictive values of these methods in epidemic settings. The MEE and PFGE data were previously published (23). The 16S rRNA gene typing and MLST data were obtained in this study.
MLST.
MLST was performed by sequencing gene fragments of the abcZ, adk, aroE, fumC, gdh, pdhC, and pgm genes as described previously (17; http://www.mlst.net). The detailed MLST results and sources of the isolates have been deposited in a public database (http://www.mlst.net).
PCR.
The 16S rRNA genes of N. meningitidis strains were amplified by PCR with the modified primers 8F (5′AGTTGATCCTGGCTCAG3′) and 1492R (5′ACCTTGTTACGACTT3′) (11). Whole-cell suspensions were used for PCR. Cells were harvested from growth on Trypticase soy agar (Difco Laboratories, Detroit, Mich.) containing 5% (vol/vol) sheep blood and transferred into 1.0 ml of 10 mM Tris buffer (pH 8) with 0.5% (vol/vol) Tween 20 and 0.5% (vol/vol) Nonidet P-40 and then heat killed at 56°C for 10 min. Each final PCR mixture (100 μl) contained 5 U of Expand DNA polymerase (Boehringer Mannheim, Mannheim, Germany); 2 μl of whole-cell suspension; 10 mM Tris-HCl (pH 8.0); 50 mM KCl; 1.5 mM MgCl2; 200 μM (each) dATP, dCTP, dGTP, and dTTP; and 0.4 μM each primer. Reaction mixtures were first incubated for 5 min at 95°C. The mixtures then underwent 35 cycles of 15 s at 94°C, 15 s at the annealing temperature of 50°C, and then 90 s at 72°C. Finally, the reaction mixtures were incubated at 72°C for 5 min. PCR products were purified with Qiaquick PCR purification kit (Qiagen, Valencia, Calif.) according to the manufacturer's protocol.
16S rRNA gene sequence determination.
The amplified product of approximately 1,480 bp was sequenced by using the previously described primers 357 (5′TACGGGAGGCAGCAG3′), 530 (5′CAGCAGCCGCGGTAATAC3′), 790 (5′ATTAGATACCCTGGTAG3′), and 981 (5′CCCGCAACGAGCGCAACCC3′) in the forward and reverse orientations, as well as primers 8F and 1492R (described above) (11, 30, 33). Sequencing was performed with the BigDye terminator cycle sequencing kit (Applied BioSystems, Foster City, Calif.). Sequencing products were purified with Centri-Sep spin columns (Princeton Separations, Adelphia, N.J.) and were resolved on a 4.25% acrylamide-8 M urea gel with an Applied BioSystems model 377 automated DNA sequencing system (Applied BioSystems). An inner fragment of 1,471 bp with four- to sevenfold sequence coverage (corresponding to positions 30 to 1500 of the 16S rRNA of Neisseria gonorrhoeae strain NCTC 83785 and N. meningitidis strain MC58 [accession no. X07714 and AE002551, respectively]) was obtained for each strain and analyzed by using the GCG (Wisconsin) Package, v10.1 (Genetics Computer Group, Madison, Wis.). A number was assigned to each different 16S rRNA gene sequence; a single base change or a mixed base (more than one nucleotide called at a single position) was considered a new 16S type. There is a continuous numbering order of 16S rRNA gene sequence types in our database; however, not all are included in this study. When a novel 16S type, mixed bases, or discrepancies in the alignment were obtained, the 16S rRNA gene amplification and sequencing of the entire gene or parts containing the problematic region were repeated.
The 16S rRNA gene sequence of N. gonorrhoeae (accession no. X07714) was included as an out-group sequence for the phylogenetic tree. A total of 100 bootstrapped trees were sampled to measure the support for each clade on the tree (12). Evolutionary distances were predicted by the method of Jukes and Cantor, which allowed consideration of partial matches between degenerate symbols (15, 27). Therefore, positions with mixed bases were considered in the analysis (a mixed-base mismatch received less value than a base mismatch). Phylogenetic dendrograms were generated by the neighbor-joining method. We also looked at the phylogenetic relationships among different N. meningitidis 16S types by analyzing the sequences by split decomposition (14).
Nucleotide sequence accession numbers.
The 264 16S rRNA gene sequences determined in this study have been deposited in the GenBank database under the following accession numbers: AF310342, AF310343, AF310345 to AF310379, AF310383 to AF310405, AF310409 to AF310416, AF310434 to AF310441, AF310443 to AF310445, AF310451 to AF310454, AF310456 to AF310459, AF310477 to AF310496, AF310501 to AF310532, AF310540 to AF310545, AF310547 to AF310550, AF310552 to AF310597, AF310613 to AF310614, AF337915 to AF337923, AF310926 to AF310946, AF377924, AF377925, AF409913 to AF409946, and AJ239307.
RESULTS
16S rRNA gene sequence diversity.
The nucleotide sequence of the 16S rRNA gene fragments (1,471 bp) from all 264 strains were aligned and compared. Thirty-one differences were found at single positions (not 2 adjacent nucleotides, etc.), and no gaps were present. In 11 of these positions, more than one base was consistently detected (mixed bases). N. meningitidis has four rRNA operons (33), and theoretically the PCR products could have a combination of different nucleotides at any single position in the consensus sequence. There was no particular region of the 16S rRNA gene primary sequence where the positions of differences were located; they were distributed throughout the gene. Differences among the 16S types ranged from 0.07 to 0.95% (a single mixed base to 13 bases and a mixed base, respectively). Among all 31 positions of differences, 28 were transitions (58% C/T and 32% A/G), and two positions were transversions (C/A and G/C at positions 625 and 1100, respectively). At the remaining position (position 755), only a C or Y (mixed nucleotide C or T) was found.
We found 49 different 16S types among the 264 sequences analyzed (Table 2). Forty-six 16S types were found to be associated with a single serogroup. Serogroup A included four 16S types, serogroup B included 18 16S types, serogroup C included 18 16S types, and serogroup Y included 6 16S types. The remaining three 16S types were seen in strains of more than one serogroup; 16S type 1 was present in strains of serogroups B and C; 16S type 13 was present in strains of serogroups B, C, and Y; and 16S type 35 was present in strains of serogroups C and Y (Table 2). The differences found in the 16S rRNA gene between N. gonorrhoeae and different N. meningitidis 16S types ranged from 17 to 23 bases (1.16 to 1.56%). Because the largest difference found among N. meningitidis 16S rRNA gene sequences analyzed was <0.95% (13 of 1,471 bases), we express the differences among the 16S types as the number of nucleotide differences rather than the percentage of nucleotides differing among sequences.
TABLE 2.
16S rRNA gene types among 264 N. meningitidis strains analyzed
| 16S type | MEE grouping | No. of strains | Sero- group | Strain no.a | GenBank accession no. |
|---|---|---|---|---|---|
| 5 | Subgroup III | 15 | A | M7139 | AF310591 |
| 33 | 4 | A | M2781 | AF310563 | |
| 34 | 2 | A | M2786 | AF310565 | |
| 48 | 2 | A | M2727 | AF310582 | |
| 4 | ET-5 complex | 34 | B | M4247 | AF310573 |
| 83 | ET-5 complex | 1 | B | M1417 | AF409941 |
| 11 | 1 | B | NZ3966 | AF310559 | |
| 25 | 4 | B | NZ3968 | AF310555 | |
| 39 | 1 | B | N218/89 | AF310550 | |
| 40 | 1 | B | N206/89 | AF310614 | |
| 41 | 2 | B | N117/89 | AF310556 | |
| 42 | 1 | B | N111/88 | AF310549 | |
| 43 | 1 | B | N112/88 | AF310558 | |
| 65 | 1 | B | M4646 | AF409937 | |
| 69 | 1 | B | M4644 | AF409938 | |
| 81 | 1 | B | M4647 | AF409939 | |
| 82 | 1 | B | M4643 | AF409940 | |
| 84 | 1 | B | M574 | AF409942 | |
| 86 | 1 | B | M1141 | AF409943 | |
| 87 | 1 | B | M2671 | AF409944 | |
| 88 | 1 | B | M4645 | AF409945 | |
| 89 | 1 | B | M690 | AF409946 | |
| 2 | ET-37 complex | 1 | C | M6304 | AF310347 |
| 9 | ET-37 complex | 2 | C | M1533 | AF310553 |
| 12 | ET-37 complex | 33 | C | M1536 | AF310358 |
| 13 | ET-37 complex | 59 | C | M4027 | AF310393 |
| 8 | Y | ||||
| 1 | B | ||||
| 28 | ET-37 complex | 1 | C | M2656 | AF310346 |
| 30 | ET-37 complex | 1 | C | M4015 | AF310544 |
| 35 | ET-37 complex | 4 | C | M2829 | AF310386 |
| 1 | Y | ||||
| 36 | ET-37 complex | 2 | C | M680 | AF310396 |
| 37 | ET-37 complex | 1 | C | M3689 | AF310385 |
| 38 | ET-37 complex | 2 | C | M3790 | AF310384 |
| 44 | ET-37 complex | 1 | C | M810 | AF310345 |
| 46 | ET-37 complex | 1 | C | M540 | AF310496 |
| 1 | 2 | C | M28 | AF310343 | |
| 2 | B | ||||
| 8 | 1 | C | M128 | AF310613 | |
| 10 | 1 | C | M1531 | AF310557 | |
| 16 | 1 | C | M26 | AF310547 | |
| 17 | 1 | C | M1532 | AF310548 | |
| 22 | 2 | C | M414 | AF310543 | |
| 29 | 1 | C | M4648 | AF310545 | |
| 45 | 1 | C | M239 | AF310541 | |
| 47 | 1 | C | M649 | AF310501 | |
| 19 | 49 | Y | M5372 | AF310516 | |
| 32 | 2 | Y | M2905 | AF337915 | |
| 49 | 1 | Y | M6199 | AF310540 | |
| 50 | 1 | Y | M2377 | AF337919 | |
| 51 | 1 | Y | M2907 | AF337918 | |
| 52 | 1 | Y | M2459 | AF337917 |
Representative strains for each 16S type.
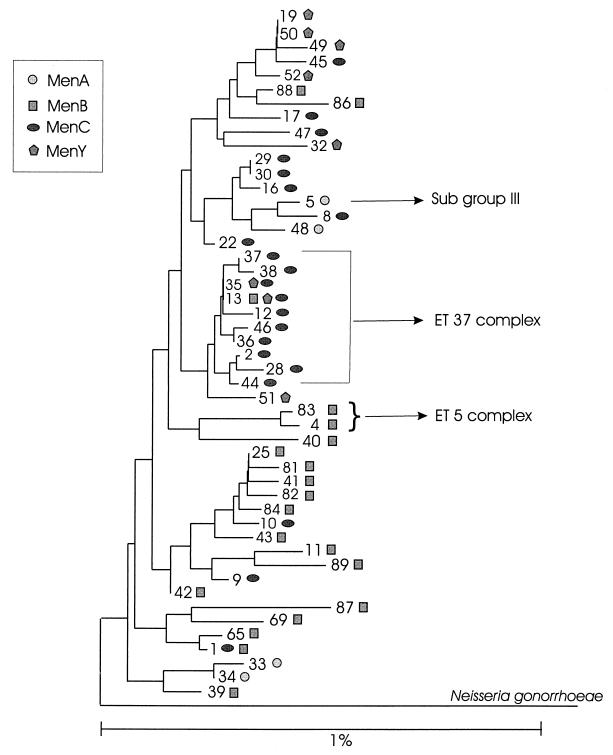
All 49 16S rRNA gene sequences and a 16S rRNA gene sequence of an N. gonorrhoeae strain were aligned. Using those sequences, we generated a phylogenetic dendrogram. However, because of the high similarity of the sequences, the bootstrap values for the tree branches were small: many were below 50%. The low bootstrap values indicated that there were conflicting phylogenetic signals in the 16S rRNA gene data. Therefore, more than one possible tree configuration fit the data. One of the possible configurations is presented in Fig. 1.
FIG. 1.
Consensus neighbor-joining phylogenetic tree constructed with 49 identified N. meningitidis 16S rRNA sequence types. The tree was rooted by using an N. gonorrhoeae 16S rRNA sequence as an out-group (accession no. X07714). The respective meningococcal serogroups are represented by different symbols: circles for serogroup A, squares for serogroup B, ovals for serogroup C, and pentagons for serogroup Y. In some cases, a particular 16S type was identified in more than one serogroup. The number on the branches indicates the 16S rRNA type. The 16S types associated with subgroup III and the ET-37 and ET-5 complexes are indicated. The scale bar represents an expected nucleotide substitution rate of 1%.
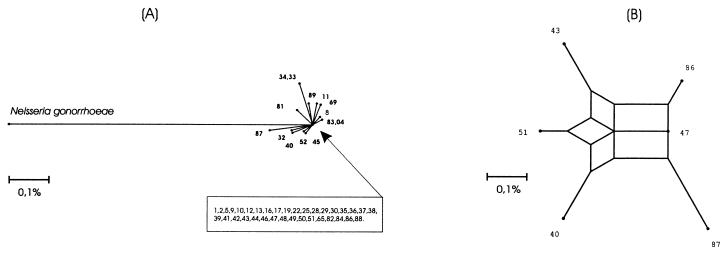
When examining the phylogenetic relationships among 16S rRNA genes of N. meningitidis strains by analyzing sequences through split decomposition (14, 28), we found evidence for networked evolution among the 16S types (Fig. 2). The split graph network supports the low bootstrap values obtained above and shows different possible phylogenies among the 16S types.
FIG. 2.
Split graphs of the Neisseria meningitidis 16S types. In panel A, 49 different 16S types were used to construct the graph. The arrow points to the unresolved part of the split graph, in which 35 of the sequence types are located. In panel B, a split graph of 6 of the 49 16S types, including four types (43, 47, 51, and 86) from the unresolved part of the split graph in panel A, is presented. Each scale bar represents a 0.1% difference among the 16S type sequences.
16S rRNA gene as a molecular marker. (i) Serogroup A strains.
Four different 16S types were found among the 23 representative MenA strains of subgroup I, II, and III. All 15 of the MenA subgroup III strains had the same 16S type, type 5. The strains of subgroups I and II differed from strains of subgroup III, but could not be differentiated from each other because they shared 16S types 33, 34, and 48.
(ii) Serogroup B strains.
Twenty different 16S types were found among the 58 MenB strains tested. Thirty-five of these serogroup B strains belonged to the ET-5 complex, a single strain belonged to the ET-37 complex, and the remaining 22 strains belonged to none of the major hypervirulent clonal groups. Thirty-four of 35 MenB strains of the ET-5 complex had 16S type 4. The only exception was a 16S type 83 strain that differs from type 4 only by a single base and one mixed base. These two 16S types were not seen in any strains outside of the ET-5 complex. Only a MenB strain in the ET-37 complex had 16S type 13. The remaining 22 MenB strains that were outside of these two hypervirulent clonal groups had 17 different 16S types.
Serogroup C strains.
Twenty-one 16S types were found among the 119 MenC strains. Two very similar 16S types were identified among the 66 strains epidemiologically associated with the investigated MenC outbreaks (Table 1): 16S type 12 in the Arizona and New Mexico outbreaks and 16S type 13 in the Texas and California outbreaks (20, 24, 32). However, 25 sporadic case isolates and 1 carrier isolate also had 16S type 12 or 13.
Serogroup Y strains.
Eight different 16S types were found among 64 MenY strains. Type 19 was identified in 49 (76%) strains, and type 13 was identified in 8 (12%) strains, while the remaining 7 strains were of six different 16S types. All 15 isolates from the two MenY outbreaks (Massachusetts and Los Angeles, Calif.) including isolates from outbreak-associated and sporadic cases were 16S type 19. However, 80% (30 of 37) of the MenY strains collected through both active laboratory-based, population-based surveillance periods (1972 to 1975 and 1992 to 1998) were also of 16S type 19; all 16 strains isolated in the period from 1972 to 1975 were of 16S type 19. Among the 12 MenY strains isolated from military personnel during 1971 to 1974, 8 and 4 strains, respectively, were of 16S types 13 and 19.
Four 16S types (16S types 4, 5, 12, and 13) among the 264 strains tested were associated with meningococcal disease outbreaks and/or hypervirulent clones (Table 1). As demonstrated in Fig. 1, these four 16S types are distributed throughout the phylogenetic tree. Two of these four 16S types, types 12 and 13, differ by a single base; furthermore, they differ from types 4 and 5 by six to seven bases. The 117 strains of the ET-37 complex that were analyzed in this study (108 MenC, 8 MenY, and 1 MenB) were previously identified as belonging to 24 distinct ETs (23, 26); 12 different 16S types were identified among these strains.
MLST typing.
We found seven MLST types among the 93 MenC strains isolated during four well-defined outbreaks. Sixty-six of these 93 MenC strains were defined as outbreak associated (63 cases and three carriers), and only two MLST types (types 11 and 1149) were found among them. All outbreak-associated isolates from California, New Mexico, and Texas were MLST type 11. Among the isolates from the Arizona outbreak, 18 were MLST type 1149 and 1 was MLST type 11. MLST types 11 and 1149 were also found in isolates not epidemiologically associated with the outbreaks, but isolated from sporadic cases during the outbreak investigations. Among isolates from sporadic cases, all nine in Texas and five of six in California were MLST type 11, and one of four in Arizona was MLST type 1149.
In New Mexico, all sporadic isolates were of different MLST types from the outbreak isolates. MLST types 11 and 1149 were only found among strains within the ET-37 complex. The sensitivity, specificity, and predictive values of 16S typing, MLST, MEE, and PFGE in identifying outbreak-associated case isolates and discriminating them from sporadic case isolates are presented in Table 3.
TABLE 3.
Comparison of sensitivities and specificities of three different methods of subtyping N. meningitidis serogroup C strains isolated during four meningococcal disease outbreaks
| Outbreak and parametersa | % of strains with result by: |
|||
|---|---|---|---|---|
| 16S | MLST | MEE | PFGE | |
| Texas (26/9) | ||||
| Sensitivity | 100 | 100 | 81 | 85 |
| Specificity | 44 | 0 | 44 | 56 |
| PPV | 84 | 100 | 81 | 85 |
| NPV | 100 | 0 | 44 | 56 |
| New Mexico (7/5) | ||||
| Sensitivity | 100 | 100 | 100 | 100 |
| Specificity | 100 | 100 | 40 | 100 |
| PPV | 100 | 100 | 70 | 100 |
| NPV | 100 | 100 | 100 | 100 |
| Arizona (19/4) | ||||
| Sensitivity | 95 | 95 | 74 | 74 |
| Specificity | 75 | 75 | 75 | 100 |
| PPV | 95 | 95 | 93 | 100 |
| NPV | 75 | 75 | 38 | 44 |
| California (14/6)b | ||||
| Sensitivity | 100 | 100 | 100 | 86 |
| Specificity | 50 | 17 | 50 | 50 |
| PPV | 82 | 69 | 82 | 80 |
| NPV | 100 | 100 | 100 | 60 |
PPV, positive predictive value; NPV, negative predictive value. The first number in parentheses represents the number of outbreak-associated strains, and the second number represents the number of sporadic strains.
Three California carrier isolates epidemiologically linked to that outbreak have been removed for these calculations.
DISCUSSION
We found a large degree of diversity among 16S rRNA genes of 264 N. meningitidis strains, suggesting that 16S rRNA gene sequencing might be a useful molecular typing tool for epidemiological investigations. We consequently focused on two major public health-oriented objectives. First, we investigated the potential association between certain 16S types and well-defined hypervirulent clones; we identified 16S types that were exclusively associated with strains of hypervirulent clones: e.g., 16S type 5 with MenA subgroup III, 16S type 4 with MenB of the ET-5 complex, and 16S types 12 and 13 with MenC and MenW135 (18) of the ET-37 complex. The second objective was to assess the ability of 16S typing to identify strains associated with outbreaks and/or an increase in incidence of sporadic meningococcal disease. The ability to rapidly and consistently differentiate outbreak-associated from sporadic strains is crucial for defining the occurrence of an outbreak. Consequently, timely public health measures, such as vaccination campaigns, a major, and costly component of effective outbreak control, can be implemented. In this study, we found that for MenC strains, 16S typing was the best discriminatory tool for outbreak-associated versus sporadic strains.
Diversity of 16S rRNA genes and genetic relationship among meningococcal strains of different serogroups.
We found more diverse 16S types among strains of serogroups B and C than among serogroups A and Y, which supports previous results obtained with other molecular typing methods (6, 7, 21). We also found associations between particular 16S types and individual serogroups. Mutation, transformation, and/or recombination of 16S rRNA or capsule genes may be responsible for the presence of strains of a particular serogroup on different branches of the tree (Fig. 1). When 16S rRNA gene sequences were analyzed by split decomposition, we found evidence for networked evolution among the 16S rRNA sequences showing different possible phylogenies among 16S types (Fig. 2). The interspecies phylogenetic relationships in the genus Neisseria, defined by the 16S rRNA gene sequence, have been shown to be affected by horizontal DNA transfer and interspecies recombination (28). By analyzing 264 N. meningitidis strains, we have demonstrated that the diversity of 16S rRNA gene sequences and the evidence for interconnected networked evolution indicate intraspecies recombination. However, the range of evolutionary distances among all 49 N. meningitidis 16S types found in our study were small (from a single mixed base to 13 bases plus a mixed base). This small number of differences suggests that the 16S rRNA gene may recombine as frequently as those encoding the housekeeping genes that are the basis for MEE and MLST analysis; however, the pool of functional 16S rRNA gene sequences appears smaller than that for the housekeeping genes.
16S types and meningococcal hypervirulent clones.
MEE has identified hundreds of different ETs, yet most invasive isolates are represented by a limited number of ETs, some of which are only marginally different from one another. Differences among assayed enzymes (6, 29) have led to the concepts of clones, complexes, subgroups, clusters, and lineages. These groups of ETs can be differentiated from one another, but are still closely related enough to recognize a common origin (6). Three major hypervirulent clonal groups defined by MEE, epidemiologically associated with large meningococcal disease epidemics and outbreaks caused by MenA, MenB, and primarily MenC, are subgroup III, the ET-5 complex, and the ET-37 complex, respectively (7, 21, 37). Identification of a strain as a member of one of these major hypervirulent clonal groups could be used as a predictor of an upcoming outbreak. Association with these hypervirulent clonal groups does not necessarily correlate with hypervirulence, because not all members of these hypervirulent clonal groups are associated with outbreaks. Moreover, the same ETs that are most frequently associated with meningococcal disease outbreaks are also frequently identified among sporadic strains (25). We found a consistent and reliable association between distinct 16S types and outbreak-associated members of each of these three major clonal groups.
Most of the recent pandemics of meningococcal disease were caused by MenA strains, and those of subgroup III have been shown to be very homogenous by PFGE (5, 36). Our 16S typing results clearly support these observations; 16S type 5 was identified in all subgroup III strains in our collection. Contrary to that homogeneity, MenB strains are extremely diverse when analyzed by a number of different molecular approaches (5, 7, 24). MenB strains of the ET-5 complex (which includes over a hundred different, but related ET types) tend to be hypervirulent by their epidemiological association with major global transmissions and large meningococcal disease outbreaks. At least six distinct MLST sequence types have been identified in strains of the ET-5 complex (http://neisseria.mlst.net). We found that all but one of the ET-5 complex strains analyzed in this study shared a single 16S type, type 4, not identified in any of the 229 remaining strains in this study. The single exception was a strain with 16S type 83 that differed from 16S type 4 by only a single base and one mixed base, clearly indicating close genetic relationship between these two 16S types.
16S typing and identification of virulent strains associated with outbreaks of meningococcal disease and/or increase in incidence of sporadic disease.
The ability of 16S typing to identify outbreak-associated isolates and discriminate them from sporadic isolates was compared with other methods on 93 MenC strains of which 66 strains were epidemiologically defined as outbreak associated. 16S rRNA gene sequencing provided high sensitivity and specificity and the best overall correlation with the epidemiologic definition of an outbreak-associated strain. Our data are in agreement with those of previous studies that show the low power of discrimination of MLST and MEE between non-outbreak and outbreak-associated isolates. Tzanakaki et al. (34) compared MLST and MEE data, and in most cases, the MLST sequence types of patient and carrier isolates were identical, and the level of discrimination was no greater than that of serotyping. Achtman et al. (1) found that MLST alone could not provide the level of discrimination needed to determine the epidemiologic relatedness of the MenA strain isolated in Moscow during several outbreaks from 1969 and 1997. In a recent study of an outbreak of MenW135 during the Hajj, we found that MLST and MEE provided no discrimination between strains that were epidemiologically associated with the outbreak and those that were not, but had been isolated within the same period (18).
While 12 16S types were identified in 51 sporadic strains of the ET-37 complex, only 2 16S types (types 12 and 13) were identified in 66 outbreak-associated strains from four well-described MenC outbreaks. However, 16S types 12 and 13 were not exclusively seen among outbreak-associated strains; they were also present in 37% of the sporadic cases. MEE analysis of 728 MenC sporadic strains collected through the ABCS from 1992 to 1996 showed that 67% of them belong to ETs related to outbreaks in the United States (ET-24 or -17 or ETs with ≤2 enzymes different from them) and, therefore, could not have been differentiated from the outbreak-associated strains (data not shown). Twenty-six of the 728 MenC strains were analyzed in that study, and 73% of them were of ET-24 or -17 or ETs with ≤2 enzymes different from them. Only 38% of them had 16S types 12 or 13, which qualified them as outbreak associated. Thus, 16S typing reduced the misclassification of strains as outbreak associated by 50%.
Over the past 2 decades, we have witnessed changes in the epidemiology of meningococcal disease in the United States. For example, we have seen an increase in the proportion of sporadic disease caused by MenY, from 11% in 1992 to 33% in 1996, as well as an increase in the number of outbreaks primarily caused by MenC (25). Molecular characterization of these strains has primarily relied on MEE and has indicated that the majority of ETs associated with MenC and MenY outbreaks are the same ones that are identified in sporadic strains, prompting researchers to look for other tools that might provide better differentiation.
The 16S typing results demonstrated that changes in the MenY population within the United States have occurred since the 1970s when great 16S heterogeneity was detected among the 28 MenY strains analyzed: seven different 16S types were identified, one-half of which were type 19. However, all 21 strains from the period 1992 to 1996 were 16S type 19. Thus, increased incidence of MenY disease in the United States appears to have been associated exclusively with 16S type 19 strains. Similar homogeneity was recently detected by serotyping or serosubtyping of a much larger sample of over 300 ABCS MenY isolates (CDC, unpublished data), and by MEE, where only three ETs were identified in one-half of strains (data not shown).
Only six small outbreaks caused by MenY have been reported in the United States (25), and strains from two of them were analyzed in this study; all strains were also 16S type 19. There appears to be a link between the 16S type 19 and MenY strains, because among over 1,200 Men strains of different serogroups in our database, only 7 MenW135 strains were also identified with 16S type 19 (data not shown).
The evolution of N. meningitidis strains during meningococcal disease outbreaks that we have investigated in the past decade was demonstrated by MEE, MLST, and PFGE. It is therefore likely that similar changes occurred with 16S rRNA genes. Indeed, in our most recent international study of MenW135 strains associated with the Hajj pilgrimage, we have seen such evolution within the (W)ET-37 clone (18). This conclusion was only possible with the use of several molecular approaches together. Therefore, it is unlikely that a single method could allow for the precise monitoring that is now possible with the combination of several molecular approaches. While striving for new approaches, we should not ignore established ones. 16S rRNA typing offers some advantages over MEE and MLST in particular situations. Further studies are needed to evaluate the usefulness of 16S rRNA typing for investigations of outbreaks caused by other meningococcal serogroups.
REFERENCES
- 1.Achtman, M., A. van der Ende, P. Zhu, I. S. Koroleva, B. Kusecek, G. Morelli, I. G. Schuurman, N. Brieske, K. Zurth, N. N. Kostyukova, and A. E. Platonov. 2001. Molecular epidemiology of serogroup A meningitis in Moscow, 1969 to 1997. Emerg. Infect. Dis. 7:420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alber, D., M. Oberkötter, S. Suerbaum, H. Claus, M. Frosch, and U. Vogel. 2001. Genetic diversity of Neisseria lactamica strains from epidemiologically defined carriers. J. Clin. Microbiol. 39:1710-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. 1997. Control and prevention of meningococcal disease and control and prevention of serogroup C meningococcal disease: evaluation and management of suspected outbreaks. Morb. Mortal. Wkly. Rep. 46:1-21. [Google Scholar]
- 5.Bygraves, J. A., and M. C. Maiden. 1992. Analysis of the clonal relationships between strains of Neisseria meningitidis by pulsed-field gel electrophoresis. J. Gen. Microbiol. 138:523-531. [DOI] [PubMed] [Google Scholar]
- 6.Caugant, D. A. 1998. Population genetics and molecular epidemiology of Neisseria meningitidis. Acta Pathol. Microbiol. Immunol. Scand. 106:505-525. [PubMed] [Google Scholar]
- 7.Caugant, D. A., L. O. Froholm, K. Bovre, E. Holten, C. E. Frasch, L. F. Mocca, W. D. Zollinger, and R. K. Selander. 1986. Intercontinental spread of a genetically distinctive complex of clones of Neisseria meningitidis causing epidemic disease. Proc. Natl. Acad. Sci. USA 83:4927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caugant, D. A., L. O. Froholm, C. T. Sacchi, and R. K. Selander. 1990. Genetic structure and epidemiology of serogroup B Neisseria meningitidis. p. 37-42. In M. Achtman (ed.), Proceedings of the Seventh International Pathogenic Neisseria Conference, Berlin, Germany.
- 9.Cruz, C., G. Pavez, E. Aguilar, L. Grawe, J. Cam, F. Mendez, J. Garcia, S. Ruiz, P. Vicent, and I. Canepa. 1990. Serotype-specific outbreak of group B meningococcal disease in Iquique, Chile. Epidemiol. Infect. 105:119-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diermayer, M., K. Hedberg, F. Hoesly, M. Fischer, B. Perkins, M. W. Reeves, and D. Fleming. 1999. Epidemic serogroup B meningococcal disease in Oregon: the evolving epidemiology of the ET-5 strain. JAMA 281:1493-1497. [DOI] [PubMed] [Google Scholar]
- 11.Eden, P. A., T. M. Schmidt, R. P. Blakemore, and N. R. Pace. 1991. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int. J. Syst. Bacteriol. 41:324-325. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1985. Confidence limits on phylogeny and approach using the bootstrap. Evolution 39:738-791. [DOI] [PubMed] [Google Scholar]
- 13.Hillis, D. M., C. Moritz, C. A. Porter, and R. J. Baker. 1991. Evidence for biased gene conversion in concerted evolution of ribosomal DNA. Science 251:308-310. [DOI] [PubMed] [Google Scholar]
- 14.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 15.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 16.Liefting, L. W., M. T. Andersen, R. E. Beever, R. C. Gardner, and R. L. Forster. 1996. Sequence heterogeneity in the two 16S rRNA genes of Phormium yellow leaf phytoplasma. Appl. Environ. Microbiol. 62:3133-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer, L. W., M. W. Reeves, N. Al-Hamdan, C. T. Sacchi, M. K. Taha, G. W. Ajello, S. E. Schmink, C. Noble, M. L. Tondella, A. M. Whitney, Y. Al-Mazrou, M. Al-Jefri, A. Mishkhis, S. Sabban, D. A. Caugant, J. Lingappa, N. E. Rosenstein, and T. Popovic. 2002. Outbreak of W135 meningococcal disease in 2000: not emergence of a new W135 strain, but clonal expansion within the electrophoretic type-37 complex. J. Infect. Dis. 185:1596-1605. [DOI] [PubMed] [Google Scholar]
- 19.Mylvaganam, S., and P. P. Dennis. 1992. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics 130:399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson, R. K. 1995. Outbreak of serogroup C meningococcal meningitis in rural New Mexico. Epidemiology report. State of New Mexico, Department of Health, Santa Fe, N.Mex.
- 21.Olyhoek, T., B. A. Crowe, and M. Achtman. 1987. Clonal population structure of Neisseria meningitidis serogroup A isolated from epidemics and pandemics between 1915 and 1983. Rev. Infect. Dis. 9:665-692. [DOI] [PubMed] [Google Scholar]
- 22.Popovic, T., C. T. Sacchi, M. W. Reeves, A. M. Whitney, L. W. Mayer, C. A. Noble, G. W. Ajello, F. Mostashari, N. Bendana, J. Lingappa, R. Hajjeh, and N. E. Rosenstein. 2000. Neisseria meningitidis serogroup W135 isolates associated with the ET-37 complex. Emerg. Infect. Dis. 6:428-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popovic, T., S. Schmink, N. A. Rosenstein, G. W. Ajello, M. W. Reeves, B. Plikaytis, S. B. Hunter, E. M. Ribot, D. Boxrud, M. L. Tondella, C. Kim, C. Noble, E. Mothershed, J. Besser, and B. A. Perkins. 2001. Evaluation of pulsed-field gel electrophoresis in epidemiological investigations of meningococcal disease outbreaks caused by Neisseria meningitidis serogroup C. J. Clin. Microbiol. 39:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenstein, N., O. Levine, J. P. Taylor, D. Evans, B. D. Plikaytis, J. D. Wenger, and B. A. Perkins. 1998. Efficacy of meningococcal vaccine and barriers to vaccination. JAMA 279:435-439. [DOI] [PubMed] [Google Scholar]
- 25.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, L. Lefkowitz, M. L. Cartter, R. Danila, P. Cieslak, K. A. Shutt, T. Popovic, A. Schuchat, L. H. Harrison, and A. L. Reingold. 1999. The changing epidemiology of meningococcal disease in the United States, 1992-1996. J. Infect. Dis. 180:1894-1901. [DOI] [PubMed] [Google Scholar]
- 26.Sacchi, C. T., L. L. Pessoa, S. R. Ramos, L. G. Milagres, M. C. C. Camargo, N. T. Hidalgo, C. E. A. Melles, D. A. Caugant, and C. E. Frasch. 1992. Ongoing group B Neisseria meningitidis epidemic in São Paulo, Brazil, due to increased prevalence of a single clone of the ET-5 complex. J. Clin. Microbiol. 30:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sacchi, C. T., R. C. Zanella, D. A. Caugant, C. E. Frasch, N. T. Hidalgo, L. G. Milagres, L. L. Pessoa, S. R. Ramos, M. C. Camargo, and C. E. A. Melles. 1992. Emergence of a new clone of serogroup C Neisseria meningitidis in São Paulo, Brazil. J. Clin. Microbiol. 30:1282-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, N. H., E. C. Holmes, G. M. Donovan, G. A. Carpenter, and B. G. Spratt. 1999. Networks and groups within the genus Neisseria: analysis of argF, recA, rho, and 16S rRNA sequences from human Neisseria species. Mol. Biol. Evol. 16:773-783. [DOI] [PubMed] [Google Scholar]
- 29.Spratt, B. G., N. H. Smith, J. Zhou, M. O'Rourke, and E. Feil. 1995. The population genetics of the pathogenic Neisseria, p. 143-160. In S. Baumberg, J. P. W. Young, E. M. H. Wellington, and J. R. Saunders (ed.), The population genetics of bacteria. Cambridge University Press, Cambridge, United Kingdom.
- 30.Stackebrandt, E., and O. Charfreitag. 1990. Partial 16S rRNA primary structure of five Actinomyces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J. Gen. Microbiol. 136:37-43. [DOI] [PubMed] [Google Scholar]
- 31.Stackebrandt, E., W. Liesack, and D. Witt. 1992. Ribosomal RNA and rDNA sequence analyses. Gene 115:255-260. [DOI] [PubMed] [Google Scholar]
- 32.Tappero, J. W., R. Reporter, J. D. Wenger, B. A. Ward, M. W. Reeves, T. S. Missbach, B. D. Plikaytis, L. Mascola, and A. Schuchat. 1996. Meningococcal disease in Los Angeles County, California, and among men in the county jails. N. Engl. J. Med. 335:833-840. [DOI] [PubMed] [Google Scholar]
- 33.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 34.Tzanakaki, G., R. Urwin, M. Musilek, P. Kriz, J. Kremastinou, A. Pangalis, C. C. Blackwell, and M. C. J. Maiden. 2001. Phenotypic and genotypic approaches to characterization of isolates of Neisseria meningitidis from patients and their close family contacts. J. Clin. Microbiol. 39:1235-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ueda, K., T. Seki, T. Kudo, T. Yoshida, and M. Kataoka. 1999. Two distinct mechanisms cause heterogeneity of 16S rRNA. J. Bacteriol. 181:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, J. F., D. A. Caugant, X. Li, X. Hu, J. T. Poolman, B. A. Crowe, and M. Achtman. 1992. Clonal and antigenic analysis of serogroup A Neisseria meningitidis with particular reference to epidemiological features of epidemic meningitis in the People's Republic of China. Infect. Immun. 60:5267-5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, J. F., D. A. Caugant, G. Morelli, B. Koumare, and M. Achtman. 1993. Antigenic and epidemiologic properties of the ET-37 complex of Neisseria meningitidis. J. Infect. Dis. 167:1320-1329. [DOI] [PubMed] [Google Scholar]
- 38.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]