Abstract
Twenty-one Escherichia coli O157:H7 strains isolated in northern Italy from sporadic cases of hemolytic-uremic syndrome and from cattle and food were characterized by virulence gene analysis, pulsed-field gel electrophoresis (PFGE) of XbaI-digested DNA, enterobacterial repetitive intergenic consensus (ERIC) sequence-based PCR (ERIC-PCR), and antibiotic resistance patterns and compared to 18 strains isolated in France from human cases of diarrhea, cattle, and the environment. Strains isolated in Sicily (southern Italy) from a local farm (one strain) and from calves just imported from France (11 strains) and Spain (six strains) were also typed. Whereas the eae and hlyA genes were always detected, Shiga toxin gene (stx) analysis showed some differences related to geographic areas. Isolates from northern Italy showed a high frequency of stx1 and stx2, while strains isolated in France and from French and Spanish calves imported to Sicily more frequently possessed the stx2c gene. The majority of the strains isolated in northern Italy were also resistant to one or more antibiotics, while most of the strains isolated in France and Sicily were fully susceptible. ERIC-PCR analysis was not able to differentiate the strains. PFGE typing after XbaI DNA digestion produced a total of 54 distinct restriction endonuclease digestion profiles (REDPs) among the 57 strains. Phylogenetic analysis was unable to cluster REDPs according to geographic origin. All epidemiologically related isolates showed either identical or ≥91% similar REDPs. Our findings suggest a peculiar circulation of antibiotic-resistant, genetically unrelated strains in northern Italy.
Different human diseases, from mild diarrhea to hemorrhagic colitis, hemolytic-uremic syndrome, and thrombotic thrombocytopenic purpura, can be caused by the O157:H7 serovar of Shiga toxin-producing Escherichia coli (13). The pathogenicity of E. coli O157:H7 strains depends on different virulence factors, including the production of one or more of the Shiga-like toxins stx1, stx2, and other stx variants, the ability to cause attaching-and-effacing lesions in the intestinal mucosa (depending on the eae gene), and the possession of large plasmids encoding adhesins and hemolysins (5, 19, 23). Human infections due to this serovar have been observed with increasing frequency in different countries, and contaminated food, mainly undercooked ground beef, is most often implicated (1, 5, 8, 13).
The principal reservoir of the microorganism has been recognized in infected cattle, especially young animals (8, 11, 14). The prevalence of E. coli O157 carriage in North American and European cattle varies according to the country and the detection method, ranging from 0 to 10% (1, 8, 11, 12, 13). In Italy, strains of E. coli belonging to serogroup O157 have been isolated from cattle in some northern regions (4, 9), where human cases of hemolytic-uremic syndrome have been observed (7, 21). In southern Italy, isolations from neither cattle of local farms nor humans have been reported so far. In 1998 to 1999, the occurrence of E. coli O157:H7 was investigated in 35 dairy farms located in Sicily (12). In all, 207 rectal swabs taken from calves at different farms were examined. All but one were negative for E. coli of the O157 serogroup. The only E. coli O157:H7 isolate was recovered at a farm where calves imported from France had been stalled in the preceding weeks. On the contrary, 11 of 196 calves just imported to Sicily from France and 6 of 150 just imported from Spain were positive for Shiga toxin-producing E. coli O157:H7. The calves were imported for slaughter, and swabs were taken just after their arrival during 1998 to 1999.
In this study, E. coli O157:H7 isolates from cattle and human cases isolated in France and in northern Italy and from French and Spanish cattle imported to Sicily were studied by the analysis of virulence genes, pulsed-field gel electrophoresis (PFGE) of digested DNA, and ERIC sequence-based PCR (ERIC-PCR) with the aim of investigating the genetic relationships among isolates from these different origins. In addition, the susceptibility of the isolates to antibiotics was tested and compared with the genotyping results.
MATERIALS AND METHODS
Strains.
A total of 57 E. coli O157:H7 strains were studied: 21 strains were isolated in northern Italy, four from human sporadic cases of hemolytic-uremic syndrome and 17 from rectal swabs of cattle and from food; 18 strains were isolated in Sicily (southern Italy), one from a rectal swab from a calf from a local farm, 11 from rectal swabs of French imported calves, and six from rectal swabs of Spanish imported calves; and 18 strains were isolated in France, nine strains from rectal swabs of calves and the environment and nine strains from human cases of diarrhea.
Biochemical tests and serotyping.
Biochemical identification of isolates was performed with the API 20E (bioMérieux, Marcy-l'Etoile, France). Serological typing for O157 and H7 antigens was performed by slide agglutination with polyvalent and monovalent anti-E. coli O and H sera (Biogenetics Diagnostics, Padua, Italy).
Antibiotic susceptibility testing.
Susceptibility to antibiotics was tested by the disk diffusion method on brain heart infusion agar with the zone size criteria recommended by the disk manufacturer (Bio-Rad Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France). The following antibiotics were used: amoxicillin, ampicillin, ceftriaxone, chloramphenicol, ciprofloxacin, imipenem, kanamycin, amdinocillin, nalidixic acid, streptomycin, sulfonamide, tetracycline, and trimethoprim.
Virulence genes.
The detection and subtyping of stx genes were performed by the Linn PCR system as previously described (2). PCR procedures were also used to detect eae (3) and enterohemorrhagic E. coli hlyA genes (17). DNA was extracted and purified with an Autogen 540 automated DNA extraction system (AutoGen Instruments, Beverly, Mass.). Bacterial lysates were subjected to two phenol-chloroform purifications and ethanol precipitation, and DNA was resuspended in 100 ml of TE buffer (10 mM Tris base [pH 8], 1 mM disodium EDTA).
PFGE fingerprinting.
The PFGE technique of contour-clamped homogeneous electric fields (CHEF) was used for the fingerprinting of isolates. XbaI (Pharmacia LKB Biotechnology AB, Uppsala, Sweden) was used for digestion of genomic DNA as described by Faith et al. (11). Following digestion, the genomic DNA fragments were resolved by CHEF-PFGE with a CHEF-DRIII apparatus (Bio-Rad Laboratories, Richmond, Calif.) at 6 V/cm for 30 h at 10°C with a pulse time of 20 to 65 s and linear ramping and an electrical field angle of 120°. Multimers of phage lambda (New England Biolabs, Inc., Beverly, Mass.) were used as molecular size standards. The gels were stained with ethidium bromide, and the bands were visualized and photographed with UV transillumination.
The similarities among restriction endonuclease digestion profiles (REDPs) were calculated by the Dice similarity index (6, 10) with the Taxotron software Restrictotyper module (Taxolab Software, Institut Pasteur, Paris, France). Bands whose molecular weights differed by less than 4% were considered identical. Fragments smaller than 40 kb in length were not used in REDP comparisons. The REDPs were considered identical when the Dice similarity index was 100%, closely related if the index was ≥91%, and not related if it was ≤90%. A dendrogram tree was constructed with the Adanson and Dendrograf modules of the Taxotron software (Taxolab Software) and applying the unweighted pair group method of averages (UPGMA) algorithm to the distance matrix resulting from the comparison of the REDP patterns.
ERIC-PCR.
Analysis by ERIC-PCR was performed according to Versalovic et al. (22). Amplification with primer ERIC1 was performed in a Gene Amp PCR system 9700 thermal cycler (PE Biosystems, Foster City, Calif.) with the following temperature program: 95°C to denature the template; four cycles of 94°C for 1 min, 26°C for 1 min, and 72°C for 2 min; 40 cycles of 94°C for 30 s, 40°C for 30 s, and 72°C for 1 min; and finally 72°C for 10 min. To separate amplified products, 25 μl was electrophoresed on a 1.6% agarose gel in TBE (Tris-borate-EDTA) buffer at 50 V for 16 h. The gels were stained with ethidium bromide, and the bands were visualized and photographed with UV transillumination. Patterns were defined by visual inspection on the basis of the number and position of bands.
RESULTS
Antibiotic susceptibility.
The majority (13 of 21) of the strains isolated in northern Italy from diseased children, calves, and food were resistant to one or more antibiotics (Tables 1 and 2). Resistance patterns ranged from resistance to sulfonamide alone to resistance to seven different antibiotics (Table 1). In contrast, the majority of the strains isolated in Sicily from Spanish calves (five of six) and from French calves (eight of eleven) were susceptible to all antibiotics tested. All but one of the strains isolated in France from calves, diseased children, and the environment were also susceptible to all antibiotics tested. The only resistant strain from Spanish calves showed a pattern including resistance to sulfonamide, streptomycin, and tetracycline. The three resistant strains isolated in Sicily from French calves showed different patterns, including resistance to two, three, and seven antibiotics, while the resistant strain isolated in France showed resistance only to chloramphenicol.
TABLE 1.
Results of typing of E. coli O157:H7 isolates from different origins
| Origin | Isolatea | Presence of virulence genes |
REDPb | Resistotypec | ||||
|---|---|---|---|---|---|---|---|---|
| stx1 | stx2 | stx2c | eae | hlyA | ||||
| Northern Italy | ||||||||
| Humans | 21 (ED 10.88) | − | + | − | + | + | NR | Sensitive |
| 22 (ED 177.94) | + | + | − | + | + | NR | Sul, Tri, Tet | |
| 30 (D 39.90) | − | − | + | + | + | NR | Sensitive | |
| 36 (D 297.98) | − | + | + | + | + | NR | Sul, Str, Tet | |
| Calves | 23 (ED 243.97) | + | + | − | + | + | NR | Sul, Str |
| 24 (ED 244.97) | + | + | − | + | + | NR | Amp, Sul, Str, Tri | |
| 25 (ED 275.97) | + | + | − | + | + | NR | Sensitive | |
| 31 (D 217) | − | − | + | + | + | NR | Sensitive | |
| 32 (D 222) | + | − | + | + | + | CR1 | Sensitive | |
| 33 (D 248.98) | + | − | + | + | + | CR1 | Sensitive | |
| 35 (D 290.98) | + | − | + | + | + | NR | Sul, Str, Tet | |
| 38 (D 316.98)d | + | + | − | + | + | CR2 | Sul, Str, Tet | |
| 40 (D 318.98)d | + | + | − | + | + | CR2 | Sul, Str, Tet | |
| 42 (D 351.98)e | − | + | − | + | + | CR3 | Sul | |
| 43 (D 352.98)e | − | + | − | + | + | CR3 | Sul | |
| 44 (D 404.99)f | − | + | + | + | + | CR4 | Sul, Str, Tet | |
| 45 (D 405.99)f | − | + | + | + | + | CR4 | Sensitive | |
| 46 (D 406.99) | + | − | + | + | + | NR | Amp, Sul, Str, Tet, Chl, Kan, Amo | |
| Minced meat | 41 (D 350.98)e | − | + | − | + | + | CR3 | Sul |
| Milk | 34 (D 282.97) | − | − | + | + | + | NR | Sensitive |
| Beets | 37 (D 300.98) | + | + | − | + | + | NR | Sul |
| Sicily (southern Italy) | ||||||||
| Calf from local farm | 06 (3A.98) | − | − | + | + | + | NR | Sensitive |
| Imported Spanish calves | 01 (01.97) | − | + | − | + | + | NR | Sensitive |
| 07 (05.99) | − | − | + | + | + | NR | Sensitive | |
| 13 (13.99) | − | − | + | + | + | CR5 | Sensitive | |
| 19 (20.99) | − | − | + | + | + | NR | Sensitive | |
| 20 (21.99) | − | − | + | + | + | NR | Sensitive | |
| 57 (27.99) | + | − | − | + | + | NR | Sul, Str, Tet | |
| Imported French calves | 02 (01.99) | − | − | + | + | + | CR5 | Sensitive |
| 03 (02.98) | − | − | + | + | + | NR | Sensitive | |
| 04 (03.4.97) | + | + | − | + | + | NR | Sensitive | |
| 08 (08.2.99) | − | − | + | + | + | NR | Sensitive | |
| 09 (08.4.99) | − | − | + | + | + | NR | Sensitive | |
| 10 (09.99) | − | + | + | + | + | NR | Sul, Str, Tet | |
| 11 (10.98) | + | + | − | + | + | NR | Amp, Sul, Tri, Str, Kan, Chl, Tet | |
| 15 (14.99) | − | − | + | + | + | NR | Sensitive | |
| 16 (16.98) | + | + | − | + | + | NR | Sul, Str | |
| 55 (14.99) | − | − | + | + | + | NR | Sensitive | |
| 56 (23.99) | − | − | + | + | + | NR | Sensitive | |
| France | ||||||||
| Calves | 47 (HSVR 03) | − | − | + | + | + | NR | Sensitive |
| 48 (HSVR 04) | − | − | + | + | + | NR | Sensitive | |
| 49 (HSVR 07)g | − | − | + | + | + | ID1 | Sensitive | |
| 50 (HSVR 08)g | − | − | + | + | + | ID1 | Sensitive | |
| 51 (HSVR 09)g | − | − | + | + | + | ID1 | Sensitive | |
| Environment | 52 (HSVR 11)h | − | − | + | + | + | ID2 | Sensitive |
| 53 (HSVR 12)h | − | − | + | + | + | ID2 | Sensitive | |
| 54 (HSVR 13)h | − | − | + | + | + | CR to ID2 | Sensitive | |
| 69 (EC 99/083) | − | − | + | + | + | CR6 | Sensitive | |
| Humans | 60 (EC 97/413) | − | + | − | + | + | CR6 | Sensitive |
| 61 (EC 98/433) | − | + | + | + | + | NR | Sensitive | |
| 62 (EC 98/475) | + | − | − | + | + | NR | Chl | |
| 63 (EC 98/491) | + | − | − | + | + | NR | Sensitive | |
| 65 (EC 98/498) | − | + | − | + | + | CR7 | Sensitive | |
| 66 (EC 98/499) | − | + | − | + | + | CR7 | Sensitive | |
| 67 (EC 99/074) | − | − | + | + | + | NR | Sensitive | |
| 68 (EC 99/081) | − | + | − | + | + | NR | Sensitive | |
| 70 (EC 99/095) | − | − | + | + | + | NR | Sensitive | |
Strains related on the basis of epidemiological data.
NR, not related REDP; ID, identical REDP; CR, closely related REDP.
Sul, sulfonamides; Str, streptomycin; Tet, tetracycline; Amp, ampicillin; Kan, kanamycin; Chl, chloramphenicol; Tri, trimethoprim; Amo, amoxicillin.
Strain 38 was isolated from both the feces and carcass of the same calf, while strain 40 was isolated from the feces of another calf at the same farm.
Srains isolated from minced meat (n = 41), the carcass of a calf (n = 42), and a rectal swab of another calf (n = 43) at the same farm.
Strains isolated from calves slaughtered in the same period in the same slaughterhouse.
Strains isolated from calves slaughtered on the same day in the same slaughterhouse.
Strains isolated from the environment of a slaughterhouse.
TABLE 2.
stx genotypes and resistotypes of E. coli O157:H7 isolates from different origins
| Isolate origin (no. of isolates) | stx genotype | No. of isolates | Resistotypea | No. of isolates |
|---|---|---|---|---|
| Northern Italy | ||||
| Humans (4) | stx1stx2 | 1 | Susceptible | 2 |
| stx2 | 1 | Resistant | 2 | |
| stx2stx2c | 1 | |||
| stx2c | 1 | |||
| Calves and food (17) | stx1stx2 | 6 | Susceptible | 6 |
| stx1stx2c | 4 | Resistant | 11 | |
| stx2 | 3 | |||
| stx2stx2c | 2 | |||
| stx2c | 2 | |||
| Sicily (southern Italy) | ||||
| Calf from local farm (1) | stx2c | 1 | Susceptible | 1 |
| Imported Spanish calves (6) | stx1 | 1 | Susceptible | 5 |
| stx2 | 1 | Resistant | 1 | |
| stx2c | 4 | |||
| Imported French calves (11) | stx1stx2 | 3 | Susceptible | 8 |
| stx2stx2c | 1 | Resistant | 3 | |
| stx2c | 7 | |||
| France | ||||
| Calves and environment (9) | stx2c | 9 | Susceptible | 9 |
| Humans (9) | stx1 | 2 | Susceptible | 8 |
| stx2 | 4 | Resistant | 1 | |
| stx2stx2c | 1 | |||
| stx2c | 2 |
Susceptible to all the antibiotics tested or resistant to one or more of the antibiotics tested.
Virulence genes.
The eae and hlyA genes were detected in all 57 strains independently of their origin (Table 1). On the contrary, six different genotypes, stx1, stx2, stx2c, stx1 stx2, stx1 stx2c, and stx2 stx2c, were observed for the verocytotoxin genes, and their distribution differed according to the geographic area of isolation of the strains (Tables 1 and 2). In fact, the strains isolated in northern Italy showed a higher frequency of the stx1 and stx2 genes, while the strains isolated in Sicily from imported calves and the strains isolated in France more frequently possessed the stx2c gene. In particular, the stx1 gene, alone or associated with stx2, was observed in 11 of 21 strains from northern Italy, but only 6 of 36 strains from Sicily and France. On the contrary, stx2c alone was observed in 23 of 36 strains from Sicily and France, but only 3 of 21 strains from northern Italy.
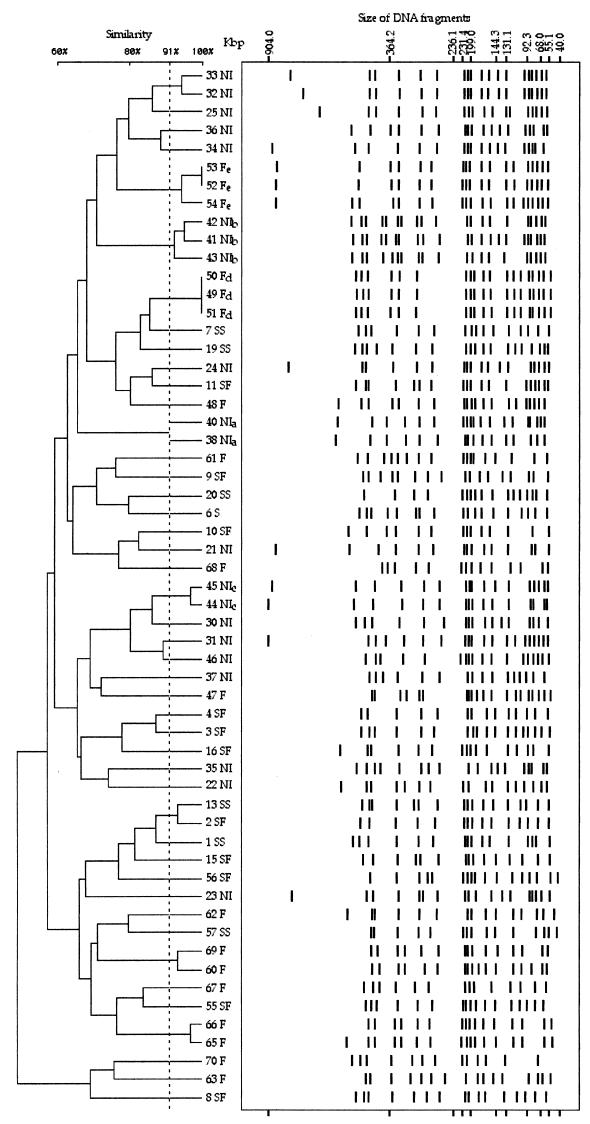
PFGE analysis.
The number of DNA fragments obtained by XbaI digestion ranged between 13 and 22, with sizes of approximately 43 to 915 kb. Bands smaller than 40 kb were not considered in the calculation of the similarity index. A total of 54 different REDPs on PFGE patterns were recognized (Fig. 1 and Table 1). Only five strains ranged in two groups of identical REDPs, ID1 and ID2. Fifteen other strains clustered in seven groups of closely related (≥91% similar) REDPs, from CR1 to CR7. Finally, one strain was closely related to the ID2 group. The strains belonging to the ID1 group were from cows slaughtered on the same day in the same slaughterhouse, while the strains with the ID2 and the CR to ID2 REDPs were isolated from different points within a slaughterhouse.
FIG. 1.
Dendrogram showing distance calculated by the Dice similarity index of PFGE REDPs among the 57 E. coli O157:H7 strains from different origins. F, France; NI, northern Italy; S, Sicily; SF, Sicily from French imported calves; SS, Sicily from Spanish imported calves. Strains related on the basis of epidemiological data are noted in Table 1 (see footnote a).
Epidemiological correlation was also found for the strains isolated in northern Italy that belonged to the CR2, CR3, and CR4 groups (Table 1). No apparent epidemiological correlation was found for strains within the CR1, CR5, CR6, and CR7 groups. No peculiarity was observed in the REDP patterns that could be correlated to the origin of the isolates. In the dendrogram produced by the UPGMA algorithm, the isolates were clustered in four groups (3 to 28 strains per group) of >60% similar strains according to the Dice similarity index. In each of the three major groups of 28, 12, and 14 strains, isolates from different geographic areas were present (Fig. 1).
ERIC-PCR analysis.
By the ERIC-PCR technique, three major and two minor bands in the 200- to 1,300-bp size range were observed with all the strains.
DISCUSSION
Strains of E. coli O157:H7 isolated in different geographic areas were studied by one phenotypic (resistance to antibiotics) and three genotypic (virulence gene profiles, XbaI PFGE patterns, and ERIC-PCR analysis) typing methods. The study of antibiotic resistance patterns (resistotyping) showed some differences between the strains isolated in northern Italy and those isolated in Sicily (southern Italy) and in France. Resistant and multiresistant strains were most frequently found in both human and animal strains isolated in northern Italy. The great majority of strains isolated from imported French and Spanish calves in Sicily and all but one of the strains isolated in France from human cases, calves, and the environment were susceptible to the antibiotics tested.
Among the genotypic typing methods, the subtyping of stx genes by the Linn PCR system confirmed the differences observed with resistotyping, since the frequency of some of the six different stx genotype patterns was different according to the origin of the strains (Table 2). In particular, the stx2c gene was more frequently observed in the strains isolated in France and in those isolated in Sicily from French and Spanish calves, while the stx1 and stx2 genes were more frequently found in northern Italian isolates. On the contrary, by the ERIC-PCR technique, all the strains gave the same result independently of their origin, confirming that this method is unable to distinguish epidemiologically unrelated strains of E. coli O157:H7 (20).
Because of its high discriminatory power, PFGE is now considered the “gold standard” for the molecular typing of O157:H7 strains (15, 16) and is accepted as the standard technique in epidemiological investigations for local events. The use of this technique has been precious for demonstrating horizontal transmission of a single strain type among animals within a farm (14) as well as characterizing outbreak-related isolates (16) and comparing sporadic isolates with epidemic strains at the local scale (15). The high discriminatory ability of PFGE analysis was clearly confirmed by our study, since we had as many as 54 different REDPs among the 57 strains from different origins (Fig. 1).
As expected, all the epidemiologically related strains (Table 1) showed either identical or ≥91% similar REDPs. On the contrary, the great majority of epidemiologically unrelated strains exhibited distinct patterns. Considering the great variety of REDPs found among strains within the same country, our results are not encouraging for the usefulness of PFGE typing for large-scale epidemiological investigations. In fact, in our study, PFGE typing was unable to characterize the strains isolated in Sicily from imported calves according to their geographic origin, since no similarities were observed between strains isolated in Sicily from French calves and strains isolated in France. However, the circulation of a great variety of endemic clones in all the geographic areas studied has been confirmed.
The data obtained from resistotyping and from the genetic analysis of stx genes were of great interest. In fact, the differences observed in the strains isolated in northern Italy compared to those isolated in France and from French cattle in Sicily suggest that the circulation of O157:H7 strains in the two geographic areas was quite independent in the period studied. Attention should be paid to the higher rate of strains resistant to antibiotics, including two human isolates, that we observed in northern Italy. The genetic and phenotypic variety observed among antibiotic-resistant isolates from northern Italy demonstrates that they do not have a single clonal origin. Although we have no history of antibiotic treatments for the calves sampled in northern Italy, a higher selective pressure linked to different attitudes about the use of antibiotics on farms is likely to be present in this area. Special care should be used in controlling the selection of new resistotypes in animals, since multiresistant clones selected in animals may spread to the human population.
Acknowledgments
Thanks are due to Bruno Andral (AFSSA, Lyon, France) for supplying some of the strains used in this study.
REFERENCES
- 1.Armstrong, G. L., J. Hollingsworth, and J. G. Morris, Jr. 1996. Emerging foodborne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol. Rev. 18:29-51. [DOI] [PubMed] [Google Scholar]
- 2.Bastian, S. N., I. Carle, and F. Grimont. 1998. Comparison of 14 PCR systems for the detection and subtyping of stx genes in Shiga-toxin-producing Escherichia coli. Res. Microbiol. 149:457-472. [DOI] [PubMed] [Google Scholar]
- 3.Beaudry, M., C. Zhu, J. M. Fairbrother, and J. Harel. 1996. Genotypic and phenotypic characterization of Escherichia coli isolates from dogs manifesting attaching and effacing lesions. J. Clin. Microbiol. 34:144-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonardi, S., E. Maggi, A. Bottarelli, M. L. Pacciarini, A. Ansuini, G. Vellini, S. Morabito, and A. Caprioli. 1999. Isolation of verocytotoxin producing Escherichia coli O157:H7 from cattle at slaughter in Italy. Vet. Microbiol. 67:203-211. [DOI] [PubMed] [Google Scholar]
- 5.Boyce, T. G., D. L. Swerdlow, and P. M. Griffin. 1995. Escherichia coli O157:H7 and the hemolytic-uremic syndrome. N. Engl. J. Med. 333:364-368. [DOI] [PubMed] [Google Scholar]
- 6.Brosch, R., J. Chen, and J. B. Luchansky. 1994. Pulsed-field fingerprinting of listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl. Environ. Microbiol. 60:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caprioli, A., I. Luzzi, F. Rosmini, P. Pasquini, R. Cirrincione, A. Gianviti, M. C. Matteucci, and G. Rizzoni. 1992. Hemolytic uremic syndrome and verocytotoxin-producing Escherichia coli infection in Italy. J. Infect. Dis. 166:154-158. [DOI] [PubMed] [Google Scholar]
- 8.Chapman, P. A., D. J. Wright, P. Norman, J. Fox, and E. Crick. 1993. Cattle as a possible source of verocytotoxin-producing Escherichia coli O157 infection in man. Epidemiol. Infect. 111:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conedera, G., P. A. Marangon, A. Chapman, A. Zuin, and A. Caprioli. 1997. Atypical strains of verocytotoxin producing Escherichia coli O157 in beef cattle at slaughter in Veneto region. J. Vet. Med. B 44:301-307. [DOI] [PubMed] [Google Scholar]
- 10.Dice, L. R. 1945. Measures of the amount of ecological association between species. Ecology 26:297-302. [Google Scholar]
- 11.Faith, N. G., J. A. Shere, R. Brosch, K. W. Arnold, S. E. Ansay, M. S. Lee, J. B. Luchansky, and C. N. Kaspar. 1996. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giammanco, G. M., S. Pignato, A. M. Marino, R. P. Giunta, G. Faro, S. Caracappa, and G. Giammanco. 2001. Rischio di introduzione in Sicilia di Escherichia coli O157:H7, mediante bovini importati per la macellazione. Ann. Ig. 13:87-92. [PubMed] [Google Scholar]
- 13.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 14.Heuvelink, A. E., F. L. A. M. Van Den Biggelaar, and J. T. M. Zwartkruis-Nahuis. 1998. Occurrence of verocytotoxin-producing Escherichia coli O157 on Dutch dairy farms. J. Clin. Microbiol. 36:3480-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izumiya, H., J. Terajima, A. Wada, Y. Inagaki, K.-I. Itoh, K. Tamura, and H. Watanabe. 1997. Molecular typing of enterohemorrhagic Escherichia coli O157:H7 isolates in Japan by using pulsed-field gel electrophoresis. J. Clin. Microbiol. 35:1675-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Preston, M. A., W. Johnson, R. Khakhria, and A. Borczyk. 2000. Epidemiologic subtyping of Escherichia coli O157 strains isolated in Ontario by phage typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 38:2366-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt, H., L. Beutin, and H. Karsch. 1997. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect. Immun. 63:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, B. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tesh, V. L., and A. D. O'Brien. 1991. The pathogenic mechanisms of Shiga toxin and the Shiga-like toxins. Mol. Microbiol. 5:1817-1822. [DOI] [PubMed] [Google Scholar]
- 20.Thompson, C. J., C. Daly, J. P. Getchell, M. J. R. Gilchrist, and M. J. Loeffelholz. 1998. Insertion element IS3-based PCR method for subtyping Escherichia coli O157:H7. J. Clin. Microbiol. 36:1180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tozzi, A. E., A. Niccolini, A. Caprioli, I. Luzzi, G. Montini, G. Zacchello, A. Gianviti, F. Principato, and G. Rizzoni. 1994. A community outbreak of haemolytic-uraemic syndrome in children occurring in a large area of northern Italy over a period of several months. Epidemiol. Infect. 113:209-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Versalovic, J., T. Kocuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu, J., and J. B. Kaper. 1992. Cloning and characterization of the eae gene of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 6:411-417. [DOI] [PubMed] [Google Scholar]