Abstract
We developed and evaluated a real-time fluorescence PCR assay for detecting the A and B subunits of diphtheria toxin (tox) gene. When 23 toxigenic Corynebacterium diphtheriae strains, 9 nontoxigenic C. diphtheriae strains, and 44 strains representing the diversity of pathogens and normal respiratory flora were tested, this real-time PCR assay exhibited 100% sensitivity and specificity. It allowed for the detection of both subunits of the tox gene at 750 times greater sensitivity (2 CFU) than the standard PCR (1,500 CFU). When used directly on specimens collected from patients with clinical diphtheria, one or both subunits of the tox gene were detected in 34 of 36 specimens by using the real-time PCR assay; only 9 specimens were found to be positive by standard PCR. Reamplification by standard PCR and DNA sequencing of the amplification product confirmed all real-time PCR tox-positive reactions. This real-time PCR format is a more sensitive and rapid alternative to standard PCR for detection of the tox gene in clinical material.
Although the diphtheria epidemic that occurred in the countries of the former Soviet Union in the early and mid-1990s is largely under control, diphtheria is likely to continue to be a presence in the public health arena due to problems such as waning immunity in the adult population, the need for optimizing immunization schedules for children and for decennial boosters (1). Consequently, rapid and reliable methods are needed for identifying cases imported to the United States, thus aiding in appropriate and timely patient management and improved surveillance of domestic cases.
Laboratory confirmation of diphtheria cases is currently dependent on culturing Corynebacterium diphtheriae and performing the Elek test (2) to determine the organism's toxigenicity. However, viable organisms are not always present in clinical specimens or are below the limit of culture detection. Consequently, molecular detection of the diphtheria toxin gene, tox, is often the only means by which a laboratory confirmation of diphtheria can be made. The standard PCR assay that detects the C. diphtheriae tox gene continues to be the “gold standard” for the molecular diagnosis of diphtheria (10, 11). We present here the development and evaluation of a new 5′ nuclease PCR-based method (real-time PCR) for the detection of the A and B subunits of the tox gene. With the emergence of new fluorescent probe gene amplification technologies, it has been possible to improve substantially upon the standard PCR assay, by providing quantitative results, eliminating postamplification handling, and increasing sensitivity (5, 7). The aim of the present study was to develop and validate a real-time PCR assay that exceeds the limit of detection obtainable by the standard PCR for the detection of the diphtheria toxin gene directly in clinical specimens.
MATERIALS AND METHODS
Control strains.
A total of 76 strains from the Centers for Disease Control (CDC) strain collection were included in the present study for evaluation of the sensitivity and specificity of the real-time PCR assay: 23 toxigenic and 9 nontoxigenic C. diphtheriae clinical isolates of diverse geographic and temporal origin (Table 1) and 44 strains representing a diversity of respiratory pathogens and normal flora (Table 2). All strains were maintained in sterile defibrinated sheep blood at −70°C until needed. C. diphtheriae NCTC 10648 (CDC510) and NCTC 10356 (CDC511) strains were used as the positive and negative controls, respectively, for both real-time and standard PCR assays. All strains were identified by using standard microbiological methods (2, 3).
TABLE 1.
C. diphtheriae clinical isolates used in evaluation of the specificity of real-time PCR assays for detection of the tox gene
| Origina | Strain | Biotypeb | Toxigenicityc (reference) | tox real-time PCR assay result |
|---|---|---|---|---|
| Canada | C65 | M | − (8) | − |
| Canada | C64 | B | − (8) | − |
| Canada | C61 | M | + (8) | + |
| Canada | C73 | I | − (8) | − |
| Canada | C78 | B | − (8) | − |
| Kazakhstan | C50 | B | −d | − |
| Kazakhstan | C52 | M | −d | − |
| Russia | 496 | M | + (12) | + |
| Russia | G4174 | G | + (12) | + |
| Russia | 749 | G | + (12) | + |
| Russia | 722 | G | + (12) | + |
| Russia | G4212 | M | + (12) | + |
| Russia | 1899 | M | + (12) | + |
| Russia | 711 | M | + (12) | + |
| Russia | 718 | B | + (12) | + |
| Russia | 713 | M | + (12) | + |
| Russia | 765 | G | + (12) | + |
| Russia | 724 | M | + (12) | + |
| Russia | G4182 | B | + (12) | + |
| Russia | 760 | M | − (12) | − |
| Russia | 1709 | M | + (12) | + |
| Russia | 750 | M | + (12) | + |
| US (Pine Ridge, SD) | G4221 | I | + (8) | + |
| US (Pine Ridge, SD) | PR75 | G | + (8) | + |
| US (Pine Ridge, SD) | C5276 | M | + (8) | + |
| US (Pine Ridge, SD) | PR110 (UT6-96) | M | + (8) | + |
| US (Pine Ridge, SD) | G4217 | M | + (8) | + |
| US (Pine Ridge, SD) | G4219 | M | + (8) | + |
| US (Pine Ridge, SD) | E8277 | G | + (8) | + |
| US (Pine Ridge, SD) | PR26 | G | − (8) | − |
| US (Pine Ridge, SD) | PR120 | G | − (8) | − |
| US (VA horse) | A12 | G | + (4) | + |
With a single strain exception all strains were isolated from humans: strain A12 was isolated from a neck wound on a horse (4). US, United States; SD, South Dakota; VA, Virginia.
M, mitis; B, belfanti; G, gravis; I, intermedius.
The toxigenicity of all strains was confirmed previously by both standard PCR and the Elek test.
Toxigenicity was determined in by the Elek test as previously described (2).
TABLE 2.
Type, reference, and standard strains (n = 44) representing respiratory pathogens and flora used in the evaluation of specificity and sensitivity of the real-time PCR assays for detection of the tox gene
| Speciesa | Isolate | Real-time PCR assay result |
|---|---|---|
| C. accolens | CDC1455 | − |
| C. diphtheriae | NCTC 10648 (tox positive) | + |
| C. diphtheriae | NCTC 10356 (tox negative) | − |
| C. diphtheriae | NCTC 3984 (weakly toxigenic) | + |
| C. jeikeium | CDC1457 | − |
| C. minutissimum | CDC536 | − |
| C. mycetoides | CDC1460 | − |
| C. pseudodiphtheriticum | CDCG2486 | − |
| C. striatum | CDC530 | − |
| C. ulcerans | NCTC 12077 | − |
| C. xerosis | NCTC 12078 | − |
| H. haemolyticus | CDC3442 | − |
| H. influenzae, biogroup aegyptius | CDC5575 | − |
| H. influenzae, nontypeable | CDC4420 | − |
| H. influenzae serotype a | CDC3956 | − |
| H. influenzae serotype b | CDC3957 | − |
| H. influenzae serotype c | CDC3958 | − |
| H. influenzae serotype d | CDC3959 | − |
| H. influenzae serotype e | CDC3960 | − |
| H. influenzae serotype f | CDC3961 | − |
| H. parainfluenzae | CDC3438 | − |
| M. catarrhalis | CDC4419 | − |
| M. catarrhalis | M6452 | − |
| N. cinerea | M6451 | − |
| N. gonorrhoea | M6450 | − |
| N. lactamica | M6454 | − |
| N. meningitidis non groupable | CDC4631 | − |
| N. meningitidis serogroup A | CDC318 | − |
| N. meningitidis serogroup B | CDC321 | − |
| N. meningitidis serogroup C | CDC323 | − |
| N. meningitidis serogroup W135 | CDC327 | − |
| N. meningitidis serogroup Y | CDC326 | − |
| N. meningitidis serogroup Z | CDC329 | − |
| N. meningitidis serogroup Z′ | CDC330 | − |
| N. sicca | M6453 | − |
| N. subflava | M6449 | − |
| S. aureus | ATCC 1258 | − |
| S. aureus | ATCC 25923 | − |
| Streptococcus group A | CDC2373-96 | − |
| Streptococcus group B | CDCSS615 | − |
| Streptococcus group C | CDCSS498 | − |
| Streptococcus group D | CDCSS1344 | − |
| Streptococcus group G | CDCSS175 | − |
| Streptococcus pneumoniae | ATCC 49619 | − |
C., Corynebacterium; H., Haemophilus; M., Moraxella; N., Neisseria; S., Staphylococcus.
Clinical specimens.
A total of 36 clinical specimens (35 throat swabs and 1 throat pseudomembrane) from patients with clinical diphtheria were collected during the period from 1997 to 2000 in the Dominican Republic, Russia, and seven U.S. states (Georgia, Minnesota, Mississippi, New York, Nevada, North Carolina, and Washington; Table 3). The throat pseudomembrane was cut into three sections (CDC a773 to a775), each of which was pulverized to homogeneity. DNA was extracted from each pseudomembrane portion and all throat swabs by a modification of the method described by Nakao and Popovic (10). Briefly, the QiaAmp blood kit (Qiagen, Inc., Santa Clara, Calif.) was used according to the manufacturer's protocol with the following changes: the swab was placed in a 1.5-ml microcentrifuge tube containing 1 ml of sterile water and then vortexed for 5 min. The organisms were collected by centrifugation at 16,000 × g for 5 min. After removal of the supernatant, the cell pellet was suspended in 180 μl of Tris-EDTA buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA), and 5 μl of lysozyme (100 mg/ml) was added to the suspension. After 30 min of incubation at 37°C, 25 μl of Qiagen protease stock solution and 200 μl of buffer AL (Qiagen) were added. The solution was mixed by vortexing and incubated first at 70°C for 2 h and then at 95°C for 30 min. After purification by using a QIAamp spin column, the resulting 200-μl DNA solution was either used immediately or stored at −20°C.
TABLE 3.
Origin and designations of U.S. clinical specimens used for evaluation of the real-time PCR assay for detection of the A and B subunits of the tox gene
| CDC no. | Origin | Date collected (mo-yr) | Initial standard PCRa | Final standard PCRb | Real-time PCR | Reamplification by standard PCR | tox sequence confirmed by DNA sequencingf |
|---|---|---|---|---|---|---|---|
| a22 | Nevada | 4-97 | B | −g | − | ||
| a27 | Nevada | 4-97 | B | − | A/B | Yes | Yes |
| a28 | Nevada | 4-97 | B | − | A/B | Yes | Yes |
| a33 | Oregon | 4-97 | A/B | A | A | Yes | Yes |
| a34 | Oregon | 4-97 | A/B | A/B | B | Yes | Yes |
| a35 | Oregon | 4-97 | B | − | B | Yes | Yes |
| a36 | Oregon | 4-97 | A/B | − | B | Yes | Yes |
| a40 | Nevada | 4-97 | A/B | − | − | ||
| a45 | Minnesota | 4-97 | B | tr/− | Ac | Yes | Yes |
| a46 | Minnesota | 4-97 | B | tr/− | Ac | Yes | Yes |
| a53 | New York | 4-97 | B | − | Ac | Yes | Yes |
| a85 | Nevada | 4-97 | A | − | A/B | Yes | Yes |
| a180 | Nevada | 5-97 | A | − | A | Yes | Yes |
| a319 | Nevada | 6-97 | A/B | − | A/B | Yes | Yes |
| a629 | Oregon | 5-98 | A/B | − | A | Yes | Yes |
| a772d | North Carolina | 12-99 | − | A/B | Yes | Yes | |
| c1 | Dominican Republic | 1-00 | − | A/B | Yes | Yes | |
| c2 | Dominican Republic | 1-00 | − | A | Yes | Yes | |
| a773e | Georgia | 1-00 | − | B | Yes | Yes | |
| a774e | Georgia | 1-00 | − | B | Yes | Yes | |
| a775e | Georgia | 1-00 | − | A/B | Yes | Yes | |
| a780 | North Carolina | 3-00 | − | A/B | Yes | Yes | |
| c10 | Dominican Republic | 5-00 | − | A/B | Yes | Yes | |
| c11 | Dominican Republic | 5-00 | A/B | A/B | NDh | ND | |
| c12 | Dominican Republic | 5-00 | A/B | A/B | ND | ND | |
| c13 | Dominican Republic | 5-00 | A/B | A/B | ND | ND | |
| c14 | Dominican Republic | 5-00 | − | A | Yes | Yes | |
| c15 | Dominican Republic | 5-00 | − | A | Yes | Yes | |
| c16 | Dominican Republic | 5-00 | − | A | Yes | Yes | |
| c17 | Dominican Republic | 5-00 | − | A | Yes | Yes | |
| b1 | Russia | 6-00 | A/B | A/B | ND | ND | |
| b2 | Russia | 6-00 | A/B | A/B | ND | ND | |
| b3 | Russia | 6-00 | A/B | A/B | ND | ND | |
| b4 | Russia | 6-00 | A/B | A/B | ND | ND | |
| b5 | Russia | 6-00 | A/B | A/B | ND | ND | |
| b6 | Russia | 6-00 | A/B | A/B | ND | ND | |
| a806 | Mississippi | 6-00 | − | A/B | Yes | Yes | |
| a807 | Mississippi | 6-00 | − | A/B | Yes | Yes |
Standard PCR analyses were performed at the time the specimens were collected for diagnostic purposes as described by Nakao and Popovic (10).
Result at the time of the retest (January to June 2000) or at the time the specimen was received. tr, trace.
Indicates discrepancy between the real-time PCR result and the standard PCR result.
Paraffin-embedded specimen.
a773 to a775 were sections taken from different regions of a single pseudomembrane tissue specimen.
Sequences compared to those published by Nakao et al. (9).
-, negative result.
ND, not determined.
Standard PCR.
Standard PCR assays targeting the A and B subunits of tox were performed as previously described (10). In addition, the same primer pairs that were used in the real-time assays were also used in a standard PCR format to directly compare the two PCR methods. Reaction and cycling conditions of the standard PCR in this case were the same as those used in the real-time assays, although the probes were omitted. Both standard PCR assays were carried out by using the GeneAmp PCR system 9700 (Applied Biosystems, Foster City, Calif.).
Real-time PCR.
The published sequence of the C. diphtheriae tox gene (13) was used to design appropriate forward and reverse primers and probes in regions encoding the A and B subunits of tox (K. Livak, J. Marmaro, and S. Flood, Perkin-Elmer Research News, 1995, p. 1-8 [Applied Biosystems, Foster City, Calif.]). Oligonucleotide primers and probes specific for either the A or the B subunit of tox were designed by using Primer Express software (Applied Biosystems; Table 4). Primer concentrations were optimized by testing in the range of 0.2 to 2.0 μM (final concentration). Probe concentrations were optimized by testing in the range of 100 to 500 nM. Reactions were carried out in a 96-well MicroAmp optical plate (Applied Biosystems) by using the ABI Prism 7700 sequence detector (Applied Biosystems). Each reaction contained 2 μl of template DNA, 2.5 μl each of either the primers for the A or the B subunit (1 μM final concentration), 2.5 μl of the appropriate probe (1 μM), 2.5 μl of buffer A, 5 μl of MgCl2 (25 mM), 0.5 μl each dGTP, dCTP, dATP (10 μM each), dUTP (20 μM), 1 unit uracil N-glycosylase, and 0.625 U of Taq Gold (Applied Biosystems). PCR-certified Apex water (Mo Bio Laboratories, Inc., Encinitas, Calif.) was added to bring the volume to 25 μl. Cycling conditions were as follows: 50°C for 10 min, followed by 40 cycles of 95°C for 1 min and 60°C for 1 min. The AB 7700 instrument reads each sample every few seconds and computes a mean baseline reading for early PCR cycles. A positive result, as reported by its cycle threshold value (Ct), is indicated by the cycle at which the fluorescence exceeds the mean baseline by 10 standard deviations.
TABLE 4.
Real-time primers and fluorescence-labeled probes specific for the A and B subunits of the tox gene
| Subunit | Primer sequencea (5′-3′) | Position (range) in K01722 |
|---|---|---|
| Subunit A (117-bp amplicon) | ||
| Forward primer | GGCGTGGTCAAAGTGACGTA | 546-565 |
| Reverse primer | CTTGCTCCATCAACGGTTCA | 663-644 |
| Probe | FAM-CCAGGACTGACGAAGGTTCTCGCACT-TAMRA | 568-592 |
| Subunit B (128-bp amplicon) | ||
| Forward primer | CGCCCTAAATCTCCTGTTTATGTT | 1725-1748 |
| Reverse primer | GTACCCAAGAACGCCTATGGAA | 1853-1832 |
| Probe | FAM-TTCACAGAAGCAGCTCGGAGAAAATTCATTC-TAMRA | 1783-1813 |
Primer Express software (Applied Biosystems) was used to select appropriate primers and probes for the A and B subunits of the C. diphtheriae tox gene.
Sensitivity and specificity of the tox real-time PCR assay.
Genomic DNA from all 76 test strains was extracted by using the cell boiling method. Haemophilus spp. were grown on chocolate II agar (Becton Dickinson, Cockeysville, Md.). All other strains were grown on blood agar with 5% defibrinated sheep blood for 16 to 24 h at 37°C. A single colony from each agar plate was then suspended in 1 ml of heart infusion (HI) broth (Remel, Lenexa, Kans.) and incubated for 16 to 24 h at 37°C. A total of 100 μl of this suspension was further incubated at 95°C for 10 min and then centrifuged at 14,000 × g for 2 min. The supernatant was diluted serially 10-fold to 10−10, and a 2-μl volume of the supernatant from each dilution was used in the PCR.
Comparative DNA extraction.
In addition to the cell boiling procedure, two further approaches were used to extract genomic DNA from C. diphtheriae strains NCTC 10648 (tox positive) and NCTC 10356 (tox negative): culture dilution and DNA dilution.
For the culture dilution method, 1 ml of overnight growth was diluted serially 10-fold in sterile water (10−1 to 10−10), and 100 μl of each dilution was processed by using the QiaAmp blood kit with slight modifications to the method described by Nakao and Popovic (10). A 2-μl volume from each dilution was used as a template in the real-time and standard PCRs. For the purpose of defining the number of CFU in each dilution, a 100-μl aliquot of dilutions of 10−5 to 10−10 was plated in duplicate on sheep blood agar. A standard plate count was used to determine the bacterial density, allowing the lower limit of detection to be reported in CFU.
For the DNA dilution method, DNA was extracted from 100 μl of culture grown in HI broth (10), with the modifications described as above. The DNA concentration was determined spectrophotometrically at 260 nm by using an MBA 2000 spectrophotometer (Perkin-Elmer, Norwalk, Conn.). Tenfold serial dilutions of the DNA (10−1 to 10−10) were prepared, and a 2-μl volume of each DNA dilution was used as the template in the real-time and standard PCRs.
Reamplification of real-time PCR amplicons by standard PCR.
Real-time PCR products (2 μl of a 25-μl reaction) were used in a reaction which also included 1 μl of forward and reverse primers (200 nM) specific for the A or B subunit of the tox gene (Table 4), 5 μl of Opti-Prime Buffer #6 (Stratagene, La Jolla, Calif.), 4 μl of deoxynucleoside triphosphate mix (2.5 mM concentrations of each deoxynucleoside triphosphate), and 0.25 U of AmpliTaq (Applied Biosystems). The reaction volume was adjusted to a final volume of 50 μl with PCR certified Apex water. Target sequences were amplified by treatment as follows: 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. After extension at 72°C for 7 min, the reaction products (15 μl of 50 μl, total volume) were electrophoresed for 40 min at 90 V on a 1.2% agarose gel containing ethidium bromide. The products were visualized by using the GelDoc system (Bio-Rad, Hercules, Calif.).
Sequencing of the real-time PCR amplicon.
Each clinical specimen and the positive and negative control real-time PCR products were sequenced by using the primers from the real-time PCR A and B subunit assays (Table 4). Excess primers and probe were removed from the real-time PCR by using the QIAquick 8 PCR purification kit (Qiagen). The AB Prism BigDye terminator cycle sequencing kit was used to produce fluorescence-labeled sequencing amplicons (using the tox subunit-specific real-time primers), which were then purified by the Dye Ex Spin Kit (Qiagen); both processes were done according to the manufacturer's protocol. Amplicons were electrophoresed on a 10% polyacrylamide gel by using the ABI Prism 377 DNA sequencer (Applied Biosystems). Identified sequences were queried against the GenBank database by using BLAST in the GCG package, v10.1 (Genetics Computer Group, Madison, Wis.) in order to confirm their identity as the C. diphtheriae tox gene.
RESULTS
Specificity and sensitivity of the tox real-time PCR assay.
Real-time PCR assays with primers directed against both A and B subunits and by using template DNA extracted (by boiling) from 42 strains representing diverse respiratory pathogens and normal flora, as well as from nine nontoxigenic C. diphtheriae strains, were consistently negative (Table 2). The A and B subunits of the tox gene were detected in two toxigenic type strains and in each of the 23 toxigenic clinical C. diphtheriae isolates, resulting in 100% sensitivity and specificity (confidence intervals of >95% and 85 to 100% for sensitivity and 94 to 100% for specificity).
Comparative limit of detection of the tox real-time PCR and standard PCR assays.
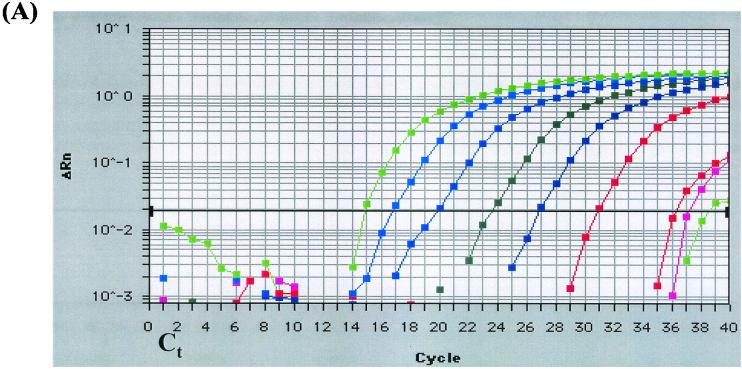
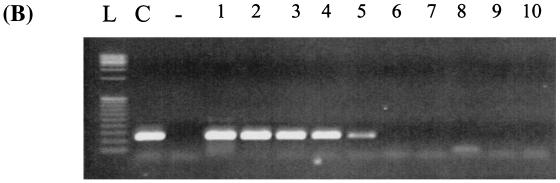
The real-time PCR assays were compared to standard PCR assays performed with the previously designed primer sets (10), as well as the primer sets developed in the present study. Real-time PCR assays reproducibly detected lower concentrations of DNA than did the standard PCR assays. Using the culture dilution method, the real-time PCR assay was 750-fold more sensitive with detection of 2 CFU (10−8 dilution) or 17 fg of genomic DNA (calculated based upon the genome size of C. diphtheriae; Fig. 1A) compared to 1,500 CFU (10−5 dilution) or 2.5 pg of DNA by standard PCR (Fig. 1B). Standard PCR with the real-time assay primers and conditions was positive at the 10−3 dilution. The cell boiling method allowed for detection of ca. 280 fg of DNA by real-time PCR based on spectrophotometric determination. Standard PCR detected 2.8 pg of DNA or 1,670 copies (10−5 dilution), 10-fold more target DNA than was required by the real-time PCR method. The DNA dilution method, whereby genomic DNA was extracted from cell culture then diluted serially 10-fold, allowed for detection of 5 pg of DNA (3340 copies) by real-time PCR and ca. 500 pg of DNA (1.34 × 105 copies) by standard PCR.
FIG. 1.
Comparative lower limit of detection of the tox real-time PCR assay and standard PCR. Corynebacterium diphtheriae (NCTC 10648) was grown on sheep blood agar at 37°C for 16 h, and serial 10-fold dilutions were prepared in HI broth. DNA was extracted from undiluted culture and from 10−1 to 10−8 dilutions and used in the tox real-time PCR assay and in the standard PCR assay. (A) Real-time PCR amplification generated by the Prism 7700 sequence detector showing amplification of target DNA from nine samples (from left to right: undiluted C. diphtheriae growth in HI broth and dilutions 10−1 to 10−8). ΔRn, difference in reporter fluorescence between the sample and the no-template controls; Ct, threshold cycle (i.e., the cycle at which a statistically significant increase in fluorescence is first detected). (B) Amplification of the A subunit of the tox gene (primers tox 1 and tox 2) by standard PCR. Lanes: L, low-mass DNA ladder; C, undiluted C. diphtheriae growth in HI broth; −, no-template control; 1 to 10, dilutions 10−1 to 10−10, respectively.
Detection of tox in clinical samples.
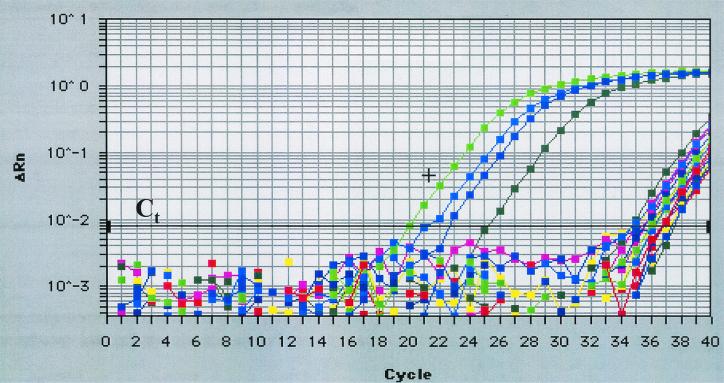
Fifteen DNA extracts from clinical specimens collected from 1997 to 1998, which had been stored at −70°C since the original time of collection (24 to 36 months), were determined to be positive by standard PCR (four were weakly positive) for either one or both of the tox subunits when tested at the time of specimen collection. Two years later, during the present study, standard PCR was repeated on these 15 extracts, and the tox gene was detected in only two samples, with trace amounts of amplicon visible in two other samples (Table 3). In comparison, when the same 15 extracts were analyzed by real-time PCR, 13 of the 15 were positive for either one or both subunits of the tox gene (Fig. 2 and Table 3).
FIG. 2.
Real-time amplification plot of 18 representative clinical samples. Positive control DNA from C. diphtheriae (NCTC 10648 toxigenic) and three clinical samples amplified early (20 to 25 cycles). The majority of samples amplified late (>34 cycles) in the 40-cycle program. +, Positive control DNA; Ct, cycle threshold (i.e., the cycle at which a statistically significant increase in fluorescence is first detected).
Since December 1999, 21 clinical specimens from the Dominican Republic (10 outbreak-associated specimens), Russia, and the United States (Georgia, Mississippi, and North Carolina) tested positive for tox by real-time PCR. Only three (CDC specimens c11, c12, and c13) from the Dominican Republic specimens were found to be positive by traditional PCR (Table 3). The specimen from Georgia was a throat pseudomembrane that had been divided into three sections, which were pulverized, and the DNA had been extracted (CDC sections a773 to a775). Two of the sections (CDC a772 and a773) yielded inconsistent results by real-time PCR, but the third (CDC a775) was consistently positive for both subunits. All three extracts were below the limit of detection by standard PCR. All six of the Russian specimens were positive by both standard PCR and real-time PCR (Table 3).
Reamplification of real-time PCR products by standard PCR.
For the tox-positive control and all clinical samples, a band of the appropriate size (117 bp for the A subunit and 128 bp for the B subunit) was observed, confirming the presence of the target DNA.
Sequencing of real-time PCR products.
Real-time amplification products from positive control and clinical specimen reactions were sequenced and identified to be the tox gene of C. diphtheriae (9).
DISCUSSION
In the present study, we developed and evaluated a highly sensitive and specific real-time PCR assay that exceeded the limit of detection of the existing standard PCR assay. As few as 2 CFU were detected by real-time PCR compared to 1,500 CFU detected by standard PCR. By this approach, real-time PCR exhibited 10-fold-greater sensitivity than the traditional PCR results reported in the study by Nakao and Popovic (10), wherein the lowest limit of detection was 25 CFU. Real-time PCR showed 750-fold-greater sensitivity than the standard PCR in the present study. Likewise, when the real-time method was compared to standard PCR with the same primer sets and cycling conditions, it exhibited enhanced sensitivity over the standard method by 5 orders of magnitude, suggesting that the internal probe and optimized conditions of the real-time assay are responsible for the improvement.
It was also observed that the culture dilution method of extraction, whereby overnight culture was serially diluted and DNA from each dilution was independently extracted, allowed for greater sensitivity in the real-time assay over either the cell boiling extraction method or the DNA dilution method. Extraction by use of the QiaAmp kit most likely resulted in a higher-purity product, explaining the enhanced sensitivity over the cell-boiling method. Furthermore, sample manipulation in the DNA dilution method could diminish target integrity, accounting for the decreased sensitivity observed when this method was used.
For comparison purposes, we tested 36 specimens collected from patients with clinical diphtheria: 15 specimens were received at the CDC (April 1997 to May 1998) and at that time were found by standard PCR to be tox positive. Two years later, upon the initiation of this project, these specimens were retested by standard PCR, as well as by real-time PCR. In all but two cases (Table 3, specimens a22 and a40) real-time PCR confirmed the original standard PCR result or detected the tox gene when standard PCR could not. The discrepant results are most likely attributable to DNA degradation due to suboptimal storage or handling and manipulation.
Since December 1999, an additional 21 specimens from patients with clinical diphtheria were prospectively examined by standard PCR and real-time PCR. All of these specimens were found to be positive for either one or both subunits of tox by real-time PCR, whereas nine were found to be positive by standard PCR (c11, c12, c13, and b1 to b6; Table 3). Real-time PCR products in these nine specimens were detected in an earlier amplification cycle than most of the other specimens, indicating a higher initial concentration of DNA. Recently, it was reported that late amplification (>30 cycles) may be a result of carryover contamination (6); however, our findings show that trace amounts of DNA can be detected and appear after 34 cycles on the amplification plot. Because of the late cycle amplification, however, we wanted to confirm the reactivity of these samples by reamplification and sequencing. All of the clinical specimens which showed a positive real-time PCR result, but which amplified >34 cycles, were reamplified by standard PCR and were also confirmed by DNA sequencing as the A or B subunit of the C. diphtheriae tox gene. Dissociation curve analysis with the Light Cycler instrument (Roche Molecular Biochemicals, Indianapolis, Ind.) confirmed the presence of a single amplicon in the real-time reactions that served as a template in the sequencing reactions (data not shown). Although internal controls were not routinely used when testing clinical specimens, previous data has shown that Qiagen-extracted DNA shows no PCR inhibition in either standard or real-time assays (6).
In the present study, the real-time PCR assay enabled detection of the tox gene in clinical specimens when standard PCR proved inadequate. Real-time PCR is sensitive, specific, and rapid and obviates the need for postamplification handling. We have established that, although many clinical specimens contain only trace amounts of DNA, real-time PCR is able to detect tox when two to three copies of the target gene are present. A nested PCR approach may likewise improve upon the sensitivity of the standard PCR method; however, a major advantage of the real-time approach is that all postamplification handling steps are eliminated and it offers high-throughput analysis, which is critical in the event of an outbreak. The real-time assay provides substantial improvement over the existing standard PCR assay for tox detection and consequently may have important epidemiological ramifications for the rapid detection of domestic and imported cases, as well as in defining the true burden of diphtheria worldwide.
Acknowledgments
We thank Frances Jamieson, Laboratory Services Branch, Ontario Ministry of Health, Toronto, Ontario, Canada; Izabella Mazurova, GN Gabrichevsky Institute of Epidemiology and Microbiology, Moscow, Russia; and Vasilij Kim, Kazakhstan Republican Sanitary-Epidemiological Station, Almaty, Kazakhstan, for providing clinical isolates. We also thank the physicians, laboratorians, and public health officials at the U.S. state health departments who collected and supplied clinical specimens. We are grateful to the Public Health Laboratory of the Dominican Republic for collecting clinical specimens. We thank our colleagues from various CDC reference laboratories for providing control strains. We especially thank Claudio Sacchi, Meningitis and Special Pathogens Branch, CDC, for DNA sequencing assistance.
REFERENCES
- 1.Dittmann, S., M. Wharton, C. Vitek, M. Ciotti, A. Galazka, S. Guichard, I. Hardy, U. Kartoglu, S. Koyama, J. Kreysler, B. Martin, D. Mercer, T. Ronne, C. Roure, R. Steinglass, P. Strebel, R. Sutter, and M. Trostle. 2000. Successful control of epidemic diphtheria in the states of the Former Union of Soviet Socialist Republics: lessons learned. J. Infect. Dis. 181(Suppl. 1):S10-S22. [DOI] [PubMed] [Google Scholar]
- 2.Efstratiou, A., and P. A. Maple. 1994. WHO manual for the laboratory diagnosis of diptheria, p. 69-71. Document ICP-EPI 038(C). World Health Organization, Geneva, Switzerland.
- 3.Funke, G., and K. A. Berhard. 1999. Coryneform gram-positive rods, p. 319-345. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 4.Henricson, B., M. Segarra, J. Garvin, J. Burns, S. Jankins, C. Kim, T. Popovic, A. Golaz, and B. Akey. 2000. Toxigenic Corynebacterium diphtheriae associated with an equine wound infection. J. Vet. Diagn. Investig. 12:253-257. [DOI] [PubMed] [Google Scholar]
- 5.Higgins, J. A., J. Ezzell, B. J. Hinnebusch, M. Shipley, E. A. Henchal, and M. S. Ibrahim. 1998. 5′ nuclease PCR assay to detect Yersinia pestis. J. Clin. Microbiol. 36:2284-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Killgore, G. E., B. Holloway, and F. C. Tenover. 2000. A 5′ nuclease PCR (TaqMan) high-throughput assay for detection of the mecA gene in staphylococci. J. Clin. Microbiol. 38:2516-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura, B. 1999. Evaluation of TaqMan PCR assay for detecting Salmonella in raw meat and shrimp. J. Food Protect. 62:329-335. [DOI] [PubMed] [Google Scholar]
- 8.Marston, C. K., F. Jamieson, F. Cahoon, G. Lesiak, A. Golaz, M. Reeves, and T. Popovic. 2001. Persistence of a distinct Corynebacterium diphtheriae clonal group within two communities in the United States and Canada where diphtheria is endemic. J. Clin. Microbiol. 39:1586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakao, H., I. K. Mazurova, T. Glushkevich, and T. Popovic. 1997. Analysis of heterogeneity of Corynebacterium diphtheriae toxin gene, tox, and its regulatory element, dtxR, by direct sequencing. Res. Microbiol. 148:45-54. [DOI] [PubMed] [Google Scholar]
- 10.Nakao, H., and T. Popovic. 1997. Development of a direct PCR assay for detection of the diphtheria toxin gene. J. Clin. Microbiol. 35:1651-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pallen, M. J., A. J. Hay, and A. Efstratiou. 1994. Polymerase chain reaction for screening clinical isolates of corynebacteria for the production of diptheria toxin. J. Clin. Pathol. 47:353-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popovic, T., S. Y. Kombarova, M. W. Reeves, H. Nakao, I. K. Mazurova, M. Wharton, and I. K. Wachsmuth. 1996. Molecular epidemiology of diphtheria in Russia, 1985-1994. J. Infect. Dis. 174:1064-1072. [DOI] [PubMed] [Google Scholar]
- 13.Ratti, G. 1983. The complete nucleotide sequence of the gene coding for diphtheria toxin in the corynephage omega (tox+) genome. Nucleic Acids Res. 11:6589-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]