Abstract
We describe a replication-independent, cell cycle-regulated chromatin assembly pathway in budding yeast. The activity of this pathway is low in S phase extracts but is very high in G2, M, and G1 cell extracts, with peak activity in late M/early G1. The cell cycle regulation of this pathway requires a specific pattern of posttranslational modification of histones H3 and/or H4, which is distinct for H3/H4 present in S phase versus M and G1 phase cell extracts. Histone H3/H4 modification is therefore important for the reciprocal control of replication-dependent and -independent chromatin assembly pathways during the cell cycle.
Keywords: yeast, chromatin assembly factor, nucleosome, H3, H4, remodeling
Under physiological conditions nucleosome assembly is mediated by proteins called chromatin assembly factors (CAFs) (1–5). CAFs operate in two general pathways of nucleosome reconstitution in vivo, one coupled to DNA replication, and one that occurs independently of replication. The “replication-dependent” pathway is cell cycle regulated, being maximally active in S phase. Although this pathway accounts for the bulk of chromatin assembly in dividing cells, it seems likely that during normal cell division nucleosome deposition also occurs by a “replication-independent” mechanism. Chromatin remodeling associated with transcriptional regulation can involve nucleosome deposition that is independent of DNA synthesis (6) and therefore perhaps involves replication-independent assembly factors. When nucleosomes are lost during G2, M, and G1 as a result of histone degradation, new nucleosome deposition is likely to occur before the next S phase (7), and this reaction also potentially involves replication-independent CAFs.
Although these observations suggest significant physiological functions for replication-independent CAFs in dividing cells, the biochemical properties of such factors, and the regulation of replication-independent assembly pathways in relation to global changes in cellular metabolism, are poorly understood. We are exploring the regulation of replication-independent chromatin assembly by a biochemical approach in budding yeast (8, 9). Here we focus on the cell cycle regulation of replication-independent chromatin assembly in mitotically dividing cells. We have discovered a replication-independent assembly pathway that is cell cycle regulated in yeast. The activity pattern of this pathway mirrors that of the replication-dependent pathway responsible for bulk assembly in dividing cells, that is, replication-independent assembly is repressed in S phase, during which time bulk replication-coupled assembly is activated (2, 10). We show that posttranslational modification of histones H3 and/or H4 plays an important role in the cell cycle control of assembly by this pathway. Posttranslational modification of histones H3 and H4 is also associated with the activation of replication-coupled chromatin assembly during S phase (2, 10). The regulation of histone H3 and/or H4 is therefore an important component of a reciprocal control mechanism that represses replication-independent assembly when bulk replication-dependent assembly is activated. This mechanism may serve to limit interference by replication-independent CAFs of the coordination between DNA synthesis and bulk nucleosome deposition mediated by replication-dependent CAFs.
MATERIALS AND METHODS
Strains.
W303–1a (11) a ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1; 17017 (12) cdc15–1; 13–53 (12) cdc15–2; cdc17–1 (12) a ade1 ade2 ura1 lys2 gal1 ade4 cdc17–1; STX448–5A (Yeast Genetic Stock Center, University of California, Berkeley) α ade1 ade2 lys2 gal1 cdc20–1; STX326–8B (Yeast Genetic Stock Center) α ade1 gal1 lys2 met14 his7 tyr1 cdc28–1; RMY102 (13) a ade2–101 his3-Δ200 lys2–801 trp1Δ901 ura3–52 hht1, hhf1∷LEU2, hht2, hhf2∷HIS3 plus pRM102 [CEN4 ARS1 URA3, P(GAL10)-HHT2, P(GAL1)-HHF2]. Cells were grown at 30°C unless otherwise noted.
Cell Synchronization; Flow Cytometry.
W303–1a cells were arrested at Start by using α-factor (Sigma) treatment for 1.5 hr (0.5 μg/ml added to cells in YP (1% yeast extract, 2% bactopeptone)/2% glucose at OD600 = 0.5, and again 45 min after the first addition). Most (95%) cells were unbudded after α-factor treatment. Cells were released by washing three times in water and seeding into fresh prewarmed (30°C) medium (modified from ref. 14). cdc15–2 cells were grown in YP/2% glucose at 26°C to OD600 = 0.5 and then arrested in M phase by incubation at 37°C for 1.5–2 hr. The medium was then rapidly cooled to 26°C, and samples were removed every 15 min for extract preparation (15). Cell cycle position was assessed by morphological criteria (14, 15) and by FACScan (Becton Dickinson) analysis of 20,000 events per sample with gating out of doublets (16).
Cell Cycle Arrest.
Cell division cycle (cdc) mutants were grown in YP/2% glucose to OD600 = 0.5 at the permissive temperature (26°C) and transferred to 37°C for 1.5–2 hr, and then the cells were frozen for extract preparation. Note that the temperature shift protocol did not affect activity in extracts from wild-type cells (not shown). W303–1a cells were grown in synthetic complete medium (17) to OD600 = 0.5 and then treated with 6 mg/ml hydroxyurea or 20 μg/ml nocodazole (18) for 1.5 hr, at which point cells were collected and frozen. cdc phenotypes are as described (19, 20).
Depletion of H3 and H4 in Vivo.
Strain RMY102 was grown in YP/2% galactose to OD600 = 1.0 and then transferred to YP/2% glucose for 12 hr (GAL-driven H3/H4 expression off) before extract preparation.
Extract and Histone Preparation: In Vitro Assembly Assay.
Extracts were prepared from frozen cells after grinding in a coffee mill and extraction in low salt buffer (245 mM KCl; refs. 8 and 9). Twenty microliter assembly reactions were performed with internally labeled, relaxed pBluescript (refs. 8 and 9; conditions in figure legends 1–4). For histone reconstitution, H3/H4-depleted extract (from strain RMY102, above) was incubated with purified histones (ref. 8; protein quantitation by Bio-Rad protein assay, BSA standard) at 22°C for 5 min. All additional reaction components (including yeast dialysis buffer I, YDBI, to bring the volume to 20 μl) were then added. DNA products were resolved by either 0.8% Tris-actetate/EDTA or 1.5% Tris/glycine agarose gel electrophoresis, and detected by autoradiography. Quantitation was performed by PhosphorImager analysis (Fujix Bas 1000 analyzer, Fuji macbas software) with subtraction of backgrounds.
Preparation of H2B Antiserum: Western Blot Analysis.
A fusion of residues 1–35 of yeast H2B (HTB2) and glutathione S-transferase [plasmid p297.1, based on pGEX-2T (Pharmacia); a generous gift of M. Grunstein (University of California, Los Angeles)] was purified from Escherichia coli strain BL21 and used to immunize rabbits as described (21). Proteins were resolved by SDS/PAGE (15% gel) or Triton X-100-acid urea gel electrophoresis (22). Antiacetylated H4 and antiphospho-H3 antibodies from Upstate Biotechnology (Lake Placid, NY) were used for Western blot analysis according to the supplier’s instructions. The anti-H2B serum was diluted 1:1000 and the enhanced chemiluminescence system (Amersham) was employed for detection. Used under these conditions to probe purified bovine (Boehringer Mannheim) and yeast histones, the antiserum specifically (compared with preimmune) detected a band of the expected size of H2B.
RESULTS
Replication-Independent Assembly in Extracts from Synchronized Wild-Type Cells.
We used a yeast extract that provides all soluble proteins required for replication-independent chromatin assembly (8, 9). Assembly as detected by plasmid supercoiling (see below) is very rapid, reaching equilibrium in 2–10 min in wild-type extracts. The reaction is ATP- and histone-dependent and is sensitive to mutation of the conserved lysines in the tail of histone H4 that are important for function in vivo (23). RNA removal from the extract has little effect on supercoiling (8). Micrococcal nuclease digestion analysis reveals the generation of a prominent mononucleosome in this system as well as correctly spaced (albeit not prominent) disomes and trisomes (8, 9). The system therefore reproduces a physiologically relevant, protein-mediated assembly pathway.
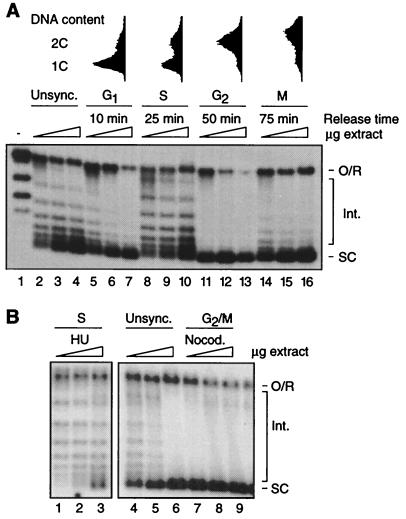
We prepared extracts from cells harvested at different stages in the cell cycle after release from α-factor arrest (Fig. 1A). Cell cycle position was determined by morphological criteria and FACScan analysis. Assembly was assayed by plasmid supercoiling. Because the degree of supercoiling of the deproteinized template is directly related to the number of nucleosomes deposited, decreased assembly is evident as the appearance of intermediate topoisomers migrating between the open circular/relaxed species and the highly supercoiled products (8). As shown in Fig. 1A (Upper), assembly activity is not uniform in extracts from different cell types; G1, G2, and M extracts support a high level of assembly, whereas assembly activity is relatively low in S extract and in extract from unsynchronized cells. Note that the proportion of assembly template migrating at the position of open circular DNA (includes relaxed closed circular DNA of linking number 0) does not vary substantially between extracts. Therefore, changes in supercoiling capacity are unlikely because of changes in nuclease activity during the cell cycle. The gross structure of nucleosomes, as determined by micrococcal nuclease digestion analysis, also did not vary between extracts (data not shown). We conclude that replication-independent chromatin assembly is cell cycle regulated in yeast whole cell extracts.
Figure 1.
Replication-independent chromatin assembly is cell cycle regulated in extracts from synchronized wild-type cells. (A Upper) FACScan analysis of α-factor synchronized cells. (Lower) Assembly-driven supercoiling of a relaxed plasmid (lane 1) is compared in extracts from unsynchronized cells (Unsync., lanes 2–4), and extracts from cells synchronized in G1 (lanes 5–7), S (lanes 8–10), G2 (lanes 11–13), and M (lanes 14–16) by release from α-factor arrest. The time of harvest after arrest is indicated. Reactions were performed for 30 min at 30°C by using 25, 50, and 100 μg of protein for each extract. (B) Assembly in extracts from cells arrested in S phase by culture in the presence of hydroxyurea (HU, lanes 1–3) and in G2/M phase by exposure to nocodazole (Nocod., lanes 7–9). Assembly in unsynchronized extract is also shown (Unsync., lanes 4–6). Reactions were performed as in A. The migration of open circular/relaxed DNA (linking number 0) [O/R], highly supercoiled species (SC), and intermediate topoisomers (Int.) is indicated.
To confirm that assembly activity in wild-type extracts is high during G2/M but low in S phase, W303–1a cells were arrested in S with hydroxyurea and in G2/M with the microtubule depolymerizing drug nocodazole (Fig. 1B). Extracts were prepared from arrested and untreated (control) cells grown to the same density. Compared with the control (lanes 4–6), the extract from nocodazole-arrested cells is highly active (lanes 7–9). Extract from hydroxyurea-arrested cells (lanes 1–3), on the other hand, is less active than extracts from unsynchronized and G2/M-arrested cells. The relative differences in assembly capacity are similar to those observed between extracts from α-factor synchronized cells, confirming that replication-independent chromatin assembly is relatively high in G2/M and low in S phase.
Replication-Independent Assembly in Extracts from cdc Mutants.
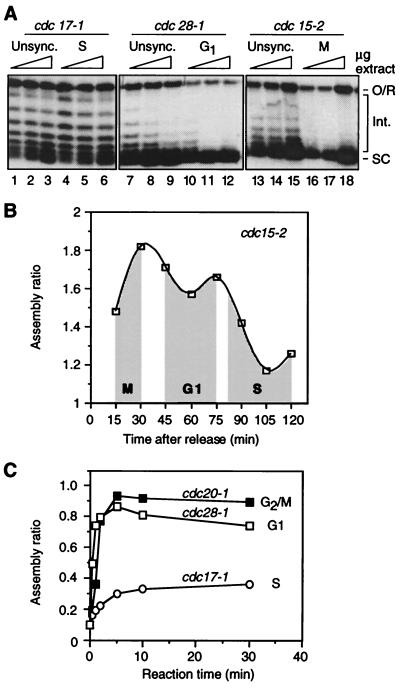
The cell cycle regulation of replication-independent chromatin assembly was analyzed further by using extracts from temperature sensitive (ts) cdc mutants. Extracts from unsynchronized cdc strains, regardless of the ts mutation, do not support a high level of assembly; the band corresponding to fully supercoiled species is somewhat diffuse and intermediate topoisomers are prominent (Fig. 2A, lanes 1–3, 7–9, and 13–15). Extract from G1 cells (cdc28–1 cells shifted to 36°C; lanes 10–12) is substantially more active than extracts from asynchronous cells (note the accumulation of highly supercoiled products and relative lack of intermediate topoisomers). A highly active extract is also obtained from M phase cells (cdc15–2 cells shifted to 36°C; lanes 16–18). However, extract from cdc17–1 cells arrested in S (lanes 4–6) has about the same assembly capacity as extract from unsynchronized cdc17–1 cells (lanes 1–3), and the activity of asynchronous and S phase cdc17–1 extracts is substantially lower than that of M and G1 extracts. cdc17–1 cells grown at the permissive temperature spend longer in S phase than wild-type cells and yield extracts with somewhat lower activity than wild-type (data not shown).
Figure 2.
Regulation of replication-independent chromatin assembly in extracts from cdc mutants. (A) Assembly is compared in extracts from unsynchronized cells growing at the permissive temperature and from cells arrested at the indicated phases of the cell cycle by temperature shift. Reactions were performed for 30 min at 22°C by using 25, 50, and 100 μg of protein for each extract. The migration of open circular/relaxed DNA (O/R), highly supercoiled species (SC), and intermediate topoisomers (Int.) is indicated. (B) Quantitation of replication-independent chromatin assembly in extracts from cdc15–2 cells synchronized in M phase, released into the cell cycle by shifting to the permissive temperature, and harvested at the indicated time points thereafter. Unshaded regions correspond to transitions between cell cycle stages. Reactions were performed for 30 min at 22°C by using 25 μg of protein for each extract. (C) Time course of supercoiling in extracts from cdc mutants arrested in G2/M, G1, and S phase. Reaction conditions as in A.
Assembly was also assayed in extracts from cdc15–2 cells synchronized in M, G1, and S by a temperature shift-release protocol. Assembly activity, in this case quantitated as [signal from highly supercoiled products] ÷ [signal from intermediate topoisomers] (24, 25), varies according to cell cycle position, namely, activity is high in M and G1 but low in S (Fig. 2B). Experiments with cdc mutants therefore confirm the pattern of cell cycle control of replication-independent chromatin assembly observed in extracts from cells synchronized with α-factor (Fig. 1A).
We performed time course experiments to test whether the kinetics of assembly is different among G2/M, G1, and S phase extracts (Fig. 2C; quantitation as in B). Assembly is efficient and very rapid in the G1 and G2/M extracts, reaching completion within 10 min. By comparison, assembly occurs at a much slower rate during the first 10 min of the S phase reaction, and the level of assembly attained at 10 and 30 min is substantially lower than the levels for G2/M and G1 extracts. The same difference in assembly between S phase and G1 and G2/M extracts was observed at 2 hr (data not shown). We conclude that, compared with G1 and G2/M extracts, replication-independent chromatin assembly in S phase extracts is significantly repressed in terms of its rate and absolute capacity for nucleosome reconstitution.
Our results suggest a peak of assembly activity at the M–G1 transition. Thus, comparing extracts from arrested cdc mutants, assembly activity is slightly higher in G2/M than in G1 (Fig. 2A). In experiments using synchronized cdc15–2 cells, activity peaks late in M, declining somewhat as cells enter G1 (Fig. 2B). Although assays of extracts from α-factor synchronized cells suggest higher activity in G2 than M (Fig. 1A), the apparent decline in M should be interpreted cautiously because the M population includes a greater proportion of cells in G1/S than does the G2 population (owing to loss of synchronization as the experiment proceeds; Fig. 1A). The presence of S phase cells is expected to reduce the assembly capacity of this “M phase” extract (see Fig. 3).
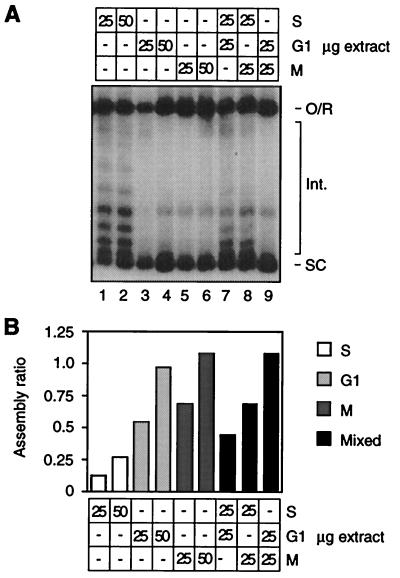
Figure 3.
Cell cycle regulation of assembly is not caused by fluctuations in the activity of a dominant inhibitor. (A) Assembly reactions using the indicated amounts of extract protein from S, G1, and M phase arrested cdc mutants were performed at 22°C for 30 min. The migration of open circular/relaxed DNA (O/R), highly supercoiled species (SC), and intermediate topoisomers (Int.) is indicated. (B) Quantitation of autoradiograph in A.
Fluctuations in Assembly Activity During the Cell Cycle Are Not Caused by a Dominant Inhibitor.
The above results could stem from fluctuations in the activity of a dominant inhibitor of assembly. We performed mixing experiments to test this possibility. In this approach “active” and “inactive” extracts are mixed and compared in activity to the same total amount of the active and inactive extracts assayed separately. If the activity of the mixture is the same as inactive extract alone (μg amount = total of mixture), then the regulatory effect is likely caused by changes in the activity of a dominant inhibitor (26). For this experiment (Fig. 3) “active” extracts are from G1 (lanes 3, 4) and M (lanes 5, 6) arrested cdc28–1 and cdc15–2 mutants, respectively. The “inactive” extract is from S phase arrested cdc17–1 cells (lanes 1, 2). As is most evident by the intensity of intermediate topoisomers, the level of assembly in a 50-μg mixture of G1 and S extracts (lane 7) falls between the levels observed with 50 μg of G1 (lane 4) and S (lane 2) extract alone. A similar result was obtained by using a mixture of M and S extracts (lane 8). Quantitation (Fig. 3B) supports the interpretation based on visual inspection of Fig. 3A. We conclude that the extract does not contain a dominant inhibitor of assembly. The regulatory mechanism underlying fluctuations in assembly activity during the cell cycle therefore involves changes in the activity/abundance of a positively acting component of the assembly machinery, or (for example) changes in the abundance of an inhibitor whose free active form is limiting in S phase extract.
The regulation of replication-independent chromatin assembly in extracts could reflect differential extraction of proteins at different stages of the cell cycle. We do not favor this possibility because (i) the assembly capacity of log extracts was not significantly increased by increasing the salt concentration of the extraction buffer; (ii) the abundance of histones and indeed all similarly migrating proteins in SDS/PAGE gels is similar between extracts (not shown), and slight variations in histone abundance did not correlate with activity; (iii) histone recovery from whole cell extract (by acid extraction) was very similar for all cell types; and (iv) we obtained evidence that histone modification plays an important role in the cell cycle regulation of replication-independent assembly. Note that Western blot analysis using various antihistone antibodies was not useful for quantifying histones in whole cell extracts because the histone content of the extract is relatively low and the specific affinity of antihistone antibodies is generally low. Indeed to our knowledge antihistone antibodies have not been used to quantitate histone levels in crude yeast extracts. Rather, such antibodies are typically used to probe nuclear proteins or histones purified by acid extraction of spheroplasted cells or isolated nuclei (e.g., ref. 27).
Cell Cycle Regulation of Histone H3/H4 Activity and Posttranslational Modification.
We performed tests to determine whether replication-independent assembly fluctuates during the cell cycle as a result of changes in histone activity. Test extracts depleted of both H3 and H4 were used for this analysis (24). Such extracts are severely defective for assembly, and the defect is complemented by total histones purified from wild-type extract. The complementation reflects the restoration of wild-type H3/H4 levels, because H3 and H4 are depleted in the test extract and because histone H2B does not play a role in supercoiling (8).
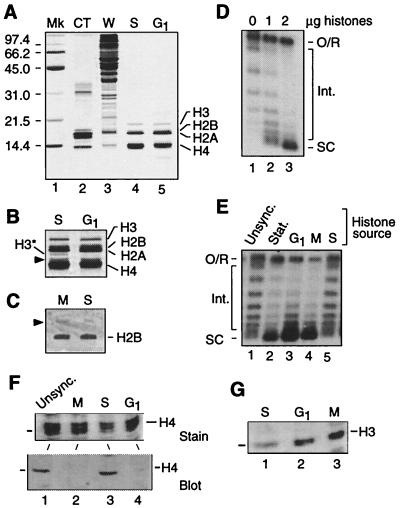
We compared the activity of histones purified from G1, S, and M phase extracts. Silver staining (Silver Stain Plus, Bio-Rad) revealed that the histone preparations are very uniform in composition (Figs. 4 A and B show representative examples) and Western blotting readily detected H2B in these preparations (Fig. 4C). Importantly no cell cycle-dependent degradation of H3 or H4 is evident in stained gels (compare S phase with G1 histones, Fig. 4A, lanes 4 and 5). Although the abundance of a likely breakdown product of H3 varies slightly (H3* in Fig. 4B; see ref. 28), intact H3 is equally represented in all preparations. A minor band running slightly above H4 is similarly abundant in all samples. Because bovine H3 is poorly stained (Fig. 4A, lane 2), the apparent under-representation of intact H3 in our preparations is a staining artifact (see also figure 1C in ref. 29). Two micrograms of histones (roughly the amount in 50 μg of wild-type extract) from unsynchronized cells complements the test extract (Fig. 4D). The activity of 2 μg of histones from cells in G1 (cdc28–1), S (cdc17–1), and M (cdc15–2) was therefore assayed by using the reconstitution system (Fig. 4E). Histones from unsynchronized, log phase cells (lane 1) and from S phase cells (lane 5) did not support a high level of assembly, whereas both G1 and M histones were highly active (lanes 3 and 4). We conclude that the capacity of H3 and/or H4 to participate in replication-independent chromatin assembly fluctuates during the cell cycle in parallel with the changes in activity of assembly extracts. We propose that histone H3/H4 modification plays an important role in the cell cycle regulation of replication-independent chromatin assembly in yeast whole cell extract. It follows that H3 and/or H4 must be differentially modified in extracts from cells at different stages of the division cycle. To test this prediction, equivalent amounts of histones purified from assembly extracts were characterized by Western blot analysis using an antiserum against the K5/8/12/16 tetra-acetylated form of H4 (Fig. 4F). This analysis reveals that tail-acetylated isoforms of H4 are abundant in unsynchronized (log) and S phase extracts but depleted in G1 and M phase extracts. Our evidence also suggests differential phosphorylation of H3 in the extracts, the relative levels being low in S phase, high in M, and moderate in G1 (Fig. 4G). These data demonstrate a complex pattern of posttranslational modification of histones H3 and H4 in the assembly extract, consistent with the hypothesis that posttranslational histone modification is important for the cell cycle regulation of replication-independent chromatin assembly in yeast.
Figure 4.
Modification of histones H3 and/or H4 regulates replication-independent chromatin assembly. (A) Analysis of histones isolated from cdc mutants arrested in S phase (cdc17–1) and G1. Acid-extracted yeast histones (1 μg per lane) were resolved by SDS/PAGE and visualized by silver staining. The protein profile of calf thymus histones (CT) and whole cell extract (W, 20 μg) is shown for comparison. Note the faint staining of H3 in calf thymus and yeast histones. The migration of protein standards (Mk) in kDa is indicated on the left. (B) Detail of lanes 4 and 5 in (A) after prolonged staining. H3* is a presumed degradation fragment of H3. The arrowhead indicates an unidentified protein. (C) Immunostaining of H2B in histones isolated from M (cdc15) and S phase (cdc17–1) extracts. Proteins (20 μg) were resolved by triton-acid-urea gel electrophoresis and probed with anti-H2B antibody. The arrowhead indicates a cross-reacting protein also detected by the preimmune serum. (D) Add-back of histones purified from late log phase assembly extract stimulates assembly in test extract (50 μg). Reactions were performed at 30°C for 30 min. The migration of open circular/relaxed DNA (O/R), highly supercoiled species (SC), and intermediate topoisomers (Int.) is indicated. (E) Direct comparison of assembly capacity of purified histones from assembly extracts of unsynchronized log phase cells (Unsync.), stationary phase cells (Stat.), and cdc mutants arrested at specified points in the cell cycle. Reactions performed as in C. Posttranslational modifications of histones H3 and H4 in yeast chromatin assembly extract. (F) Histones (20 μg) isolated from M (cdc15–1), S (cdc17–1), and G1 (cdc28–1) extracts were resolved by SDS/PAGE and detected by Coomassie blue staining [Upper; only proteins similar to H4 (the middle band of the triplet) in size are shown] or characterized by Western blot analysis using an antibody raised against the tetra-acetylated isoform of the amino-terminal tail of H4 (Lower). (G) Western blot analysis as in F but with an antibody raised against the phospho-Ser 10 isoform of the amino-terminal tail of H3.
DISCUSSION
A Replication-Independent Assembly Pathway That Is Activated During G2, M, and G1.
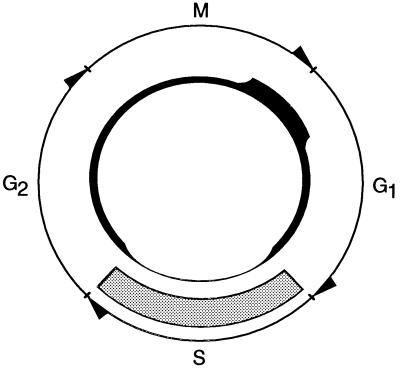
We have discovered a replication-independent, cell cycle-regulated chromatin assembly pathway in yeast (Fig. 5). In extracts from cells synchronized by α-factor arrest or by temperature shift of cdc mutants, this pathway is elevated in G2, M, and G1, and is repressed in S phase. This pattern is reproduced in extracts from cells treated with cell cycle-arresting drugs. Histones H3 and/or H4 are critical targets of this regulatory mechanism. Thus, in a reconstitution assay using a H3/H4-dependent test extract, the core histones isolated from M and G1 extracts are significantly more active in assembly than S phase histones. This result is in contrast to the preferential use of newly synthesized S phase histones in bulk replication-coupled assembly mediated by CAF-I (2).
Figure 5.
Activity of the replication-independent assembly pathway described in this report (■), and the replication-dependent pathway responsible for bulk chromatin assembly (░⃞), during the yeast cell cycle. In this model line thickness indicates the level of assembly. Replication-dependent assembly coupled to nucleotide excision repair is not shown.
The cell cycle-controlled, replication-independent assembly pathway in our extract is expected to involve a histone chaperone/CAF and an activity that plays a role in the posttranslational modification of H3 and/or H4. Maximal supercoiling activity in log phase whole cell extract requires one known assembly protein, the Msi1p subunit of CAF-I (ref. 30; T. Harkness, T. Arnason and M.C.S., unpublished data), but not the Nap1p (8, 32) or Spt6p (data not shown; reagents kindly provided by F. Winston, ref. 28) chaperones. Therefore, either a subpopulation of CAF-I serves in the assembly pathway defined here or the pathway is mediated by a distinct CAF sharing Msi1p as a subunit. Both possibilities are reasonable considering the presence of Msi1p in multiple complexes in mammalian cells (33, 34), the existence of distinct subpopulations of CAF-I in mammalian cells (34), and the observation that CAF-I has replication-independent assembly activity (35). Purification of the assembly machinery and identification of functionally relevant histone modifications will be required to fully characterize the participants in the assembly pathway defined in this study.
Reciprocal Control of Chromatin Assembly Pathways in Yeast.
The replication-dependent chromatin assembly machinery likely has evolved to function efficiently in the context of ongoing DNA synthesis during S phase (1). We speculate that the cell cycle-regulated, replication-independent pathway is inactivated in S phase to prevent it from interfering with the coordination of DNA replication and new nucleosome deposition by the replication-dependent assembly machinery. This reciprocal control mechanism (Fig. 5) would ensure rapid reestablishment of normal bulk chromatin organization in S phase, whereas induction of replication-independent assembly outside of S phase would provide a mechanism to counteract nucleosome loss in G2, M, and G1.
CAF-I plays an important role in replication-dependent chromatin assembly coupled to nucleotide excision repair (36). According to our model, when initiated outside of S phase, repair/replication-coupled assembly could be subject to interference by the replication-independent pathway described here. Presumably, mechanisms exist to limit such interference. To explore this idea, it will be necessary to determine the details of the replication-independent and repair/replication-coupled reactions, in particular the respective requirements for histone modification and possible repression of replication-independent activity in response to DNA damage.
H3/H4 Modification as a Mechanism for the Reciprocal Control of Chromatin Assembly Pathways.
Changes in histone abundance partly govern the activity of the replication-dependent chromatin assembly pathway in vivo, and acetylation of lysine residues in the tails of H3 and H4 is associated with replication-dependent nucleosome deposition during S phase (2, 10, 37–39). We demonstrate that histones are also important targets for the regulation of replication-independent chromatin assembly by a mechanism involving posttranslational modification of H3 and/or H4. Because our assembly system is sensitive to deletion of the amino-terminal tails of H3 and H4, as well as to mutation of lysines in the tail of H4 that are known to be acetylated in vivo (23, 24), it is attractive to consider that histone acetylation status might be important for histone recognition and/or utilization by the CAFs involved in the cell cycle-regulated replication independent pathway, or that a component of this pathway might itself regulate histone acetylation. Because acetylation is also associated with replication-dependent assembly (2, 37–39) and the replication-dependent and -independent pathways appear to be under reciprocal control, different patterns of acetylation might serve to target histones to one pathway or the other. For example, H4 molecules diacetylated on K5 and K12 might be preferentially recruited by the replication-dependent machinery, whereas those monoacetylated on K8 might preferentially serve for replication-independent assembly. This would be consistent with the heavier acetylation of H4 in S phase as opposed to M and G1 extracts (Fig. 4F). The redundancy of K5, K8, and K12 of H4 in vivo (23) could be explained in part by the ability of replication-independent assembly factors to use, albeit relatively inefficiently, histones targeted to the replication-dependent machinery; in this way the replication-independent machinery could compensate for the reduced assembly capacity of the replication-dependent machinery when the pattern of histone acetylation is not optimal for replication-dependent assembly. It is also possible that other modifications, such as H3 phosphorylation, play a role in the regulation of this pathway.
Function of Replication-Independent Assembly During the Cell Cycle.
The role of the cell cycle-regulated, replication-independent assembly pathway is likely be to replacement of nucleosomes lost as a result of histone degradation or displacement during G2, M, and G1. Presumably the pathway’s most critical function would be to replace nucleosomes lost soon after bulk replication-dependent assembly has ceased, otherwise the persistent exposure of transcriptional control elements until the next S phase might lead to inappropriate transcription of genes whose untimely expression would be detrimental to survival. Replication-independent chromatin assembly likely serves a similar function in terminally differentiated metazoan cells, in which there is no opportunity for nucleosome replacement by the replication-dependent pathway except in response to DNA damage.
Is the cell cycle-regulated, replication-independent assembly pathway involved in the establishment and/or maintenance of heterochromatin-like domains in yeast? A primary role seems unlikely with respect to the silent HM mating type loci because, during phases of the cell cycle when we observe high activity of the replication-independent assembly machinery, silencerless minichromosomes generated in vivo lose supercoiling (40). The replication-independent assembly machinery therefore cannot maintain chromatin states initially established by the HM silencing machinery. On the other hand the replication-independent assembly machinery may function in the metabolism of telomeric heterochromatin. Telomeric heterochromatin is relatively unstable during G2/M, when it is proposed that silencing factors can compete with transcription factors to establish the silenced state at a telomere (31). We find that the cell cycle-regulated, replication-independent assembly pathway is most active in late M/early G1 extracts. Perhaps the activation of replication-independent assembly in late M phase is a component of the mechanism involved in establishing heterochromatin at telomeres, such that promoters placed in their vicinity have become refractory to activation by the time the cell is in G1.
Acknowledgments
We are grateful to Michael Grunstein for strain RMY102 and the H2B–GST fusion construct, and to Troy Harkness and Dorothy Rutkowski for assistance with flow cytometry. This study was supported by the Medical Research Council of Canada (scholarship and operating grants to M.C.S.) and the Alberta Heritage Foundation for Medical Research (Senior Scholar award and establishment grant to M.C.S.).
ABBREVIATIONS
- CAF
chromatin assembly factor
- cdc
cell division cycle
- YP
yeast extract/peptone
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Kaufman P D, Botchan M R. Curr Opin Genet Dev. 1994;4:229–235. doi: 10.1016/s0959-437x(05)80049-8. [DOI] [PubMed] [Google Scholar]
- 2.Roth S Y, Allis C D. Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 3.Ito T, Tyler J K, Kadonaga J T. Genes Cells. 1997;2:593–600. doi: 10.1046/j.1365-2443.1997.1500348.x. [DOI] [PubMed] [Google Scholar]
- 4.Cairns B R. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- 5.Grunstein M. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 6.Cavilli G, Thoma F. EMBO J. 1993;12:4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson V. Biochemistry. 1990;29:719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- 8.Schultz M C, Hockman D J, Harkness T A A, Garinther W I, Altheim B A. Proc Natl Acad Sci USA. 1997;94:9034–9039. doi: 10.1073/pnas.94.17.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz, M. C. (1999) Methods17, in press. [DOI] [PubMed]
- 10.Krude T. Curr Biol. 1995;5:1232–1234. doi: 10.1016/s0960-9822(95)00245-4. [DOI] [PubMed] [Google Scholar]
- 11.Thomas B J, Rothstein R. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 12.Hartwell L H, Mortimer R K, Culotti J, Culotti M. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann R K, Grunstein M. EMBO J. 1992;11:3297–3306. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wittenberg C, Sugmoto K, Reed S I. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]
- 15.Oehlen L J W M, Cross F R. Genes Dev. 1994;8:1058–1070. doi: 10.1101/gad.8.9.1058. [DOI] [PubMed] [Google Scholar]
- 16.Nash R, Tokiwa G, Anand S, Erickson K, Futcher A B. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherman F. Methods Enzymol. 1991;194:16–18. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 18.Din S U, Brill S J, Fairman M P, Stillman B. Genes Dev. 1990;4:968–977. doi: 10.1101/gad.4.6.968. [DOI] [PubMed] [Google Scholar]
- 19.Guacci V, Hogan E, Koshland D. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Culotti J, Hartwell L H. Exp Cell Res. 1971;67:389–401. doi: 10.1016/0014-4827(71)90424-1. [DOI] [PubMed] [Google Scholar]
- 21.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. [Google Scholar]
- 22.Delcuve G P, Davies J R. Anal Biochem. 1992;200:339–341. doi: 10.1016/0003-2697(92)90475-m. [DOI] [PubMed] [Google Scholar]
- 23.Ma X-J, Wu J, Altheim B A, Schultz M C, Grunstein M. Proc Natl Acad Sci USA. 1998;95:6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling X, Harkness T A A, Schultz M C, Fisher-Adams G, Grunstein M. Genes Dev. 1996;10:686–699. doi: 10.1101/gad.10.6.686. [DOI] [PubMed] [Google Scholar]
- 25.Garinther W I, Schultz M C. Mol Cell Biol. 1997;17:3520–3536. doi: 10.1128/mcb.17.7.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White R J, Gottlieb T M, Downes C S, Jackson S P. Mol Cell Biol. 1995;15:6653–6662. doi: 10.1128/mcb.15.12.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edmondson D G, Smith M M, Roth S Y. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 28.Bortvin A, Winston F. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 29.Workman J L, Kingston R E. Science. 1992;258:1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]
- 30.Kaufman P D, Kobayashi R, Stillman B. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 31.Aparicio O M, Gottschling D E. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 32.McQuibban G A, Commisso-Cappelli C N, Lewis P N. J Biol Chem. 1998;273:6582–6590. doi: 10.1074/jbc.273.11.6582. [DOI] [PubMed] [Google Scholar]
- 33.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 34.Marheineke K, Krude T. J Biol Chem. 1998;273:15279–15286. doi: 10.1074/jbc.273.24.15279. [DOI] [PubMed] [Google Scholar]
- 35.Kamakaka R T, Bulger M, Kaufman P D, Stillman B, Kadonaga J T. Mol Cell Biol. 1996;16:810–817. doi: 10.1128/mcb.16.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martini E, Roche D M J, Marheineke K, Verreault A, Almouzni G. J Cell Biol. 1998;143:563–575. doi: 10.1083/jcb.143.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobel R E, Cook R G, Perry C A, Annunziato A T, Allis C D. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo M-H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Nature (London) 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 39.Wade P A, Pruss D, Wolffe A P. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 40.Bi X, Broach J R. Mol Cell Biol. 1997;17:7077–7087. doi: 10.1128/mcb.17.12.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]