Abstract
Mutations in an 81-bp region of the rpoB gene associated with rifampin resistance were studied in 41 rifampin-resistant clinical strains of Mycobacterium tuberculosis isolated in Turkey. Fourteen different rpoB alleles, three of which had not been reported before, were found. A reverse hybridization-based line probe assay (the Inno-LiPA Rif.TB test) for rapid detection of the mutations was evaluated with these isolates. Rifampin resistance was correctly identified in 23 of 41 isolates (56.1%) with the kit's R probes specific for these mutations. Seventeen of 41 isolates (41.5%) yielded hybridization patterns, with at least one negative signal obtained with the S probes for the wild type. One isolate was identified as rifampin sensitive by the line probe assay. The rate of concordance of the results of the line probe assay with the results of the in vitro susceptibility test was high (97.6%). These results demonstrate that the line probe assay kit may be useful for the rapid diagnosis of rifampin-resistant tuberculosis.
Mycobacterium tuberculosis remains one of the most significant causes of death from an infectious agent, annually leading to 2 million deaths worldwide (4). As the incidence of tuberculosis has increased, there has been a corresponding rise in the incidence of drug-resistant strains of M. tuberculosis. Early diagnosis of the disease and rapid identification of resistance to primary antituberculosis agents are essential for efficient treatment and control of multidrug-resistant (MDR) strains. Rifampin (RIF) is one of the most potent antituberculosis drugs; therefore, resistance to RIF often results in high clinical relapse rates, particularly if RIF resistance is associated with resistance to other antituberculosis drugs (5, 14). Moreover, more than 90% of RIF-resistant isolates are also resistant to isoniazid; therefore, detection of RIF resistance could also identify MDR strains (3, 5, 17). The incidence of pulmonary tuberculosis in Turkey was 35.5 per 100,000 population in 2000. In the Aegean region, 8.2% of M. tuberculosis strains isolated between 1999 and 2001 were found to be resistant to RIF. During the same period, the incidence of resistance to both RIF and isoniazid was 6.8% (6).
Collectively, DNA sequencing studies demonstrate that more than 95% of RIF-resistant M. tuberculosis strains have a mutation within the 81-bp hot-spot region (codons 507 to 533) of the RNA polymerase B subunit (rpoB) gene (9, 10, 12, 13, 15, 16, 18, 20, 21, 23, 24). In addition, other studies reveal that mutations associated with RIF resistance can also occur in other regions of the rpoB gene, although these occur less frequently (7, 8). Several molecular methods have been developed in recent years to evaluate the rpoB gene for RIF resistance mutations, including DNA sequencing, line probe assay, and analysis with DNA microarrays (19). These molecular tests can also serve as a means of detecting molecular epidemiological markers, since the relative frequencies of the alleles associated with resistance can vary geographically (10).
The aim of the present study was to determine the drug resistance profiles of 41 RIF-resistant M. tuberculosis isolates obtained in western Turkey and to detect and identify mutations present in the rpoB gene. Two molecular assays were used in this study. In the first one, rpoB mutations were determined by a commercially available rapid test, the PCR-based reverse hybridization line probe assay (LiPA; Inno-LiPA Rif.TB test; Innogenetics N.V., Ghent, Belgium). The results obtained by LiPA were then compared with the results obtained by automated DNA sequence analysis.
MATERIALS AND METHODS
M. tuberculosis isolates.
The 41 RIF-resistant clinical M. tuberculosis isolates obtained from 41 different patients used in this study were isolated in the Region Tuberculosis Laboratory and Ege University Mycobacteriology Laboratory, Izmir, Turkey. These laboratories serve about 8 million people in the Aegean region of Turkey. The proportional method with 7H10 medium had been used to test resistance to the primary antituberculosis drugs (11). All isolates were resistant to 1 μg of RIF per ml. Isolates were considered resistant to isoniazid when they were resistant to at least 0.2 μg of isoniazid per ml. They were considered resistant to streptomycin and ethambutol when they were resistant to 2 μg of streptomycin per ml and 5 μg of ethambutol per ml, respectively (11). The drug susceptibility patterns of the isolates are shown in Table 1.
TABLE 1.
Resistance patterns of RIF-resistant M. tuberculosis strains isolated in Turkey
| No. of strains | Resistance patterna |
|||
|---|---|---|---|---|
| RIF | INH | STR | EMB | |
| 15 | R | R | R | S |
| 13 | R | R | R | R |
| 5 | R | R | S | S |
| 4 | R | R | S | R |
| 2 | R | S | S | R |
| 2 | R | S | S | S |
| Total | 41 | 37 | 28 | 19 |
INH, isoniazid; STR, streptomycin; EMB, ethambutol; R, resistant; S, susceptible.
Detection of mutations.
DNA sequencing was performed with an automated DNA sequencer (model 310; Applied Biosystems, Foster City, Calif.). DNA was extracted from cultures and amplified in a PCR. A 394-bp product was generated by using primers LIPA OP1 (outer primer; 5′-GAGAATTCGGTCGGCGAGCTGATCC-3′) and LIPA OP2 (outer primer; 5′-GGAAGCTTGACCCGCGCGTACACC-3′), and a 256-bp fragment of the rpoB gene was sequenced by using primers LIPA IP1 (inner primer; 5′-GGTCGGCATGTCGCGGATGG-3′) and LIPA IP2 (inner primer; 5′-GCACGTCGCGGACCTCCAGC-3′) (22).
The Inno-LiPA Rif.TB LiPA kit was used according to the instructions of the manufacturer. The RIF resistance-determining region of the rpoB gene was amplified with specific biotin-labeled primers by using 1 U of Taq DNA polymerase per reaction mixture in a thermocycler (model 9600; Perkin-Elmer). The biotinylated 256-bp PCR product was then denatured and hybridized to a strip with 10 specific oligonucleotide probes. M. tuberculosis was detected in a sample by use of the M. tuberculosis complex-specific probe. The reactivities of an amplified fragment with the S-type probes for the wild type (probes S1 through S5) were used to detect the mutations that lead to RIF resistance in M. tuberculosis. Furthermore, four probes (R-type probes) were specifically designed to hybridize to the sequences of the four most frequently observed mutations: R2 (Asp-516-Val), R4a (His-526-Tyr), R4b (His-526-Asp), and R5 (Ser-531-Leu).
In conclusion, when all the wild-type S probes gave a positive signal and all the R probes reacted negatively, the M. tuberculosis isolate was considered susceptible to RIF. When at least one negative signal was obtained with the wild-type S probes, the isolate was considered RIF resistant (Inno-LiPA Rif.TB S patterns). When the resistance to RIF was due to one of the four most frequently observed mutations described above, a positive reaction was obtained with one of the four R probes and was always accompanied by a negative reaction with the corresponding S probe (Inno-LiPA Rif.TB R patterns). M. tuberculosis strain H37RV was used as a control.
Nucleotide sequence accession numbers.
The sequences with novel mutations found in this study are deposited in EMBL under accession numbers AF515787, AF5157888, and AF515789.
RESULTS AND DISCUSSION
Fourteen different types of mutations were identified in 41 RIF-resistant M. tuberculosis isolates. The codons most frequently involved in mutations were codon 531 (56.1%) and codon 526 (19.5%). In concordance with previous reports (9, 10, 12, 13, 15, 16, 18, 20, 21, 23, 24) but not that of Barfai et al. (1), 19 (46.3%) isolates carried the most common mutation, Ser-531-Leu. Two (4.9%) isolates had an His-526-Tyr mutation, one (2.4%) isolate had an His-526-Asp mutation, and another (2.4%) isolate had an Asp-516-Val mutation. These mutations were also correctly detected by LiPA (Table 2). By comparison of these data with those in the literature (1, 9, 13, 16, 21, 23, 24), it was observed that three mutations occurring less frequently in Turkish strains (526-TAC, 526-GAC, and 516-GTC) had different relative frequencies in strains from different countries (Table 3). While mutation of CAC to GAC at codon 526 occurred at frequencies of 24.3% in Italian isolates (16) and 17.6% in Greek isolates (13), the mutation of CAC to TAC at codon 526 was dominant in American isolates (23) and Brazilian isolates (21) and occurred at frequencies of 27.9 and 11%, respectively. In Australian isolates (24), the two mutations at codon 526 occurred at frequencies of 12.1% each. In addition, Barfai et al. (1), who analyzed Hungarian isolates, and Hirano et al. (9), who analyzed strains mostly isolated in Asian countries, found high frequencies of mutation of GAC to GTC at codon 516 (Table 3). In the present study, however, the most frequently encountered mutation at codon 526 was CAC to CGC and the most frequently encountered mutation at codon 516 was GAC to TAC (Table 2). The mutation of TCG to TGG at codon 531 also occurred at a relatively high frequency, 9.8% (Table 2). These findings demonstrate that the frequencies of particular mutations in RIF-resistant M. tuberculosis isolates from western Turkey are different from those that have been reported for isolates from other parts of the world.
TABLE 2.
Relative frequencies of mutated rpoB alleles in Turkish RIF-resistant isolates of M. tuberculosis
| Allele | LiPA profile | Amino acid change(s) | No. (%) of strains |
|---|---|---|---|
| 531-TCG→TTG | R5 | Ser→Leu | 19 (46.3) |
| 531-TCG→TGG | ΔS5 | Ser→Trp | 4 (9.8) |
| 533-CTG→CCG | ΔS5 | Leu→Pro | 1 (2.4) |
| 526-CAC→TAC | R4a | His→Tyr | 2 (4.9) |
| 526-CAC→GAC | R4b | His→Asp | 1 (2.4) |
| 526-CAC→CGC | ΔS4 | His→Arg | 4 (9.8) |
| 526-CAC→TGC | ΔS4 | His→Cys | 1 (2.4) |
| 522-TCG→TGG | ΔS3 | Ser→Trp | 2 (4.9) |
| 516-GAC→GTC | R2 | Asp→Val | 1 (2.4) |
| 516-GAC→TAC | ΔS2 | Asp→Tyr | 2 (4.9) |
| 513-CAA→CCA | ΔS1 | Gln→Pro | 1 (2.4) |
| 515-ATG→ATC, 533 CTG→CCG | ΔS2, ΔS5 | Met→Ile, Leu→Pro | 1 (2.4) |
| CGG insertion between 514 and 515 | ΔS1, ΔS2 | Arg | 1 (2.4) |
| 490-CAG→CAT | S | Gln→His | 1 (2.4) |
TABLE 3.
Frequency of mutations in RIF-resistant M. tuberculosis isolates from different geographic regions
| Geographic region (reference; no. of isolates) | Frequency (%) of mutated codons |
|||||
|---|---|---|---|---|---|---|
| Ser-531-Leua | His-526-Aspa | His-526-Tyra | His-516-Vala | Other mutations | No mutation within hot-spot region | |
| Asia (9; n = 77) | 44.2 | 5.2 | 10.4 | 15.6 | 18.1 | 6.5 |
| Australia (24; n = 33) | 48.5 | 12.1 | 12.1 | 6.1 | 18.2 | 3.0 |
| Brazil (21; n = 82) | 50.0 | 1.2 | 11.0 | 6.2 | 28.0 | 3.6 |
| Greece (13; n = 17) | 41.2 | 17.6 | 11.8 | 23.5 | 5.9 | |
| Hungary (1; n = 29) | 31.0 | 6.9 | 37.9 | 13.9 | 10.3 | |
| India (12; n = 44) | 54.5 | 2.3 | 4.5 | 36.4 | 2.3 | |
| Italy (16; n = 37) | 56.7 | 24.3 | 2.7 | 2.7 | 13.6 | |
| United States (23; n = 61) | 34.4 | 8.2 | 27.9 | 4.9 | 16.4 | 8.2 |
| Turkey (n = 41) | 46.3 | 2.4 | 4.9 | 2.4 | 41.5 | 2.4 |
Includes frequency of mutations only at a single codon.
LiPA did not detect the correct type of mutation in 17 (41.5%) isolates. However, it indicated the presence of a genetic alteration. Moreover, one isolate that had a mutation outside the 81-bp region of the rpoB gene was identified as RIF sensitive by LiPA. Three novel mutations were also recognized in this study. A mutation from ATG (Met) to ATC (Ile) at codon 515 and a mutation from CTG (Leu) to CCG (Pro) at codon 533 in one isolate and insertion of CGG between codons 514 and 515 in one isolate have not been reported previously. One new mutation (CAG to CAT at codon 490) outside the 81-bp hot-spot region was also seen in one isolate. Previous research (12, 16, 18, 24) has also reported mutations outside the hot-spot region: GGG to GAG at codon 534, CCC to CAC at codon 535, GAG to GAT at codon 504, GAG to GAT at codon 541, TCG to GCG at codon 553, and ATC to TTC at codon 572. Although the Turkish isolates exhibited the three novel mutations mentioned above, the patterns of mutations in the 81-bp hot-spot region were similar to those reported for the majority of clinical isolates in different geographical areas of the world. In this study, no association between specific rpoB mutations and multidrug resistance patterns was found, supporting the view that the mutations leading to RIF resistance are independent events unrelated to those mutations affecting the development of resistance to other antibiotics tested.
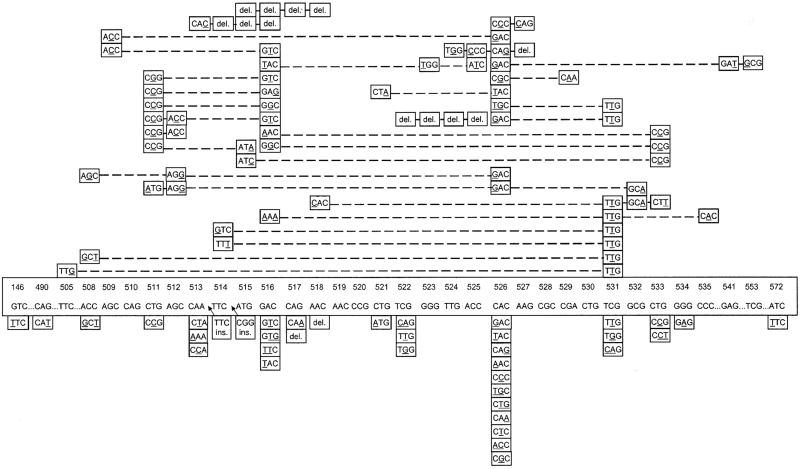
Mutations at codon 531, followed by mutations at codons 526, 516, and 511, are the most common mutations worldwide. While TTG at codon 531 and CCG at codon 511 are the dominant mutated alleles, codon 526 and codon 516 show large numbers of allelic variations (Fig. 1). Two postulates can be offered to explain this situation. (i) Billington et al. (2) observed that mutants isolated more frequently in clinical practice have higher mean relative fitness, and the prevalence of each mutant type depends on its ability to survive. This might be the reason for the higher rates of occurrence of the mutation of TCG to TTG at codon 531 and the mutation of CTG to CCG at codon 511 in isolates worldwide (12). (ii) Mutations within codons 526 and 516 have been shown to lead to high-level RIF resistance in M. tuberculosis (23); therefore, mutations continue to arise in these codons, probably due to the ability of M. tuberculosis to adapt to drug exposure.
FIG. 1.
Mutations and alleles in rifampin-resistant M. tuberculosis isolates reported by different groups (1, 9, 12, 13, 16, 20, 21, 23, 24; this study). The original sequence is boxed. The bottom panels show the mutation at a single codon; and the upper panels show the mutations involved in double, triple, and quadruple codons.
LiPA rapidly identifies clinical isolates as members of the M. tuberculosis complex and determines the presence of point mutations within the 81 bp of the rpoB gene. The test can detect the type of mutation for only the four most common mutations of the rpoB gene (Ser-531-Leu, His-526-Tyr, His-526-Asp, and Asp-516-Val), while for isolates with other mutations, it indicates only the presence of a genetic alteration. However, previous reports and this study suggest that the frequencies of particular mutations are different in different countries; therefore, further studies on how to improve the kit for the precise diagnosis of RIF-resistant M. tuberculosis infections are needed (Table 3).
In conclusion, in the present study, LiPA was able to detect a genetic alteration in 40 (97.6%) of the 41 RIF-resistant strains and to identify the particular mutation in 23 (56.1%) strains (Table 2). Since 90.2% of RIF-resistant strains examined in this study were also resistant to isoniazid, this indicates that RIF resistance is a good predictor of multidrug resistance in Turkey. Although the kit could not detect in vitro resistant isolates with the wild-type sequence in the hot-spot region of the rpoB gene, it may be especially useful for routine work in clinical laboratories that are not capable of carrying out DNA sequencing. However, the test results must always be confirmed by phenotypic methods.
Acknowledgments
This work was supported by the Ege University Medical Science Program (grant 2000/TIP/007).
REFERENCES
- 1.Barfai, Z., A. Somoskövi, C. Ködmön, N. Szabo, E. Puskas, L. Kosztolanyi, E. Frago, J. Mester, L. M. Parsons, and M. Salfinger. 2001. Molecular characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Hungary by DNA sequencing and the line probe assay. J. Clin. Microbiol. 39:3736-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billington, O. J., T. D. McHugh, and S. H. Gillespie. 1999. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 41:1866-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drobniewski, F. A., and S. M. Wilson. 1998. The rapid diagnosis of isoniazid and rifampin resistance in Mycobacterium tuberculosis—a molecular story. J. Med. Microbiol. 47:189-196. [DOI] [PubMed] [Google Scholar]
- 4.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalance, and mortality by country. World Health Organization Global Surveillence and Monitoring Project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 5.Goble, M., M. D. Iseman, L. A. Madsen, D. Waite, L. Ackerson, and C. R. Hobsburg, Jr. 1991. Treatment of 171 patients with pulmonary tuberculosis resistant to isoniazid and rifampin. N. Engl. J. Med. 328:527-532. [DOI] [PubMed] [Google Scholar]
- 6.Guneri, S., I. Unsal, A. Oztop, M. Erkut, V. Avkan Oguz, and R. Cakmak. 2002. Antituberculosis drug resistance in Aegean Region. Toraks Derg. 3:77-78. [PubMed] [Google Scholar]
- 7.Heep, M., B. Brandstatter, U. Rieger, N. Lehn, E. Richter, S. Rusch-Gerdes, and S. Niemann. 2001. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 39:107-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heep, M., U. Rieger, D. Deck, and N. Lehn. 2000. Mutations in the beginning of the rpoB gene can induce resistance to rifamycins in both Helicobacter pylori and Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:1075-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirano, K., C. Abe, and M. Takahashi. 1999. Mutations in the rpoB gene of rifampin-resistant Mycobacterium tuberculosis strains isolated mostly in Asian countries and their rapid detection by line probe assay. J. Clin. Microbiol. 37:2663-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapur, V., L.-L. Li, S. Iordanescu, M. R. Hamrick, A. Wagner, B. N. Kreiswirth, and J. M. Musser. 1994. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase β subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York and Texas. J. Clin. Microbiol. 32:1095-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology. A guide for a level III laboratory. Centers for Disease Control, Atlanta, Ga.
- 12.Mani, C., N. Selvakumar, S. Narayanan, and P. R. Narayanan. 2001. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis clinical isolates from India. J. Clin. Microbiol. 39:2987-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsiota-Bernard, P., G. Vrioni, and E. Marinis. 1998. Characterization of rpoB mutations in rifampin-resistant clinical Mycobacterium tuberculosis isolates from Greece. J. Clin. Microbiol. 36:20-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchinson, D., and A. Nunn. 1986. Influence of initial drug resistance on the response to short-course chemotherapy of pulmonary tuberculosis. Am. Rev. Respir. Dis. 133:423-430. [DOI] [PubMed] [Google Scholar]
- 15.Musser, J. M. 1995. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pozzi, G., M. Meloni, E. Iona, G. Orro, O. F. Thorensen, M. L. Ricci, M. R. Oggioni, L. Fattorini, and G. Orefici. 1999. rpoB mutations in multidrug resistant strains of Mycobacterium tuberculosis isolated in Italy. J. Clin. Microbiol. 37:1197-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossau, R., H. Traore, H. De Beenhouwer, W. Mijs, G. Jannes, P. de Rijk, and F. Portales. 1997. Evaluation of the INNO-LiPA Rif.TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob. Agents Chemother. 41:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schilke, K., K. Weyer, G. Bretzel, B. Amthor, J. Brandt, V. Sticht-Grohn, P. B. Fourie, and W. H. Hans. 1999. Universal pattern of rpoB gene mutations among multidrug-resistant isolates of Mycobacterium tuberculosis complex from Africa. Int. J. Tuber. Lung Dis. 3:620-626. [PubMed] [Google Scholar]
- 19.Soini, H., and J. M. Musser. 2001. Molecular diagnosis of mycobacteria. Clin. Chem. 47:809-814. [PubMed] [Google Scholar]
- 20.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schpfer, and T. Bodmer. 1993. Detection of rifampin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 21.Valim, A. R. M., M. L. R. Rosetti, M. O. Ribeiro, and A. Zaha. 2000. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from Brazil. J. Clin. Microbiol. 38:3119-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watterson, S. A., S. M. Wilson, M. D. Yates, and F. A. Drobniewski. 1998. Comparison of three molecular assays for rapid detection of rifampin resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 36:1969-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams, D. L., C. Wanguespack, K. Eisanach, J. T. Crawford, F. Porteals, M. Salfinger, C. M. Nolan, C. Abe, V. Sticht-Groh, and T. P. Gillis. 1994. Characterization of rifampin resistance in pathogenic mycobacteria. Antimicrob. Agents Chemother. 38:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuen, L. K. W., D. Leslie, and P. J. Coloe. 1999. Bacteriological and molecular analysis of rifampin-resistant Mycobacterium tuberculosis strains isolated in Australia. J. Clin. Microbiol. 37:3844-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]