Abstract
The direct repeat (DR) region in Mycobacterium tuberculosis complex strains is composed of multiple well-conserved 36-bp DRs interspersed with nonrepetitive DNA spacer sequences of similar size. Clinical isolates show extensive polymorphism in this DR region, and this has led to the development of a 43-spacer reversed line blot methodology: spoligotyping. Although this method has contributed significantly to the molecular epidemiology of tuberculosis in the last decade, the discriminatory power and the readability of this method were not found to be optimal. In order to improve the discriminatory power, the usefulness of 43 redesigned oligonucleotides and the usefulness of 51 new spacer oligonucleotides were evaluated. For 314 M. tuberculosis complex strains isolated in the central part of The Netherlands over a 5-year period, 264 different IS6110 RFLP types could be distinguished, and 160 different spoligotype patterns were identified by traditional spoligotyping. After the introduction of 51 new spacer oligonucleotides, 14 additional spoligotypes were recognized. This enabled us to split 11 clusters of isolates identified by the traditional spoligotyping. Furthermore, on the basis of the new spacer oligonucleotides a dichotomy was found among the Beijing genotype isolates. Among 76 Mycobacterium bovis strains, 20 patterns were found by traditional spoligotyping and 30 patterns were found by novel probe spoligotyping, respectively. Nine M. bovis subsp. caprae isolates yielded six patterns by traditional spoligotyping and eight patterns by novel probe spoligotyping. A part of the redesigned oligonucleotides slightly improved the reading of spoligotype patterns. The reproducibility of spoligotyping, based on internal control probes, invariably yielded a high score; only 4 (1%) of the 314 patient isolates gave discrepant results. Analysis of a set of 31 duplicate M. tuberculosis complex strains demonstrated a 10% error rate for the identification of blinded duplicate samples. In a redundancy analysis, 40 essential spacer oligonucleotides of the 94-spacer sequences were selected, yielding the same number of spoligotype patterns. We propose to leave the traditional commercialized first-generation membrane for spoligotyping unchanged for current applications and to introduce a second-generation spoligotyping membrane whenever extended discrimination is required, e.g., for low-copy-number IS6110 strains or for phylogenetic studies of Beijing genotype strains.
Molecular typing of Mycobacterium tuberculosis complex isolates is a useful tool for epidemiological studies on different levels. In the last decade the application of different DNA fingerprinting techniques has contributed significantly to our understanding of the transmission of tuberculosis (15, 45). Actually, these techniques provide a more sensitive means of identifying outbreaks than conventional surveillance (4).
A wide variety of DNA fingerprinting techniques has been developed to differentiate between M. tuberculosis complex isolates (11, 16, 20, 29, 30), and of these, restriction fragment length polymorphism (RFLP) typing using insertion element IS6110 as a probe has gained recognition as the standard method (38). This typing method has proven to be extremely valuable for the molecular epidemiology of tuberculosis, but this technique also has some significant disadvantages. RFLP typing requires about 2 μg of purified DNA and, hence, suffers a delay of several weeks due to culture. Furthermore, the method is technically demanding and time-consuming. In addition, sophisticated computer software is required to analyze the IS6110 RFLP patterns (18). Several of these disadvantages can be circumvented by using PCR-based DNA fingerprinting techniques such as spacer oligotyping (spoligotyping) (20), variable numbers of tandem repeats typing (12) and mycobacterial interspersed repetitive unit (MIRU) typing (35). One of the most commonly used PCR-based typing methods for M. tuberculosis isolates is spoligotyping. With this method the DNA polymorphism in the genomic direct repeat (DR) locus of M. tuberculosis complex isolates is visualized. This locus contains multiple, well-conserved 36-bp DRs interspersed with nonrepetitive 34- to 41-bp DNA spacer sequences (14). This DR locus has been designated the spacer interspersed DR (SPIDR) region and was recently renamed by Jansen et al. as the “clustered regularly interspaced short palindromic repeats” (CRISPR) region (19).
Spoligotyping involves the amplification of the whole CRISPR region using the DR as a target in PCR, followed by hybridization of the amplified DNA to a set of spacer oligonucleotides, covalently linked to a membrane. Because clinical isolates vary in the presence of spacer sequences (20), the spoligotype patterns obtained are strain specific. Differences between spoligotype patterns have been shown to be due to deletions of spacer sequences in the CRISPR region by transposition of IS6110 elements (9, 10, 24, 39) and are probably mediated by homologous recombination (14) or replication slippage (17).
Spoligotyping is easy to perform and can be used to detect and type M. tuberculosis complex bacteria simultaneously, even directly in clinical samples. However, an important drawback of the current spoligotyping method is its limited discriminatory power compared to that of IS6110 RFLP typing (20, 22, 42). On the other hand, for M. tuberculosis complex strains containing small numbers of IS6110 copies, including Mycobacterium bovis, spoligotyping is generally more discriminatory than IS6110 RFLP typing (3).
In an earlier study on the nature of genetic variations in the DR locus, 94 spacer sequences were described in total (39). Caimi et al. reported the finding of five alternative spacers by analysis of spacer sequencing performed on 20 M. tuberculosis strains and 20 M. bovis strains (5). In addition 94 spacers, including the 5 spacers described by Caimi et al., were found by van Embden et al. by analysis of the DR region of 19 M. tuberculosis strains, two M. bovis strains, two M. bovis BCG strains, one strain intermediate between M. tuberculosis and M. bovis, one Mycobacterium microti strain, and one Mycobacterium canettii strain (5, 39).
The current first-generation spoligotyping membrane consists of a set of 43-spacer oligonucleotides derived from M. tuberculosis reference strain H37Rv and M. bovis BCG strain P3 (20). We evaluated the potential value of recently described new spacer oligonucleotides as far as discriminatory power and reproducibility are concerned. Also the first-generation spacer oligonucleotides were redesigned to optimize the recognition of positive hybridization signals. The study samples comprised a set of 314 M. tuberculosis complex strains isolated during a 5-year period in a regional medical laboratory in The Netherlands, the set of blinded DNAs of the interlaboratory study of Kremer et al. (22), 93 M. bovis strains from various origins, and 78 M. tuberculosis Beijing genotype strains.
MATERIALS AND METHODS
Study population.
Eight hundred forty M. tuberculosis complex strains were isolated in the period from 1993 to 1997 from 314 patients in a central part of The Netherlands covering a population of 1 million inhabitants. All initial isolates from each of the 314 patients were subjected to IS6110 RFLP typing and spoligotyping using traditional and new spacer oligonucleotides. All follow-up isolates were tested by traditional 43-spacer oligotyping only.
The primary isolates of the 314 patients represented the following subspecies of the M. tuberculosis complex: M. tuberculosis (n = 303), M. bovis BCG (n = 4), M. bovis (n = 5), M. microti (n = 1), and M. canettii (n = 1). All isolates were cultured and identified as previously described (36). The cultures were stored for 1 to 8 years at −70°C until use.
M. bovis strains.
Sixty-eight M. bovis strains were isolated from cattle in Argentina. Furthermore, six clinical M. bovis BCG isolates and the following 10 vaccine strains were included: Glaxo, Tice, Connaught, USSR (Russian), Moreau (Brazil), P3 (The Netherlands), Armand Frappier (Canada), Tokyo (Japan), Vien (Vietnam), and Behringer (Sofia lot no. 26991, Moscow).
Nine isolates of M. bovis subsp. caprae comb. nov. were studied; six were kindly provided by Alicia Aranaz, Departamento de Patología Animal (Sandidad Animal), Facultad de Veterinaria, Universidad Complutense de Madrid, Madrid, Spain, and three were a gift of Stefan Niemann, Forschungszentrum Borstel, National Reference Center for Mycobacteria, Borstel, Germany.
M. tuberculosis Beijing genotype strains.
Seventy-eight Beijing genotype strains were selected on the basis of diversity in their IS6110 RFLP patterns. These strains represented in total 138 Beijing genotype strains isolated in The Netherlands between 1993 and 1999. Strains were designated as belonging to the Beijing genotype on the basis of their characteristic Beijing (spacers 35 to 43 present) or Beijing-like (at least four or more of the spacers 35 to 43 present) spoligotype patterns, as based on conventional spoligotyping results. In addition, these strains showed the specific insertion of IS6110 in the dnaA-dnaN region described by Kurepina et al. (23).
DNA extraction from bacterial cells.
DNA extracts for spoligotyping were prepared by suspending approximately 10 mg of wet bacterial cells in 100 μl of sterile distilled water (Sigma, Taufkirchen, Germany) and subsequently heating at 100°C for 30 min to inactivate and lyse the cells (36). Cell debris was removed by centrifugation at 13,000 × g for 2 min. The lysates were stored at −20°C until use.
Purified chromosomal DNA extracts for IS6110 RFLP analysis were prepared according to the standardized method (38).
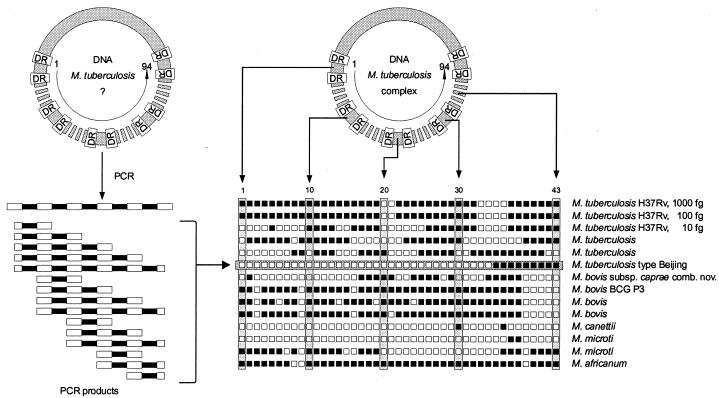
Traditional spoligotyping.
Traditional 43-spacer oligotyping was performed according to the previously described method (36). The 43-spacer oligonucleotides were derived from the DR region of M. tuberculosis reference strain H37Rv and the BCG vaccine strain P3 (20) (Fig. 1). Briefly, the CRISPR region was amplified by PCR using primers derived from the DR sequence. Ten microliters of the lysates obtained from the cultured M. tuberculosis strains was added to 50 μl of PCR mixture. The mixture was overlaid with 1 drop of mineral oil (Sigma, Steinheim, Germany) and incubated for 60 min at 37°C with uracil DNA glycosylase, 3 min at 95°C for uracil DNA glycosylase inactivation (25) and DNA denaturation, 1 min for primer annealing at 57°C, and 1 min for primer extension at 72°C. The following cycles were repeated 25 times: 1 min at 95°C, 1 min at 57°C, and 30 s at 72°C. PCR products were kept at −20°C until further analysis. PCR products were analyzed by hybridization using the reverse line blotting technique (21). After hybridization and washing of the membrane, hybridized DNA was detected with peroxidase-labeled streptavidin as described previously (36).
FIG. 1.
The DNA sequence of the CRISPR region, a synonym of the DR region, has been determined for the reference species M. tuberculosis H37Rv and M. bovis BCG P3. Ninety-four variable spacers (spacers 1 to 94) have been identified, pieces of DNA of the variable spacers have been synthesized, and 43 of them have been introduced vertically onto the membrane used for traditional spoligotyping. The conserved DRs serve as targets for the primers and thus the basis for amplification. These products can be added to the membrane of a species or samples containing a species of an unknown genotype and belonging to the M. tuberculosis complex after PCR is carried out. After hybridization and detection, the spoligotype pattern can be read from the horizontal axis of the various clinical isolates belonging to the M. tuberculosis complex, such as M. tuberculosis, M. bovis BCG, M. bovis, M. canettii, M. microti, M. africanum, and M. bovis subsp. caprae comb. nov.
Redesigned first-generation spacer oligonucleotides and new spacer oligonucleotides.
In order to optimize the spoligotyping, the usefulness of in total 104-spacer oligonucleotides (detecting 51 alternative spacers) was tested (Table 1). These oligonucleotides with aminolink were designed using Oligo software (version 4.0; MedProbe, Oslo, Norway) and were covalently bound to a membrane as previously described by Kaufhold et al. (21).
TABLE 1.
List of spacer oligonucleotide sequences of M. tuberculosis complex
| Membrane spacer no. | Genome spacer no. | Previous designationa | Oligonucleotide concnb | Oligonucleotide sequencec |
|---|---|---|---|---|
| 1 | 2 | 1 | 300 | CGCTCCCCTAGTCGT |
| 2 | 3 | 2 | 150 | TGGGCGACAGCTTTTGA |
| 3 | 4 | 3 | 25 | CTTCCAGTGATCGCCTT |
| 4 | 12 | 4 | 30 | TCATACGCCGACCAATC |
| 5 | 13 | 5 | 1000 | TTCTGACCACTTGTGCG |
| 6 | 14 | 6 | 150 | TCATTTCCGGCTT |
| 7 | 15 | 7 | 5 | TGAGGAGAGCGAGTACT |
| 8 | 18 | 8 | 600 | TGAAACCGCCCCCAG |
| 9 | 19 | 9 | 10 | ACTCGGAATCCCATGTG |
| 10 | 20 | 10 | 50 | CTCTAGTTGACTTCCGG |
| 11 | 21 | 11 | 30 | CAGGTGAGCAACGGC |
| 12 | 22 | 12 | 10 | ATGGGATATCTGCTGCC |
| 13 | 23 | 13 | 25 | ATTGCCATTCCCTCTCC |
| 14 | 24 | 14 | 300 | TTTCGGTGTGATGCGGA |
| 15 | 25 | 15 | 200 | TGAATAACGCGCAGTGAAT |
| 16 | 26 | 16 | 200 | TCGCACGAGTTCCCG |
| 17 | 27 | 17 | 30 | CCGGCAACAATCGCG |
| 18 | 28 | 18 | 1500 | TGCAGATGGTCCGGG |
| 19 | 29 | 19 | 10 | TTGCGCTAACTGGCTTG |
| 20 | 30 | 20 | 250 | ATTTCCTTGACCTCGCC |
| 21 | 31 | 21 | 50 | CGATGTCGATGTCCCAA |
| 22 | 32 | 22 | 5 | ACGGCACGATTGAGACA |
| 23 | 33 | 23 | 10 | GTCCAGCTCGTCCGT |
| 24 | 34 | 24 | 60 | GCCTGCTGGGTGAGA |
| 25 | 35 | 25 | 400 | GGAGCCGATCAGCGA |
| 26 | 36 | 26 | 80 | CTTCAGCACCACCATCA |
| 27 | 37 | 27 | 25 | TTCGTGATCTCTTCCCG |
| 28 | 38 | 28 | 5 | GATCACAACACCAACTAATG |
| 29 | 39 | 29 | 10 | GAAATACAGGCTCCACG |
| 30 | 40 | 30 | 5 | TCTTGACGATGCGGTTG |
| 31 | 41 | 31 | 5 | TTCGCGTCAGACAGGTT |
| 32 | 42 | 32 | 50 | ACTCCCGACCAAATAGG |
| 33 | 43 | 33 | 250 | TCGACACCGACATGAC |
| 34 | 44 | 34 | 100 | GAAGTCACCTCGCCC |
| 35 | 46 | 35 | 10 | AGTCCGTACGCTCGAAA |
| 36 | 47 | 36 | 50 | CGAAATCCAGCACCACA |
| 37 | 51 | 37 | 50 | TTTGAGCGCGAACTCGT |
| 38 | 52 | 38 | 75 | TGGATGGCGGATGCG |
| 39 | 53 | 39 | 10 | AAATCGGCGTGGGTAAC |
| 40 | 62 | 40 | 25 | TCATACAGGTCCAGTGC |
| 41 | 63 | 41 | 25 | GCTTTCCGGCTTCTATC |
| 42 | 64 | 42 | 5 | GACATGGACGAGCGC |
| 43 | 65 | 43 | 50 | CAGAATCGCACCGGG |
| 44 | 1 | 500 | CAACCCGGAATTCTTGC | |
| 45 | 5 | 600 | CAGGCGTGGCTAGG | |
| 46 | 6 | 500 | GTCGCCGTAAGTGCC | |
| 47 | 7 | 20 | GTTGACCACGAATTTTCAG | |
| 48 | 8 | 20 | GCTGGCGCGCATCAT | |
| 49 | 9 | 50 | CCATATCGGGGACGG | |
| 50 | 10 | 10 | GCGTCGTGCCATCAG | |
| 51 | 11 | 200 | CCGTGCACATGCCGT | |
| 52 | 16 | 25 | ACGTTAGGGCATGCAG | |
| 53 | 17 | 150 | TCTTGAGCAACGCCATC | |
| 54 | 45 | 5 | AAGTTGGCGCTGGGG | |
| 55 | 48 | 1200 | AACCGTCCCACCTG | |
| 56 | 49 | 5 | AACACTTTTTTTGAGCGTGG | |
| 57 | 50 | 10 | CGGAAACGCAGCACC | |
| 58 | 54 | 5 | CGATCATGAGAGTTGCG | |
| 59 | 55 | 5 | TTTTCGCTGTTGTGGTTCT | |
| 60 | 56e | 10 | AGCACCTCCCTTGACAA | |
| 61 | 57 | 25 | TGCTGACTTCGCCTGTA | |
| 62 | 58 | 75 | CGAGCAGCGGCATAC | |
| 63 | 59 | 100 | GCATCCACTCGTCGC | |
| 64 | 60 | 75 | TGGTAATTGCGTCACGG | |
| 65 | 61 | 100 | ACCATCCGACGCAGG | |
| 66 | 66 | 700 | CCACGCTACTGCTCC | |
| 67 | 67 | 20 | CACCGCCGATGACAG | |
| 68 | 68 | 300 | GTGTTTCGGCCGTGC | |
| 69 | 69 | 5 | GTTGCATTCGTCGACTG | |
| 70 | 70 | 400 | GGCGGCGCCGAGAA | |
| 71 | 71 | 10 | TTCCATGACTTGACGCC | |
| 72 | 72 | 100 | CGATGCGGCCACTAG | |
| 73 | 73 | 5 | GCTGACCCCATGGATG | |
| 74 | 74 | 10 | CAACAAGGTCTACGCGT | |
| 75 | 75 | 100 | GATCAGGCGAAGGCG | |
| 76 | 76 | 10 | ATTGCAGCGACGGGC | |
| 77 | 77 | 5 | CAACGACGCTGTATTGG | |
| 78 | 78 | 5 | AGCAGCATGGACGGTTT | |
| 79 | 79 | 5 | GCGGATGTGGTGGTC | |
| 80 | 80 | 250 | GTACATAGCGAGCTG | |
| 81 | 81 | 5 | GCCGCGGGTTTCGTT | |
| 82 | 82 | 10 | GGGGCGTGTGTTCGT | |
| 83 | 83 | 5 | CTGGTGTGCTTATGCCT | |
| 84 | 84 | 5 | CAAATGTTTGGACTGTGATC | |
| 85 | 85 | 10 | TTGTCGCGCGCCTTTTT | |
| 86 | 86 | 10 | GTTTCAGTTTTCTTGTCCC | |
| 87 | 87 | 25 | CTGGTTGTTGCCCGG | |
| 88 | 88 | 5 | TGTTCGGTGTTCTCCTG | |
| 89 | 89 | 250 | TCATGACGAGCCCGCA | |
| 90 | 90 | 10 | ACACGGCCTGATCGGT | |
| 91 | 91 | 20 | CGGATTGTCTGGCCC | |
| 92 | 92 | 20 | TAAGCACGCGTCTGTCA | |
| 93 | 93 | 5 | GACCACCGAATCACCAT | |
| 94 | 94 | 5 | TCTGGTAGTGGGCTTCT | |
| 95 | 11 | 1080 | 200 | ACATGCCGTGGCTCA |
| 96d | 11 | 1080 | 350 | TGAGCCACGGCATGT |
| 97 | 16 | 1377 | 100 | CACGACGTTAGGGCA |
| 98d | 16 | 1377 | 150 | ATGCCCTAACGTCGTG |
| 99 | 5 | 790 | 50 | CGGCAGGCGTGGCTA |
| 100d | 5 | 790 | 50 | TAGCCACGCCTGCCG |
| 101d | 6 | 863 | 1000 | CACTTACGGCGACGG |
| 102 | 6 | 863 | 25 | CCGTCGCCGTAAGTG |
| 103 | 17 | 1453 | 20 | GAGCAACGCCATCAT |
| 104d | 17 | 1453 | 40 | GATGATGGCGTTGCT |
Data from reference 20.
Optimized oligonucleotide concentration on the membrane. Values are in picomoles.
Sequences used for 5′ amino acid-linked oligonucleotides in spoligotyping. Except where otherwise noted, sequences given are in the 5′ to 3′ direction. For genome spacer numbers 1 to 94, the sequences given were published previously by van Embden et al. (39). For genome spacer numbers 5, 6, 11, 16, and 17, the sequences given were also published previously by Caimi et al. (5).
Sequence given is in the 3′ to 5′ direction.
Oligonucleotide is polymorphic at position 14 (39).
The first 43 oligonucleotides in the set of 104 were complementary to the same spacers used for first-generation spoligotyping, but the DNA sequences of some of the oligonucleotides were redesigned to obtain a more specific hybridization signal and to correct for errors present in the original DNA sequence data on spacer oligonucleotides one and two (39). These 43-spacer oligonucleotides were numbered according to their location on the membrane designed by Kamerbeek et al. (20) and therefore differ from their subsequent positions in the DR region of the genome of M. tuberculosis strain H37Rv and M. bovis BCG strain P3 (39).
The 51 new spacer oligonucleotide sequences, representing the oligonucleotides 44 to 95 on the evaluation blots, were derived from the study of van Embden et al. (39). The 10 oligonucleotides 95 to 104, of which five oligonucleotides (95, 97, 99, 101, and 103) had cDNA sequences to the oligonucleotides 96, 98, 100, 102, and 104, respectively, were described in the studies of both van Embden et al. and Caimi et al. (5, 39). One of the oligonucleotides of each pair was also included as a duplicate on the membrane at positions 51, 52, 45, 46, and 53, respectively (Table 1), to act as internal controls of reproducibility in each hybridization.
In order to obtain one hybridization temperature for all probes on the membrane an equal melting temperature (Tm) for all probes was calculated; a Tm (mean ± standard deviation) of 55.4°C ± 1.5°C was achieved for almost all new oligonucleotides except for oligonucleotides 80, 101, 102, 103, and 104, which had a Tm of 50.4°C; oligonucleotide 55, which had a Tm of 51.6°C; oligonucleotides 95 and 96, which had a Tm of 53.1°C; oligonucleotide 70, which had a Tm of 57.5°C; and oligonucleotides 99 and 100, which had a Tm of 58.6°C. The concentrations of all oligonucleotides on the membrane were adjusted (Table 1) to those on the traditional 43-spacer oligotyping membrane (20).
The hybridization conditions for spoligotyping with the 104-spacer oligonucleotides were modified slightly compared to the original protocol, since hybridization was performed for 45 min instead of 60 min at 57°C because a longer hybridization time is not required.
IS6110 RFLP analysis.
DNA fingerprinting was performed as described by van Embden et al. (38). Briefly, purified mycobacterial DNA from a culture was digested with PvuII, separated on an agarose gel, transferred by Southern blotting, and hybridized to a peroxidase-labeled 245-bp probe. The IS6110 RFLP DNA-containing PvuII restriction fragments were visualized using the ECL detection system (Amersham International, Amersham, Buckinghamshire, United Kingdom). Cases were referred to as belonging to clusters if the IS6110 DNA fingerprints of the isolated strains were identical.
Computer-assisted analysis.
Analysis of the banding patterns obtained with IS6110 RFLP was performed with the GelCompar software (version 4.0; Applied Maths, Kortrijk, Belgium), as previously described (18). Spoligotype patterns were entered in a word processor (Microsoft Word) file. BioNumerics software (version 4.0; Applied Maths) was used to compare IS6110 fingerprints and spoligotype patterns. A dedicated BASIC program (Statistical Analysis System Institute Inc., Cary, N.C.) was used to perform redundancy selection on the basis of the discriminatory power of the individual spacer oligonucleotides.
Analysis of discriminatory power and reproducibility.
To be able to compare the discriminatory power and reproducibility of the 104-spacer oligotyping to those of other methods used to type the M. tuberculosis complex, the set of 131 DNA samples used in the comparative study of Kremer et al. was tested (22). The numbers of the DNA samples were randomized by computer and renumbered with randomly generated numbers and analyzed on the basis of blinded codes. The discriminatory power and reproducibility were determined by methods similar to those described in the original paper (22).
RESULTS
Strain differentiation by traditional and novel probe spoligotyping.
In order to evaluate the usefulness of new spacer oligonucleotides and also to test the 43 redesigned oligonucleotides of the first-generation spoligotyping membrane, 314 isolates were subjected to traditional spoligotyping on the basis of the 43 first-generation oligonucleotides, the 104 new oligonucleotides, and IS6110 RFLP typing.
Traditional 43-spacer oligotyping of the 314 isolates resulted in 45 clusters with 196 isolates; 115 spoligotype patterns were found only once, and there were in total 160 different spoligotype patterns. The cluster sizes obtained by traditional spoligotyping varied from 2 to 31 isolates (Fig. 2, A1 to A45).
FIG. 2.
Correlation between spoligotype patterns obtained with traditional spoligotyping (43-spacer oligonucleotides) and novel probe spoligotyping (with 104-spacer oligonucleotides) and IS6110 RFLP analysis of M. tuberculosis complex isolates from 314 different patients. The spacer oligonucleotides for traditional spoligotyping were ordered from 1 to 43 as previously designated, and the new spacer sequences were numbered according to their location in the genome (Table 1). The fingerprints of clusters were determined on the basis of agreement between traditional and novel probe spoligotyping and IS6110 RFLP. Only one DNA fingerprint is depicted for each cluster. **, unique DNA fingerprints. (A) Clusters of strains determined by traditional spoligotyping. (B) Clusters of strains determined by novel probe spoligotyping. The numbers in parentheses indicate the number of strains with that specific spoligotype pattern. *, hybridization signals of the oligonucleotides 71 to 95 were negative for all clusters. (C) The clusters of IS6110 RFLP patterns were numbered from 1 to 23. **, unique DNA fingerprints. The last column (Total) indicates the strains belonging to a cluster and the strains with a unique IS6110 RFLP pattern.
The 104-spacer oligotyping novel probe spoligotyping of the same isolates detected 44 clusters (Fig. 2, B1 to B44) with 184 isolates, and 130 spoligotype patterns were found only once out of a total of 174 different spoligotype patterns. Thus, 14 additional spoligotype patterns were found. The B cluster sizes ranged from 2 to 29 isolates (Fig. 2). Novel probe spoligotyping with 104-spacer oligonucleotides identified one cluster (cluster B44) which was not identified by traditional spoligotyping. This was due to the absence of any hybridization signal from (redesigned) spacer oligonucleotide 18 in the novel probe spoligotyping procedure, whereas this spacer oligonucleotide gave a weak signal in the traditional spoligotyping procedure.
Also, after the use of additional spacer oligonucleotides, some of the spoligotype patterns predominated over others, e.g., type B42, which was found 29 times; type B39, which was found 12 times; type B17, which was found 11 times; and type B2, which was found 11 times. The spoligotypes of B42 and B39 are the most commonly found patterns worldwide today and are referred to as spoligotypes 53 and 50, respectively, by Sola et al. (34).
The clusters A6, A22, and A28, identified by traditional spoligotyping and each comprising two strains, could now (as with IS6110 RFLP) also be differentiated into unique strains by means of the novel probe spoligotyping (Fig. 2). Cluster A8, containing five strains, was subdivided into one cluster (B7) comprising two strains (IS6110 RFLP cluster 4) and three strains with unique novel probe spoligotype patterns (and unique IS6110 RFLP patterns). Cluster A37, consisting of five strains, was subdivided into one cluster (B34) comprising four strains (three strains in cluster 20 and one unique pattern in IS6110 RFLP), and one strain with a unique novel probe spoligotype pattern (and a unique IS6110 RFLP pattern). Cluster A41, containing 15 isolates, was divided into three different patterns: cluster B38, comprising two strains (with unique IS6110 RFLP patterns); cluster B39, comprising 12 strains (10 unique patterns and two strains in IS6110 RFLP cluster 16), and strains with a unique novel probe spoligotype pattern (and a unique IS6110 RFLP pattern). Cluster A42, comprising seven strains, was subdivided into one cluster (B40) comprising six strains (with unique IS6110 RFLP patterns) and one strain with a unique novel probe spoligotype pattern (and a unique IS6110 RFLP pattern). Finally, cluster A44, comprising 31 strains, was split into cluster B42 with 29 strains (26 strains with a unique IS6110 pattern, 2 strains in cluster 11, and 1 strain in cluster 20 in IS6110 RFLP) and two unique novel probe spoligotype patterns (and unique IS6110 RFLP patterns) (Fig. 2).
Differentiation of M. bovis strains.
Within the study population, nine M. bovis strains, including four M. bovis BCG isolates, were investigated. M. bovis usually harbors only one or a few copies of IS6110, and therefore such strains often cannot be differentiated by IS6110 RFLP (6). Two of the most common spoligotypes among M. bovis isolates are the spoligotypes of clusters A20 and A24 (2, 31). IS6110 RFLP typing of the nine M. bovis strains yielded the characteristic one-band fingerprint pattern, two types (cluster A20 and A24) of the traditional spoligotyping, and four types of the novel probe spoligotyping.
The traditional spoligotype cluster A20, containing four M. bovis isolates, was divided into a cluster (B19) of three strains with identical patterns and one strain with a unique pattern (Fig. 2). A single M. bovis isolate of cluster A24 showed a unique pattern with the novel probe spoligotyping technique. The four remaining M. bovis BCG strains of cluster A24, harboring the characteristic IS1081-containing PvuII fragment of 8.0 kb (31, 40), all represented the same cluster B22 obtained with novel probe spoligotyping (Fig. 2).
To further investigate this polymorphism among M. bovis BCG strains, we subjected in addition 10 BCG vaccine strains and six M. bovis BCG clinical isolates to the novel probe spoligotyping. All strains exhibited the same spoligotype pattern.
Furthermore, an additional set of 68 M. bovis strains isolated from cattle in Argentina was subsequently studied. These strains yielded four clusters of identical patterns by traditional spoligotyping with 39, 8, 5, and 3 isolates, respectively, and clusters of 36, 4, 3, 3, and 2 identical patterns by the novel probe spoligotyping technique. Using the traditional spoligotyping, 18 different patterns were identified, and with the novel probe spoligotyping 26 different patterns were obtained. Thirteen M. bovis strains yielded separate genotypes by traditional and novel probe spoligotyping.
Finally, nine M. bovis subsp. caprae comb. nov. strains yielded six different patterns by traditional spoligotyping and eight different genotypes by novel probe spoligotyping.
Extended differentiation of Beijing genotype strains.
The Beijing genotype of M. tuberculosis represents a group of strains that are closely related genetically. Because the majority of these strains originated from the province of Beijing, China, this group of strains was designated the Beijing family (42). It was shown that more than 85% of the M. tuberculosis isolates found in the Beijing area exhibited more than 66% agreement between the multibanded IS6110 RFLP patterns and identical spoligotype patterns in traditional spoligotyping (42). A Beijing genotype-specific spoligotype pattern (cluster A2, consisting of the absence of the first 34 spacer oligonucleotides and the presence of the remaining 9 [35 to 43] spacer oligonucleotides [Fig. 2]) was found 12 times among the 314 M. tuberculosis complex isolates tested in this study. Furthermore, two isolates exhibited similar hybridization, lacking a reaction with spacer oligonucleotide 37 (cluster A1 [Fig. 2]) in comparison with the Beijing-specific pattern.
Spoligotyping with the new spacer oligonucleotides revealed that 13 of the 14 Beijing or Beijing-like strains showed hybridization with spacer oligonucleotides 55 to 57 and 66 to 68. One of the 12 isolates in cluster A2 lacked hybridization of spacer oligonucleotide 57 in the extended panel of spacer oligonucleotides (Fig. 2). In addition, the set of DNAs of the comparative study of Kremer et al. (22) contained one strain with a Beijing-like pattern (lacking spacer oligonucleotides 38 and 39) and eight strains with the pattern of cluster A2 obtained by traditional spoligotyping, of which one lacked hybridization of spacer oligonucleotide 56.
To investigate this polymorphism further, 78 M. tuberculosis Beijing genotype strains with polymorphic IS6110 RFLP patterns were subjected to novel probe spoligotyping. In traditional spoligotyping 63 of these exhibited a Beijing characteristic spoligotype pattern and 15 a Beijing-like spoligotype pattern. Among the Beijing-like isolates, eight different types could be distinguished with traditional spoligotyping, including three clusters of five, three, and two isolates and five unique patterns. Novel probe spoligotyping distinguished the same number of types among these Beijing-like strains and showed that, in addition to hybridization to at least four of the spacer oligonucleotides 35 to 43, the majority of these strains showed hybridization with spacer oligonucleotides 55 to 57 and 66 to 68. One strain, lacking hybridization to spacers 37 and 38, also lacked hybridization to spacer 57, and one strain showed hybridization to only spacers 38, 39, 42, 43, 66, and 67. The isolates in the cluster of five showed hybridization to spacer oligonucleotides 37 to 43, 57, and 66 to 68. Fifty-four of the 63 strains containing spacers 35 to 43 showed hybridization to spacers 55 to 57 and 66 to 68. The nine others lacked, in comparison, spacer 56 (data not shown).
Thus, in comparison with traditional spoligotyping, in total, two additional spoligotype types were identified among the 101 Beijing genotype strains tested with the 104 new spacer oligonucleotides.
Reproducibility of spoligotyping.
In order to obtain information on the reproducibility and the stability of the first generation of spoligotyping results, serial isolates of 314 patients were tested. From the 314 patients, 68 duplicate samples, 58 triplicate samples, and once even 15 samples were available, resulting in a total number of 850 strains.
With the 104-spacer oligotyping all but four of the 314 serial isolates gave concordant results by looking for differences using 10 probes of five of the same spacer sequences and five oligonucleotides complementary to those oligonucleotides. Hybridization signals of the complementary oligonucleotides were obtained, probably due to rehybridization of the PCR products with the biotin-labeled opposite DNA strand probe. After repeating the novel probe spoligotyping technique, three of the four isolates yielded concordant results. For one isolate three-spacer hybridization signals were incorrectly identified.
In addition, the reproducibility of 104-spacer oligotyping was investigated by testing the coded DNAs of an interlaboratory study of Kremer et al. (22). Thirty-one duplicate DNA samples were included in the set of in total 131 blinded DNAs. Twenty-eight (90%) of these were identified correctly in the first test. The three sets of duplicate DNA samples that did not result in identical patterns had patterns that were highly similar, differing in one, two, and three spacer oligonucleotides, respectively. Evaluation of these differences revealed that in fact two of the patterns had not been properly read and one pattern contained three spacer oligonucleotides more than the original spoligotype pattern. In the previous study (22), in which the reproducibility of 43-spacer oligotyping was determined with the same set of DNAs, a similar observation was made for one of the two duplicate DNA sets that were not in concurrence.
The 104-spacer oligotyping missed one duplicate DNA sample more than 43-spacer oligotyping, which is not significant considering the number of duplicates investigated. Therefore, both the traditional and the novel probe spoligotyping may be considered reproducible methods, compared to other methods (22).
Discriminatory power of IS6110 RFLP and novel probe spoligotyping.
Among the 314 IS6110 RFLP patterns generated, 264 different patterns were visualized, of which 241 were found only once. The remaining 73 strains were shared by the isolates of two or more patients (clusters). The clustered isolates showed 23 different IS6110 RFLP patterns (Fig. 2). The three largest clusters comprised isolates from 11, 8, and 7 patients, respectively. The majority of the remaining clusters consisted of two cases (Fig. 2).
In comparison with IS6110 RFLP typing (264 types), spoligotyping using the new 104-spacer oligonucleotides yielded 173 types (Fig. 2). When strains belonging to the clusters B2, B17, B21, B39, B40, and B42 were excluded from the determination of the discriminatory power, traditional spoligotyping, novel probe spoligotyping, and IS6110 RFLP yielded 154, 168, and 191 different patterns, respectively. In agreement with previous observations (7), none of the IS6110 RFLP clusters, consisting of patterns with five or more bands, were subdivided by traditional spoligotyping. However, four IS6110 RFLP clusters (C20, C21, C22, and C23) with patterns containing one or two bands were further differentiated by spoligotyping with 104-spacer oligonucleotides.
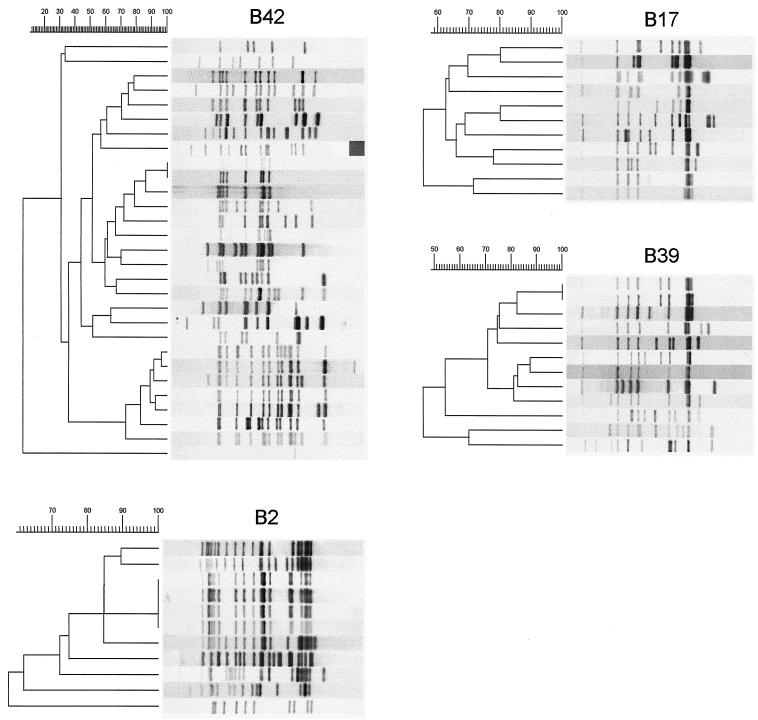
The isolates of the four major spoligotype clusters exhibited polymorphic IS6110 RFLP patterns and were subdivided into clusters of 29 (cluster B42), 12 (cluster B39), 11 (cluster B17), and 11 (cluster B2) isolates, respectively (Fig. 3).
FIG. 3.
IS6110 RFLP patterns of the isolates of the four largest novel probe spoligotyping clusters (B42, B39, B17, and B2), which contain 29, 12, 11, and 11 isolates, respectively. The relationship of the IS6110 RFLP patterns is depicted by the dendrograms to the left of the IS6110 RFLP patterns. Note that the agreement between the IS6110 RFLP patterns of the strains belonging to the largest spoligotype cluster (B42) is only 11%.
The IS6110 RFLP patterns of four isolates in cluster B2 were identical, and the overall agreement of the patterns was 61%. The agreement between the IS6110 RFLP patterns of the isolates of the spoligotype clusters B39 and B17 was 50 and 59%, respectively. The isolates of the largest novel probe spoligotype cluster B42 were the most diverse; the respective IS6110 RFLP patterns shared only 11% similarity.
Hybridization with redesigned oligonucleotides.
Sixty-one different spoligotypes were obtained with both the novel probe and traditional spoligotyping method by testing the set of 90 M. tuberculosis complex strains of the study of Kremer et al. (22). Although the number of types was the same, there were a few differences in the matches of strains, usually due to differential hybridizations of redesigned spacer oligonucleotides.
Twelve strains exhibited different hybridization patterns with the first 43-spacer oligonucleotides when traditional and novel probe spoligotyping were compared; however, these patterns were always highly similar. The hybridization patterns of seven strains differed only in a single spacer oligonucleotide, and five strains differed in two spacer oligonucleotides. Four times spacer oligonucleotide 18 was absent from the novel probe spoligotyping pattern, while it was present in the traditional spoligotype pattern, although the hybridization signal of this spacer probe was weaker. A similar observation was made for spacer oligonucleotides 30 and 36. In four cases spacer oligonucleotide 3 and in one case spacer oligonucleotide 20 was missing from a novel probe spoligotyping pattern, while clearly present in the traditional spoligotype pattern. In three cases spacer oligonucleotide hybridization was positive in the novel probe pattern and absent from the traditional pattern (spacer oligonucleotides 25, 29, and 30). Spacer oligonucleotides 38 and 39 were found positive in the novel probe spoligotyping pattern and were weakly positive according to traditional spoligotyping, but scored negative.
Also the hybridization of the 43 redesigned oligonucleotides of the first generation and the results of traditional spoligotyping were compared by testing the initial isolates of the 314 patients. The newly designed oligonucleotides of the spacers 2, 9, 17, 18, 25, 26, 32, 33, and 36 sometimes gave a strong, or weak, hybridization signal in contrast to the weak, or almost negative, hybridization signal obtained with the traditional 43-spacer oligonucleotides (data not shown). Discordance was tested again and confirmed with other isolates from the same patient. This may be due to the fact that the new spacer oligonucleotides are 5 to 12 nucleotides shorter (Table 1), resulting in a lower Tm of approximately 15°C for the oligonucleotides, enabling more-stringent hybridization conditions and leading to increased specificity.
Discriminatory power of the individual new spacer oligonucleotides.
In order to identify redundant spacer oligonucleotides among the 94-spacer (43 traditional and 51 novel probes) oligonucleotides as far as the discrimination of the 173 patterns is concerned (Fig. 4), we sequentially omitted individual spacer oligonucleotides and checked whether the remaining subset of spacer oligonucleotides would still discriminate the 173 patterns.
FIG.4.
Representative spoligotype patterns with 104 and 40 new spacer oligonucleotides of 173 M. tuberculosis strains. (A) Presence or absence of any of the 104 spacers; the spacer oligonucleotides were ordered as in Table 1. (B) Reduced hybridization patterns of 40 selected spacer oligonucleotides that provide the same level of discrimination as the 104-spacer oligonucleotides. Columns 1 to 40 contain spacer oligonucleotides in the same order as on the membrane: 1, 2, 3, 4, 5, 8, 10, 13, 14, 17, 18, 19, 20, 22, 23, 24, 25, 27, 28, 30, 31, 32, 37, 38, 39, 40, 43, 44, 50, 54, 56, 57, 60, 66, 67, 68, 69, 45, 46, and 53. Lane 1 contains an isolate that lacks the CRISPR region; lane 2 contains an M. canettii isolate; lane 3 contains an M. microti isolate; lanes 6 and 7 contain isolates of the Beijing genotype; lanes 36, 37, and 38 contain M. bovis isolates; lane 39 contains an M. bovis BCG isolate; and the remaining lanes contain M. tuberculosis strains.
In total, 54 of the 94 individual new spacer oligonucleotides together with the 10 duplicate spacer oligonucleotides could be removed from the panel without any loss of discriminatory power for the strains used in this study. Thus, 40 oligonucleotides contained all the discriminatory power of the 104 oligonucleotides in this study. The oligonucleotides of the spacers in genome order, listed in Fig. 2B from 1 to 40 were 2, 3, 4, 12, 13, 18, 20, 23, 24, 27, 28, 29, 30, 32, 33, 34, 35, 37, 38, 40, 41, 42, 51, 52, 53, 62, 65, 1, 10, 45, 49, 50, 56, 66, 67, 68, 69, 5, 6, and 17.
DISCUSSION
After the introduction of 51 new spacer oligonucleotides onto the spoligotyping membrane, the discriminatory power of this typing technique improved slightly from 160 types among 314 isolates to 173 types. The slight improvement achieved by 51 new spacer oligonucleotides cannot be attributed to the 10 duplicate probes and the 26 probes from M. canettii. The improvement in the discriminatory power was due to 25 novel probes. Spoligotyping has a significant advantage when used in areas populated with isolates with five or fewer IS6110 copies. Isolates of M. tuberculosis that possess few copies of IS6110 do not generate sufficient polymorphism to be readily distinguished by IS6110 RFLP. The spoligotyping results did not have the discriminatory power of IS6110 RFLP typing (264 types). However, when the 76 strains of the six major clusters obtained with the novel probe spoligotyping procedure—also containing the globally predominant spoligotypes—were left out, the spoligotyping results approached the discriminatory power of RFLP typing. If tested strains showed different spoligotype patterns, then the strains were almost invariably from different sources. If the spoligotype patterns were identical and did not belong to the clusters B42, B40, B39, B21, B17, and B2, the strains were usually from a common source, as shown by IS6110 RFLP typing. When spoligotype patterns belong to these clusters, then no conclusions can be drawn about the epidemiological relationship between the respective patients. In these cases, the results of IS6110 RFLP typing, which can only be performed after culturing, should be awaited in order to perform molecular epidemiological investigations.
This makes spoligotyping useful as a screening method for detection and to obtain direct epidemiological information on the identity of the M. tuberculosis complex strain. Also the evaluation of redesigned oligonucleotides of the first generation used for spoligotyping did not add much (one cluster) to the improvement of the discriminatory power of this assay. The new design of the 43 oligonucleotides used for traditional spoligotyping yielded some differences in the hybridization signals. Especially oligonucleotides 2 and 18 had weak hybridization signals with traditional spoligotyping. In the novel probe spoligotyping technique, oligonucleotide 2 was redesigned to obtain the correct DNA sequence, and the redesigned oligonucleotide 18 gave more specific hybridization conditions.
The novel probe spoligotyping showed three different patterns for the classical Beijing genotype strains that only differed in the presence of one spacer in the CRISPR region. In the study of Kremer et al. Beijing strains with different IS6110 RFLP patterns were shown to exhibit little or no polymorphism, by different other genetic markers (22).
This study confirms the previously described existence of distinct genotypic families among M. tuberculosis isolates, which can be recognized by spoligotyping and IS6110 RFLP typing. This grouping may be associated partly with the geographic origin of the patients (33). Since about 60% of the tuberculosis patients enrolled in this study originated from countries other than The Netherlands, the set of oligonucleotides selected for this study may not be the most appropriate as far as the discriminatory power of spoligotyping in other regions is concerned (7, 32, 34). Therefore, the usefulness of the second-generation spoligotyping membrane should be evaluated for other geographic areas individually.
M. bovis strains were discriminated more accurately by the novel probe spoligotyping than by traditional spoligotyping. On the other hand, IS6110 RFLP gave more discrimination between the M. bovis BCG strains. Molecular typing methods have contributed to the identification of rarely encountered M. tuberculosis complex bacteria that previously were difficult to distinguish with biochemical procedures: M. africanum, M. bovis, M. bovis BCG, M. microti, M. canettii, and M. bovis subsp. caprae comb. nov. (1, 28, 41, 43). This study of 314 primarily clinical isolates provided more information about the identification of the subspecies and provides improved strain differentiation within the subspecies of M. bovis, M. bovis BCG, M. microti, and M. canettii. The M. microti strain reacted with only two spacer oligonucleotides of the traditional spoligotyping membrane and seven additional spacer oligonucleotides of the novel probe spoligotyping membrane. M. canettii also reacted with only two spacer oligonucleotides of traditional spoligotyping and 26 additional spacer oligonucleotides used in the novel probe spoligotyping. Spacer oligonucleotides 3, 9, 16, and 39 to 43 of the traditional spoligotyping as well as the new spacer oligonucleotides are not found in M. bovis BCG and M. bovis (Fig. 3). This lack of spacers in the DR region of M. bovis was observed previously (5). We also detected M. tuberculosis strains by traditional spoligotyping with no spacer oligonucleotides between 39 and 43, which is inconsistent with a previous study (26), and no spacer oligonucleotide 34, which is inconsistent with an earlier observation that M. bovis strains differ from all M. tuberculosis strains in the presence of this specific oligonucleotide (20). We compared the spoligotype patterns with those of Viana-Niero et al. and found three strains of M. africanum subgroup A1 and one strain of M. africanum subgroup A2 (44). Spacer oligonucleotides eight and nine were generally missing, and the absence of spacer oligonucleotide 39 seems to be a traditional spoligotyping signature of the M. africanum strains, as mentioned by Viana-Niero et al. (44), but this was not observed in group 5 of M. africanum by Niemann et al. (27).
In this study we confirmed the existence of M. tuberculosis strains that are genetically divergent, as measured by IS6110 RFLP analysis, but which exhibit identical spoligotype patterns. Strains with predominant spoligotypes, such as type 50 or 53, often have highly diverse IS6110 RFLP patterns. This indicates that the CRISPR region remained unchanged during a long period of time and that molecular change due to the mobility of the insertion element IS6110 occurs more frequently than rearrangements in the CRISPR region.
With traditional spoligotyping, we found strains with spoligotype 50 and different IS6110 RFLP patterns; one strain contained a single IS6110 element, which gave a different pattern with novel probe spoligotyping. On the contrary we also found strains with one or two IS6110 copies with different spoligotype patterns. In such strains the IS6110 element seems to be “frozen” at a fixed genomic position, whereas the CRISPR region shows considerable DNA polymorphism. This indicates that solid evolutionary concepts cannot be based solely on a small incomplete set of spacer oligonucleotides from a single locus of the genome. Therefore, further efforts to improve spoligotyping will be only marginal in contrast to fingerprinting methods, which are based on more genomic loci, such as MIRU and variable numbers of tandem repeats typing (35). The availability of the whole genome sequence of M. tuberculosis strains may offer the possibility of developing new molecular markers with slower and more rapid turnovers than that of IS6110 RFLP, to visualize relationships on the basis of genome sequences and, e.g., the whole CRISPR region. Ideally, these approaches could be visualized by techniques such as multiplex PCRs on a fluorescence-based DNA analyzer, DNA microarrays, or DNA chip technology (13, 35). By using several more additional genetic markers, it becomes possible to detect and genotype M. tuberculosis complex strains directly in clinical samples or after culturing, even if the strains do not have any of the known spacers, like one strain in this study, or lack any IS6110 DNA (33, 46).
In order to avoid confusion we propose to continue the use of the traditional spoligotyping method with the oligonucleotides described by Kamerbeek et al. This assay is commercially available and contains two additional oligonucleotides, oligonucleotides 1 and 2, without the sequence errors (20).
When the reproducibility of spots was studied, it appeared that discrepancies found in the spoligotype patterns of duplicate oligonucleotides were not differences in the images but rather the result of human error in reading the results as negative and entering them as positive into the computer or vice versa. Having a second reader is a way of reducing human error in reading spoligotyping results. To avoid errors in reading low-intensity hybridization signals, it probably is better to use purified DNA, undiluted and diluted 1:10, extracted from the isolates. By reading the images based on ECL directly into the computer without a personal interpretation of the reader or by using fluor-labeled primers and a fluor imager to read the images, a better reproducibility as found by MIRU typing may be achieved (22). For specialized laboratories requiring the optimum level of discrimination, or for studies on the phylogeny of Beijing genotype isolates, a separate membrane for 43 oligonucleotides can be used with 25 oligonucleotides of the new spacers and 2 to 18 oligonucleotides from the spacer sequences of M. canettii. The conditions of this second generation of spoligotyping remain the same except for the hybridization temperature and time currently used for the first generation of spoligotyping.
In conclusion, this study has shown that the reproducibility of spoligotyping could be improved by the introduction of new spacer oligonucleotides. In our laboratory spoligotyping enabled us to identify all species of the M. tuberculosis complex and provided improved strain differentiation within each subspecies. In addition spoligotyping allowed us to assess a sample exchange between two patients in our hospital. Spoligotyping was also used to establish the diagnosis of tuberculosis and to trace the transmission chain without culturing mycobacteria from samples from sources such as mummified human bodies, patients, and health care workers; to detect cross-contamination of specimens; and to assess the treatment of tuberculosis (8, 37; A. G. M. van der Zanden, R. H. Rammeloo, F. J. Mud, T. A. Manschot, and D. van Soolingen, Letter, Int. J. Tuberc. Lung Dis. 5:784-785, 2001). Screening of samples by spoligotyping may be a fast way to obtain a first clue to possible epidemiological links with other cases of tuberculosis.
Acknowledgments
We thank Arianne te Koppele-Vije, Ria Hoentjen, Harriët Ardesch-Hobert, Ineke In 't Veld, Hetty Harmsen, Els Arents, Yvette Hondijk, Petra de Haas, and Annelies Bunschoten for their excellent technical help and Peter van Es for his assistance in the design and production of the figures.
REFERENCES
- 1.Aranaz, A., E. Liebana, E. Gomez-Mampaso, J. C. Galan, D. Cousins, A. Ortega, J. Blazquez, F. Baquero, A. Mateos, G. Suarez, and L. Dominguez. 1999. Mycobacterium tuberculosis subsp. caprae subsp. nov.: a taxonomic study of a new member of the Mycobacterium tuberculosis complex isolated from goats in Spain. Int. J. Syst. Bacteriol. 49:1263-1273. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, J., A. B. Andersen, K. Kremer, and H. Miorner. 1999. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J. Clin. Microbiol. 37:2602-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedwell, J., S. K. Kairo, M. A. Behr, and J. A. Bygraves. 2001. Identification of substrains of BCG vaccine using multiplex PCR. Vaccine 19:2146-2151. [DOI] [PubMed] [Google Scholar]
- 4.Bifani, P. J., B. Mathema, Z. Liu, S. L. Moghazeh, B. Shopsin, B. Tempalski, J. Driscol, R. Frothingham, J. M. Musser, P. Alcabes, and B. N. Kreiswirth. 1999. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA 282:2321-2327. [DOI] [PubMed] [Google Scholar]
- 5.Caimi, K., M. I. Romano, A. Alito, M. Zumarraga, F. Bigi, and A. Cataldi. 2001. Sequence analysis of the direct repeat region in Mycobacterium bovis. J. Clin. Microbiol. 39:1067-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cousins, D., S. Williams, E. Liebana, A. Aranaz, A. Bunschoten, J. van Embden, and T. Ellis. 1998. Evaluation of four DNA typing techniques in epidemiological investigations of bovine tuberculosis. J. Clin. Microbiol. 36:168-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz, R., K. Kremer, P. E. de Haas, R. I. Gomez, A. Marrero, J. A. Valdivia, J. D. van Embden, and D. van Soolingen. 1998. Molecular epidemiology of tuberculosis in Cuba outside of Havana, July 1994-June 1995: utility of spoligotyping versus IS6110 restriction fragment length polymorphism. Int. J. Tuberc. Lung Dis. 2:743-750. [PubMed] [Google Scholar]
- 8.Doveren, R. F., S. T. Keizer, K. Kremer, and D. van Soolingen. 1998. Connection between 2 tuberculosis outbreaks demonstrated after 8 years by DNA-fingerprinting of the causative mycobacteria. Ned. Tijdschr. Geneeskd. 142:189-192. [PubMed] [Google Scholar]
- 9.Fang, Z., N. Morrison, B. Watt, C. Doig, and K. J. Forbes. 1998. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J. Bacteriol. 180:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filliol, I., C. Sola, and N. Rastogi. 2000. Detection of a previously unamplified spacer within the DR locus of Mycobacterium tuberculosis: epidemiological implications. J. Clin. Microbiol. 38:1231-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman, C. R., M. Y. Stoeckle, W. D. Johnson, Jr., and L. W. Riley. 1995. Double-repetitive-element PCR method for subtyping Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 33:1383-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 13.Gingeras, T. R., G. Ghandour, E. Wang, A. Berno, P. M. Small, F. Drobniewski, D. Alland, E. Desmond, M. Holodniy, and J. Drenkow. 1998. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 8:435-448. [DOI] [PubMed] [Google Scholar]
- 14.Groenen, P. M., A. E. Bunschoten, D. van Soolingen, and J. D. van Embden. 1993. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 10:1057-1065. [DOI] [PubMed] [Google Scholar]
- 15.Gutierrez, M., S. Samper, M. S. Jimenez, J. D. van Embden, J. F. Marin, and C. Martin. 1997. Identification by spoligotyping of a caprine genotype in Mycobacterium bovis strains causing human tuberculosis. J. Clin. Microbiol. 35:3328-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas, W. H., W. R. Butler, C. L. Woodley, and J. T. Crawford. 1993. Mixed-linker polymerase chain reaction: a new method for rapid fingerprinting of isolates of the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 31:1293-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock, J. M. 1995. The contribution of slippage-like processes to genome evolution. J. Mol. Evol. 41:1038-1047. [DOI] [PubMed] [Google Scholar]
- 18.Heersma, H. F., K. Kremer, and J. D. van Embden. 1998. Computer analysis of IS6110 RFLP patterns of Mycobacterium tuberculosis. Methods Mol. Biol. 101:395-422. [DOI] [PubMed] [Google Scholar]
- 19.Jansen, R., J. D. van Embden, W. Gaastra, and L. M. Schouls. 2002. Identification of a novel family of sequence repeats among prokaryotes. OMICS 6:23-33. [DOI] [PubMed] [Google Scholar]
- 20.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufhold, A., A. Podbielski, G. Baumgarten, M. Blokpoel, J. Top, and L. Schouls. 1994. Rapid typing of group A streptococci by the use of DNA amplification and non-radioactive allele-specific oligonucleotide probes. FEMS Microbiol. Lett. 119:19-25. [DOI] [PubMed] [Google Scholar]
- 22.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurepina, N. E., S. Sreevatsan, B. B. Plikaytis, P. J. Bifani, N. D. Connell, R. J. Donnelly, D. van Sooligen, J. M. Musser, and B. N. Kreiswirth. 1998. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuber. Lung Dis. 79:31-42. [DOI] [PubMed] [Google Scholar]
- 24.Legrand, E., I. Filliol, C. Sola, and N. Rastogi. 2001. Use of spoligotyping to study the evolution of the direct repeat locus by IS6110 transposition in Mycobacterium tuberculosis. J. Clin. Microbiol. 39:1595-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longo, M. C., M. S. Berninger, and J. L. Hartley. 1990. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 93:125-128. [DOI] [PubMed] [Google Scholar]
- 26.Niemann, S., D. Harmsen, S. Rusch-Gerdes, and E. Richter. 2000. Differentiation of clinical Mycobacterium tuberculosis complex isolates by gyrB DNA sequence polymorphism analysis. J. Clin. Microbiol. 38:3231-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niemann, S., E. Richter, and S. Rusch-Gerdes. 2000. Differentiation among members of the Mycobacterium tuberculosis complex by molecular and biochemical features: evidence for two pyrazinamide-susceptible subtypes of M. bovis. J. Clin. Microbiol. 38:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niemann, S., E. Richter, and S. Rusch-Gerdes. 2002. Biochemical and genetic evidence for the transfer of Mycobacterium tuberculosis subsp. caprae Aranaz et al. 1999 to the species Mycobacterium bovis Karlson and Lessel 1970 (approved lists 1980) as Mycobacterium bovis subsp. caprae comb. nov. Int. J. Syst. Evol. Microbiol. 52:433-436. [DOI] [PubMed] [Google Scholar]
- 29.Palittapongarnpim, P., S. Chomyc, A. Fanning, and D. Kunimoto. 1993. DNA fragment length polymorphism analysis of Mycobacterium tuberculosis isolates by arbitrarily primed polymerase chain reaction. J. Infect. Dis. 167:975-978. [DOI] [PubMed] [Google Scholar]
- 30.Plikaytis, B. B., J. T. Crawford, C. L. Woodley, W. R. Butler, K. D. Eisenach, M. D. Cave, and T. M. Shinnick. 1993. Rapid, amplification-based fingerprinting of Mycobacterium tuberculosis. J. Gen. Microbiol. 139:1537-1542. [DOI] [PubMed] [Google Scholar]
- 31.Serraino, A., G. Marchetti, V. Sanguinetti, M. C. Rossi, R. G. Zanoni, L. Catozzi, A. Bandera, W. Dini, W. Mignone, F. Franzetti, and A. Gori. 1999. Monitoring of transmission of tuberculosis between wild boars and cattle: genotypical analysis of strains by molecular epidemiology techniques. J. Clin. Microbiol. 37:2766-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soini, H., X. Pan, A. Amin, E. A. Graviss, A. Siddiqui, and J. M. Musser. 2000. Characterization of Mycobacterium tuberculosis isolates from patients in Houston, Texas, by spoligotyping. J. Clin. Microbiol. 38:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soini, H., X. Pan, L. Teeter, J. M. Musser, and E. A. Graviss. 2001. Transmission dynamics and molecular characterization of Mycobacterium tuberculosis isolates with low copy numbers of IS6110. J. Clin. Microbiol. 39:217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sola, C., A. Devallois, L. Horgen, J. Maisetti, I. Filliol, E. Legrand, and N. Rastogi. 1999. Tuberculosis in the Caribbean: using spacer oligonucleotide typing to understand strain origin and transmission. Emerg. Infect. Dis. 5:404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Zanden, A. G., A. H. Hoentjen, F. G. Heilmann, E. F. Weltevreden, L. M. Schouls, and J. D. van Embden. 1998. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis complex in paraffin wax embedded tissues and in stained microscopic preparations. Mol. Pathol. 51:209-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van der Zanden, A. G. M., T. Bosje, F. G. C. Heilmann, and D. van Soolingen. 2001. Nosocomial transmission of tuberculosis to a nurse demonstrated by means of spoligotyping of a formalin-fixed bronchial biopsy. Neth. J. Med. 59:152-157. [DOI] [PubMed] [Google Scholar]
- 38.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Embden, J. D., T. van Gorkom, K. Kremer, R. Jansen, B. A. Der Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Soolingen, D., P. W. Hermans, P. E. de Haas, and J. D. van Embden. 1992. Insertion element IS1081-associated restriction fragment length polymorphisms in Mycobacterium tuberculosis complex species: a reliable tool for recognizing Mycobacterium bovis BCG. J. Clin. Microbiol. 30:1772-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Soolingen, D., T. Hoogenboezem, P. E. de Haas, P. W. Hermans, M. A. Koedam, K. S. Teppema, P. J. Brennan, G. S. Besra, F. Portaels, J. Top, L. M. Schouls, and J. D. van Embden. 1997. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int. J. Syst. Bacteriol. 47:1236-1245. [DOI] [PubMed] [Google Scholar]
- 42.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Soolingen, D., A. G. van der Zanden, P. E. de Haas, G. T. Noordhoek, A. Kiers, N. A. Foudraine, F. Portaels, A. H. Kolk, K. Kremer, and J. D. van Embden. 1998. Diagnosis of Mycobacterium microti infections among humans by using novel genetic markers. J. Clin. Microbiol. 36:1840-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viana-Niero, C., C. Gutierrez, C. Sola, I. Filliol, F. Boulahbal, V. Vincent, and N. Rastogi. 2001. Genetic diversity of Mycobacterium africanum clinical isolates based on IS6110-restriction fragment length polymorphism analysis, spoligotyping, and variable number of tandem DNA repeats. J. Clin. Microbiol. 39:57-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, Z. H., P. E. de Haas, D. van Soolingen, J. D. van Embden, and A. B. Andersen. 1994. Restriction fragment length polymorphism Mycobacterium tuberculosis strains isolated from Greenland during 1992: evidence of tuberculosis transmission between Greenland and Denmark. J. Clin. Microbiol. 32:3018-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuen, L. K., B. C. Ross, K. M. Jackson, and B. Dwyer. 1993. Characterization of Mycobacterium tuberculosis strains from Vietnamese patients by Southern blot hybridization. J. Clin. Microbiol. 31:1615-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]