Abstract
Eleven laboratories evaluated the use of dried blood and plasma spots for quantitation of human immunodeficiency virus (HIV) RNA by two commercially available RNA assays, the Roche Amplicor HIV-1 Monitor and the bioMerieux NucliSens HIV-1 QT assays. The recovery of HIV RNA was linear over a dynamic range extending from 4,000 to 500,000 HIV type 1 RNA copies/ml. The Monitor assay appeared to have a broader dynamic range and seemed more sensitive at lower concentrations. However, the NucliSens assay gave more consistent results and could be performed without modification of the kit. HIV RNA was stable in dried whole blood or plasma stored at room temperature or at −70°C for up to 1 year. Dried blood and dried plasma spots can be used as an easy and inexpensive means for the collection and storage of specimens under field conditions for the diagnosis of HIV infection and the monitoring of antiretroviral therapy.
Human immunodeficiency virus (HIV) RNA levels in blood plasma are used to monitor the response to antiretroviral drug therapy in developed countries. Since the International AIDS Conference held in Durban, South Africa, in 2000, there has been a global effort to provide access to antiretroviral drugs to all infected individuals, including those in the resource-poor countries which have been hit the hardest by the AIDS pandemic. Studies have demonstrated that the provision of treatment is not enough, however. In order for the drugs to be most effective, both for an individual and for public health reasons, it will be necessary to monitor the responses to the therapy. HIV RNA assays typically use plasma, which implies access to laboratory equipment that may not be readily available in all field settings. Therefore, alternative specimens must be considered for use in these situations.
Whole blood dried on filter paper (dried blood spots [DBSs]) has been used to qualitatively detect HIV antibodies (13, 15), HIV DNA (1, 3, 5-7, 9, 10, 11, 12, 16, 18) and HIV RNA (4, 14, 17; J. L. Gerstel and A. M. Comeau, Proc. 10th Natl. Neonatal Screening Symp., abstr. P-19, p. 64, 1994). In addition, dried plasma spots (DPSs) have also been used to quantitate the HIV load (8). Although the stabilities of antibodies and DNA in DBSs have been determined (2, 6, 13), there has been some question regarding the stability of the HIV RNA when it is dried and stored at room temperature (14, 17).
The purpose of this study was to compare two different methods for quantitatively measuring HIV RNA in DBSs and DPSs and to assess their long-term stability at room temperature and at −70C.
(This study was presented in part at the 12th World AIDS Conference, Geneva, Switzerland, 28 June to 3 July 1998 [S. Cassol, A. Comeau, S. Fiscus, G. Aldrovandi, J. Sullivan, J. Bremer, and B. Jackson, Abstr. 12th World AIDS Conf., abstr. 33166, 1998].)
MATERIALS AND METHODS
Specimens.
Three different panels were prepared by the Virus Quality Assurance Laboratory by using filter paper (903; Schleicher & Schuell, Keene, N.H.) spotted with 50 μl of plasma or whole blood. The 903 filter paper is inexpensive, available worldwide, and commonly used for assays for the screening of newborns (Guthrie cards) and has been used before for the detection of HIV antibodies (2, 15), DNA (5-7, 9-12), and RNA (8, 14, 17). The spotted filter papers were allowed to dry overnight at room temperature, placed in individual ziplock bags containing a silica desiccant (MultiSorb Technology, Inc., Buffalo, N.Y.), and then frozen at −30 or −70°C or stored at room temperature. Panels 1A and 2 were frozen at −30°C; panel 1B consisted of spots from panel 1A which were subsequently thawed and stored at room temperature for 3 months prior to retesting; panel 3 was stored at either −70°C or room temperature, with no freeze-thaw events, for various lengths of time up to 52 weeks. All specimens were coded to blind the laboratories.
Panel 1 consisted of seronegative plasma and whole blood, to which fivefold serial dilutions of a well-characterized HIV type 1 (HIV-1) stock with from 0 to 500,000 copies/ml was added (nominal concentrations). Each specimen was tested in duplicate by the participating laboratories, and each panel was tested at two different time points (panels 1A and 1B).
Panels 2 and 3 each consisted of liquid plasma, DPSs, and DBSs from three HIV-1-infected patients and one uninfected patient, all in triplicate. For the six HIV-1-infected patients (three different patients for each of the two panels), the nominal concentrations determined by the Virus Quality Assurance Laboratory by the Roche Diagnostics (Branchburg, N.J.) Amplicor HIV-1 Monitor assay with liquid plasma were as follows: for panel 2, 1,157, 16,620, and 231,040 copies/ml, respectively; for panel 3, 1,064, 99,980, and 65,332 copies/ml, respectively.
Panel 3 was designed to evaluate the stability of HIV RNA in DBSs and DPSs during storage at room temperature and −70°C. Each panel consisted of triplicate blood or plasma spots from four HIV-infected patients (12 spots for each panel). One complete set (blood and plasma) was assayed at baseline. Four sets (samples B and D) were placed in storage at room temperature, and the other four sets (samples C and E) were stored at −70°C. One set of blood and plasma spots from each storage condition was assayed after 2, 4, 26, and 52 weeks of storage.
HIV-1 RNA assays.
Assays for detection of HIV-1 RNA were conducted over a 3-year period (September 1997 to June 2000), and multiple kit lots were used. Three (panels 1A, 1B, and 2) or four (panel 3) laboratories used the NucliSens HIV-1 QT assay (bioMérieux, Inc. [formerly Organon Teknika], Durham, N.C.) with minor modifications. One DPS or DPS was placed in 9 ml of NucliSens lysis buffer and rocked at room temperature for 2 h to elute the RNA from the filter paper. Once the filter paper was removed, the manufacturer's instructions were followed for the remainder of the procedure.
Five (panels 1A and 1B) or six (panel 2) laboratories used a modified Amplicor HIV-1 Monitor assay (version 1.0; Roche Diagnostics) to quantitate the RNA in the blood and plasma spots. HIV RNA was eluted from the filter paper by treatment with a chaotrope, organic reagent, and detergent (CORD) solution consisting of 33% (vol/vol) buffered phenol (pH 4.3), 1.34 M guanidinium isothiocyanate, 0.2% (vol/vol) Sarkosyl, 0.3% (vol/vol) sodium dodecyl sulfate, 0.2% (vol/vol) mercaptoethanol, 0.6% (wt/vol) glycogen, and 8 mM sodium citrate. The DBS or DPS was cut or punched into small pieces and incubated in 0.75 ml of CORD reagent containing 0.5 μg of tRNA and 3.3 μl of the kit quantitation standard at 56°C, with shaking, for 30 min. The RNA was then extracted from the mixture with chloroform, followed by alcohol precipitation and elution with the HIV-1 Monitor specimen diluent (100 μl). Amplification and detection of the extracted RNA (50 μl) were performed according to the manufacturer's guidelines. The CORD reagent was prepared by Roche Molecular Systems and distributed to the laboratories that used the HIV-1 Monitor assay.
Statistical methods.
Data from the three panels were transformed to base 10 logarithms for analysis. The data for the HIV-negative specimens were excluded from all of the analyses. Data from the NucliSens and Monitor assays were analyzed separately. Linear regressions of the log10 estimated HIV RNA concentration on the log10 nominal HIV RNA concentration were used to analyze the data for panels 1A and 1B. The analysis was restricted to six laboratories from which results were obtained for both rounds of testing (Monitor assay, four laboratories; NucliSens assay, two laboratories). Predictor variables were added to the model to test for variations in the results between DBSs and DPSs, between the two panels, or among laboratories. More complex predictors were added to determine if differences between panels or between specimen types varied over laboratories or if any of the differences varied with nominal concentration.
Analysis of variance was used for panel 2. Indicator variables for the donor were included in all models. Hematocrit-corrected estimates of the concentrations from DBSs with those from DPSs were compared with estimates of the concentrations from plasma in separate analyses. Predictors were added to the models to determine if differences between specimen types varied among donors or laboratories.
The stability of HIV RNA in stored DBSs and DPSs was assessed by using regressions on the log10 estimated RNA concentration on time in storage and donor to model the results for panel 3. The analysis was stratified by specimen type (DPSs versus DBSs) and storage condition (room temperature versus −70°C) to form four separate regressions. Variables were added to the models to determine if the rate of change in HIV RNA concentration in storage varied among donors or laboratories.
In some cases, results that were below the limit of detection of an assay were included in an analysis. Estimates that were below the limit of detection for the NucliSens kit were set at 400 copies/ml. Estimates that were below the limit of detection for the Monitor test were set at values that were calculated by substituting an optical density of 0.2 units for the optical density for the undiluted amplified sample. This is the minimum RNA concentration that would have produced a positive result by the Monitor test, given the results for the internal quantitation standard in each assay.
RESULTS
Panels 1A and 1B were used to evaluate the technical performance of the laboratories, focusing primarily on their abilities to perform the DBS and DPS extractions and the quantitative RNA assays. One false-positive result was obtained by the NucliSens assay (estimated RNA concentration, 420 copies/ml). Another was obtained by the Monitor assay (estimated RNA concentration, 285 copies/ml).
The samples with nominal concentrations of 800 copies/ml were excluded from the statistical comparisons of estimates of the concentrations from DBSs and DPSs because nearly half of the results were below the limits of detection of the two kits (NucliSens assay, 8 of 12 from DPSs and 7 of 12 from DBSs; Monitor assay, 9 of 19 from DPSs and 7 of 21 from DBSs). Results at a nominal concentration of 4,000 copies/ml that were below the limit of detection of the assay were included in the analysis, as described earlier (Nuclisens assay, 4 of 12 from DPSs and 2 of 12 from DBSs; Monitor assay, 1 of 22 from DPSs and 3 of 21 from DBSs).
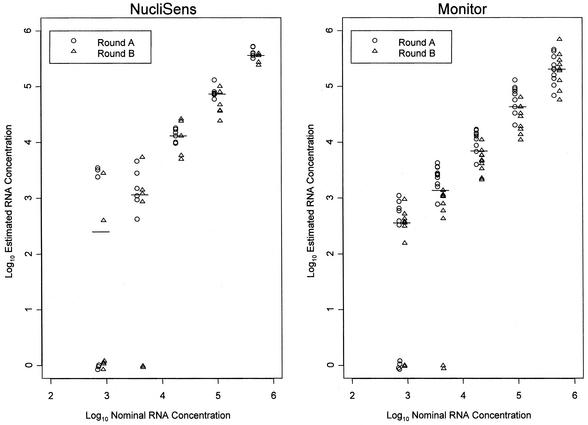
Log-log plots of the estimated HIV RNA concentrations from DBSs against the nominal RNA concentrations are shown separately for the NucliSens assay (Fig. 1A) and the Monitor assay (Fig. 1B). On average, estimates by the NucliSens assay for panel 1A were 0.11 log10 (29%) higher than estimates for panel 1B (P = 0.011), and the results from DBSs were, on average, 0.11 log10 (29%) higher than those from DPSs. There was no evidence that the differences between log-transformed estimates of the concentrations from the two panels or two specimen types varied among nominal concentrations or among laboratories.
FIG. 1.
Log10 estimated HIV RNA concentrations plotted against log10 nominal HIV RNA concentrations for the NucliSens and Monitor assays. Panel 1 specimens were tested on two different occasions (round A and round B). The horizontal line represents the median estimated concentration at each nominal concentration.
The results of the Monitor assay were more heterogeneous than those of the NucliSens assay, which made the analysis very difficult. The difference between estimates of the concentrations from DBSs and DPSs depended on both the laboratory and the panel that was considered (P = 0.016). When the results for DPSs and DBSs were analyzed separately, estimates of the concentrations from DPSs in panel 1A were, on average, 0.27 log10 (86%) higher than those from DPSs in panel 1B (P < 0.001), with no evidence that the difference between panels varied among laboratories or nominal concentrations. However, for DBSs, the differences between panels varied among laboratories (P < 0.001) and nominal concentrations (P = 0.005), making it very difficult to draw any general conclusions from the data.
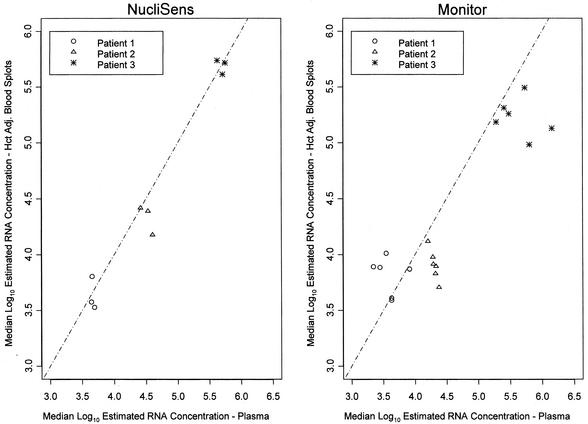
Log-log plots of median hematocrit-adjusted estimated HIV RNA concentrations from DBSs in panel 2, for each donor in each laboratory, against the median for plasma, for data combined across laboratories, are shown separately for the NucliSens assay (Fig. 2A) and the Monitor assay (Fig. 2B). By the NucliSens assay, the hematocrit-corrected estimates of the concentrations from DBSs were, on average, 0.078 log10 (16.5%) lower than those from plasma (P = 0.16), with no detectable variation in the difference among laboratories (P = 0.39) or donors (P = 0.49). Similar results for panel 2 were seen for the comparison of DPSs and plasma by the NucliSens assay (data not shown).
FIG. 2.
Results for panel 2 showing the median log10 hematocrit-adjusted (Hct Adj.) estimated HIV RNA concentrations in DBSs in each laboratory plotted against the median log10 estimated HIV RNA concentrations in plasma for all laboratories in which the same kit was used. The dotted line represents the line of equivalence.
Differences between estimates of the concentrations from DBSs and plasma by the modified Monitor assay varied among the six laboratories (P < 0.001). No differences between the two sample types were noted in two laboratories (P = 0.24 and P = 0.17), but estimates of the concentrations from DBSs were, on average, 0.22 log10 (40%) lower than those from plasma in a third laboratory (P = 0.04). In each of the other three laboratories, the difference between DBSs and plasma varied among donors. The differences (concentrations in DBS − concentrations in plasma) ranged from −0.44 to 0.82 log10 across donors and laboratories, even though the specimens from the three donors were assayed in the same run in each laboratory. For the Monitor assay, the difference between estimates of the concentrations from DPSs and plasma varied among laboratories (P = 0.002) but not among donors (P = 0.20) (data not shown).
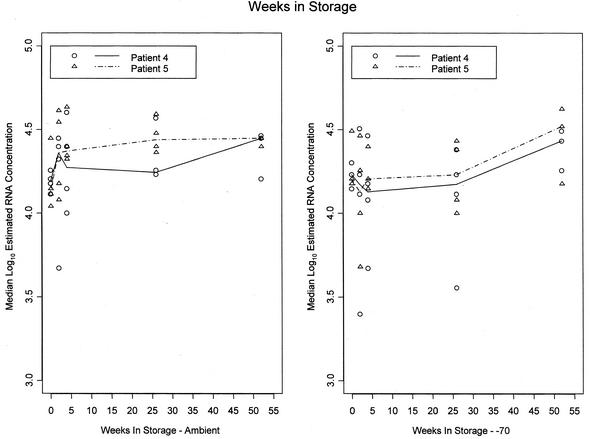
Panel 3 was designed to evaluate the effects of long-term storage on RNA levels in DBSs and DPSs (Fig. 3) and was tested only by the NucliSens assay. Data through 52 weeks of storage were obtained from three laboratories, and data through 26 weeks of storage were obtained from a fourth laboratory. The results for one of three HIV-infected donors were excluded because 40% of the values were below the limit of detection of the assay. Values below the limit of detection were obtained for DBSs and DPSs stored at both temperatures and for DBSs and DPSs at all time points for this donor.
FIG. 3.
Median log estimated RNA concentrations for DBSs versus time (weeks) in storage. The effect of storage at room temperature (left) or at −70°C (right) for DBSs was tested by the NucliSens assay. Each symbol is the median for one patient at a given time point in a given laboratory. The line segments join the medians for data from all laboratories combined.
Rates of change in the log10 RNA concentration varied among the laboratories for DBSs stored at both room temperature (P < 0.01) and −70°C (P < 0.001) and for DPSs stored at −70°C (P < 0.001), but there was little or no evidence of substantial loss of RNA from these samples in any of the laboratories. The rates of change in the RNA concentration differed from zero for only 5 of 12 samples (P < 0.02 for each versus P > 0.10 for changes that were not statistically significant). These occurred at −70°C at four laboratories and at ambient temperature at only one laboratory. The changes were actually positive at four of the five laboratories. The one statistically significant negative change indicated a 52-week loss of only 7% at −70°C. No variation in the rate of change in RNA concentration in DPSs at room temperature was detected among the laboratories. The 52-week loss averaged 2.4%, but this was not statistically significant (P = 0.18).
DISCUSSION
Both the NucliSens assay and a modified version of the Monitor assay can be used to quantitate HIV-1 RNA from DBSs or DPSs, although in this evaluation there was considerably less variation with the NucliSens assay. Detection of HIV RNA was linear over a dynamic range extending from 4,000 to 500,000 HIV-1 RNA copies/ml.
Variation in the Monitor assay made analysis difficult, and the sources of this variation were difficult to pinpoint. The variability might be due to the modified RNA extraction method that had to be used for the dried spots. Phenol-chloroform extractions are more technically demanding, and differences in the recovery of the appropriate phase during the extraction may affect HIV RNA recovery. Potential sources of variation include the retention of various levels of inhibitors such as heme and iron, errors during the RNA extraction, and reagent or kit variability. Furthermore, the reagents used for the modified Monitor extraction method were in their initial stages of development and, hence, were not completely optimized. To the best of our knowledge, Roche no longer supports this application and these reagents are not available commercially.
On the other hand, the Monitor assay appeared to be somewhat more sensitive, particularly in detecting specimens with lower viral loads. This is probably a function of the differences in the amount of the actual RNA eluate that went into the amplification steps for the two assays. For the Monitor assay, the RNA was eluted in 100 μl of diluent and half was used for amplification. The specimen input for the plasma spot was equivalent to 25 μl, a volume comparable to that used for the standard Monitor assay. The NucliSens assay elutes the RNA into 50 μl of diluent, but only 5 μl is used for amplification. The specimen input for the plasma spot was equivalent to 5 μl, which is 1/4 to 1/20 the volume used for plasma analysis by the NucliSens assay. The NucliSens assay could potentially be made more sensitive by reducing the elution volume used during the extraction.
HIV RNA was stable in DBSs and DPSs stored at room temperature and at −70°C for at least 1 year (Fig. 3), and possibly longer. The differences observed in our experiments were probably due to factors such as interassay variation and/or lot-to-lot variation in the kits and are not considered to be clinically significant. Interassay variation would account for the small positive changes if results in a given laboratory were unusually low at early time points or unusually high at later time points, as was the case in some laboratories. These data confirm those of O'Shea et al. (17) regarding the stability of HIV RNA dried on Guthrie cards. Previous results suggesting that HIV RNA was lost with increasing time of storage (14) was probably due to differences in the paper used.
Although the sensitivity of an RNA assay with DBSs may never reach the sensitivity of an assay with whole plasma, it may still be sensitive enough given recent changes in treatment guidelines (patients should be treated when viral load is >50,000 copies/ml or when the CD4 count is <350 cells/mm3). In this study we were able to reliably measure viral loads of 4,000 copies/ml or greater. The potential to improve this sensitivity exists and may be warranted. DBSs offer an easy and inexpensive means for the collection and storage of blood, since they are stable and noninfectious and can easily be sent to centralized testing laboratories for analysis. DBSs clearly provide the perfect means for the collection of specimens under field conditions for the diagnosis of HIV infection and the monitoring of antiretroviral therapy and vaccine efficacy.
Acknowledgments
This work was supported by the Pediatric AIDS Clinical Trials Group through the following grants and contracts or source: AI35712 and AI85354 (to D.B., C.J., and J.B.), AI32907 (to J.L.S.), CFAR grant AI42845 (to J.L.S.), HD-05-3328 (to A.M.C.), 97PVCL05 (to G.A.), 97PVCL06 (to S.A.F.), 97PVCL07 (to J.B.J.), and the Ontario Ministry of Health (to S.A.C.). BioMerieux and Roche Molecular Systems provided the kits for the assays.
We thank Kevin Byron, Don Decker, Jim Ellerbrock, Jacalyn Gerstel-Thompson, Michelle Janes, Richard Pilon, Jana Parsons, Jody Schock, Sal Scianna, and Art Sunhachawee for expert technical assistance and Suzanne Granger for statistical and graphic expertise.
REFERENCES
- 1.Beck, I. A., K. D. Drennan, A. J. Melvin, K. M. Mohan, A. M. Herz, J. Alarcon, J. Piscoya, C. Velazquez, and L. M. Frenkel. 2001. Simple, sensitive, and specific detection of human immunodeficiency virus type 1 subtype B DNA in dried blood samples for diagnosis in infants in the field. J. Clin. Microbiol. 39:29-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behets, F., M. Kashamuka, M. Pappaioanou, T. A. Green, R. W. Ryder, V. Batter, J. R. George, W. H. Hannon, and T. C. Quinn. 1992. Stability of human immunodeficiency virus type 1 antibodies in whole blood dried on filter paper and stored under various tropical conditions in Kinshasa, Zaire. J. Clin. Microbiol. 30:1179-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggar, R. J., W. Miley, P. Miotti, T. E. Taha, A. Butcher, J. Spadoro, and D. Waters. 1997. Blood collection on filter paper: a practical approach to sample collection for studies of perinatal HIV transmission. J. Acquir. Immune Defic. Syndr. 14:368-373. [DOI] [PubMed] [Google Scholar]
- 4.Biggar, R. J., M. Janes, R. Pilon, P. Miotti, T. E. Taha, R. Broadhead, L. Mtimivalye, N. Kumwenda, and S. Cassol. 1999. Virus levels in untreated African infants infected with human immunodeficiency virus type 1. J. Infect. Dis. 180:1838-1843. [DOI] [PubMed] [Google Scholar]
- 5.Cassol, S., T. Salas, M. Arella, P. Neumann, M. Schecter, and M. O'Shaughnessy. 1991. Use of dried blood spot specimens in the detection of human immunodeficiency virus type 1 by the polymerase chain reaction. J. Clin. Microbiol. 29:667-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassol, S., T. Salas, M. Gill, M. Montpetit, J. Rudnik, C. Sy, and M. O'Shaughnessy. 1992. Stability of dried blood spot specimens for detection of human immunodeficiency virus DNA by polymerase chain reaction. J. Clin. Microbiol. 30:3039-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassol, S., B. G. Weniger, P. G. Babu, M. O. Salminen, X. Zheng, M. T. Htoon, A. Delaney, O'Shaughnessy, and C.-Y. Ou. 1996. Detection of HIV type 1 env subtypes A, B, C, and E in Asia using dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Res. Hum. Retrovir. 12:1435-1441. [DOI] [PubMed] [Google Scholar]
- 8.Cassol, S., M. J. Gill, R. Pilon, M. Cormier, R. F. Voigt, B. Willoughby, and J. Forbes. 1997. Quantification of human immunodeficiency virus type 1 RNA from dried blood spots collected on filter paper. J. Clin. Microbiol. 35:2795-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassol, S. A., N. Lapointe, T. Salas, C. Hankins, M. Arella, M. Fauvel, G. Delage, G. M. Boucher, J. Samson, J. Charest, M. L. Montpetit, and M. V. O'Shaughnessy. 1992. Diagnosis of vertical HIV-1 transmission using the polymerase chain reaction and dried blood spot specimens. J. Acquir. Immune Defic. Syndr. 5:113-119. [PubMed] [Google Scholar]
- 10.Cassol, S. A., A. Butcher, S. Kinard, J. Spadoro, T. Sy, N. Lapointe, S. Read, P. Gomez, M. Fauvel, C. Major, and M. O'Shaughnessy. 1994. Rapid screening for early detection of mother-to-child transmission of human immunodeficiency virus type 1. J. Clin. Microbiol. 32:2641-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comeau, A. M., H. Hsu, M. Schwerzler, G. Mushinski, E. Walter, L. Hofman, and G. F. Grady. 1993. Identifying human immunodeficiency virus infection at birth: application of polymerase chain reaction to Guthrie cards. J. Pediatr. 123:252-258. [DOI] [PubMed] [Google Scholar]
- 12.Comeau, A. M., J. Pitt, G. V. Hillyer, S. Landesman, J. Bremer, B. H. Cheng, J. Lew, J. Moye, G. F. Grady, and K. McIntosh. 1996. Early detection of human immunodeficiency virus on dried blood spot specimens: sensitivity across serial specimens. J. Pediatr. 129:111-118. [DOI] [PubMed] [Google Scholar]
- 13.Evengard, B., A. Ehrnst, M. von Sydow, P. O. Pehrson, P. Lundberg, and E. Linder. 1989. Effect of heat on extracted HIV viral infectivity and antibody activity using the filter paper technique of blood sampling. AIDS 3:591-595. [DOI] [PubMed] [Google Scholar]
- 14.Fiscus, S. A., D. Brambilla, L. Grosso, J. Schock, and M. Cronin. 1998. Quantitation of human immunodeficiency virus type 1 RNA in plasma by using blood dried on filter paper. J. Clin. Microbiol. 36:258-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gwinn, M., M. Pappaioanou, J. R. George, W. H. Hannon, S. C. Wasser, M. A. Redus, R. Hoff, G. F. Grady, A. Willoughby, and A. C. Novello. 1991. Prevalence of HIV infection on childbearing women in the United States: surveillance using newborn blood samples. JAMA 265:1704-1708. [PubMed] [Google Scholar]
- 16.Nyambi, P. N., K. Fransen, H. de Beenhouwer, E. N. Chomba, M. Temmerman, J. O. Ndinya-Achola, P. Piot, and G. van der Groen. 1994. Detection of human immunodeficiency virus type 1 (HIV-1) in heel prick blood on filter paper from children born to HIV-1-seropositive mothers. J. Clin. Microbiol. 32:2858-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Shea, S., J. Mullen, K. Corbett, I. Chrystie, M. L. Newell, and J. E. Banatvala. 1999. Use of dried whole blood spots for quantitation of HIV-1 RNA. AIDS 13:630-631. [DOI] [PubMed] [Google Scholar]
- 18.Panteleefe, D. D., G. John, R. Nduati, D. Mbori-Ngacha, B. Richardson, J. Kreiss, and J. Overbaugh. 1999. Rapid method for screening dried blood samples on filter paper for human immunodeficiency virus type 1 DNA. J. Clin. Microbiol. 37:350-353. [DOI] [PMC free article] [PubMed] [Google Scholar]